Abstract
The aim of this study was to investigate chitosan/siRNA complexes formulated with various chitosan salts (CS) including chitosan aspartate (CS-Asp), chitosan glutamate (CS-Glu), chitosan acetate (CS-Ac), and chitosan hydrochloride (CS-HCl) for in vitro siRNA delivery into stable and constitutive enhanced green fluorescent protein (EGFP)-expressing HeLa cells. The CS/siRNA complexes were characterized by 2% agarose gel electrophoresis and investigated for their transfection efficiency in stable and constitutive EGFP-expressing HeLa cells. The cytotoxicity of the complexes was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. The formation of complexes CS/siRNA is mainly dependent on the weight ratio, whereas salt form and molecular weight has less effect. The particle sizes of the complete complexes were in the range of 270–373 nm except the complete complex of CS-Ac, with a slightly positive charge of less than 2 mV. The ability of CS to transfer functionally active siRNA into cell culture is mainly dependent on the weight ratio and molecular weight of CS whereas salt form of CS has less effect. The high gene-silencing efficiency was observed with low MW of CS (20 kDa) and high weight ratio of 32. Over 80% average cell viabilities were observed for CS/siRNA complexes in all weight ratios comparison to untreated cells. This study suggests CS salts have the potential to be used as safe siRNA delivery vectors.
Key words: chitosan salts, EGFP, siRNA delivery
INTRODUCTION
RNA interference (RNAi) represents a powerful tool for specific gene silencing. It is mediated through a 21–23-mer duplex siRNA with 2 nucleotide (nt) overhanged at the 3′ ends so-called small interfering (si)RNA which sequence-specifically triggers the cleavage and subsequent degradation of their target mRNA (1). As a consequence, siRNA has gained a high potential of new drug development to silence various disease genes. However, siRNA therapeutics is hindered by the poor intracellular uptake and limited blood stability of siRNA. To date, a number of gene delivery systems have been developed including viral-based vector, and non-viral-based vector; cationic lipids and cationic polymers. Although viral vectors are more powerful vehicles for gene transfer than non-viral vector, their applications are limited due to safety issues. Several non-viral vectors including lipofectamine 2000, l-alpha-dioleoyl phosphatidylethanolamine (DOPE), poly-l-lysine (PLL), polyethyleneimine (PEI), atelocollagen, and chitosan (CS) has been successfully used for gene delivery (2–6). Among these, chitosan-derived vectors have gained more increasing interest as they are natural-derived, biocompatible, biodegradable, and non-toxic.
Chitosan is a copolymer of N-acetyl-d-glucosamine (GlcNAc) and d-glucosamine (GlcN) produced by alkaline deacetylation of chitin. Chitosan is a weak base with a pKa value of the d-glucosamine residue of about 6.2–7.0; therefore, it is insoluble at neutral and alkaline pH values, but soluble in acid solution. At acidic pH below its pKa, primary amines of glucosamine repeating units are protonated conferring positive charges on chitosan backbone. These protonated amines provide the site for chitosan to bind to negatively charged DNA or RNA and self-assemble into particles by simple mixing. Although chitosan has long been studied as a gene delivery vehicle for plasmid DNA (pDNA), to our knowledge, there are few studies that have been carried out to investigate the use of chitosan to deliver siRNA in vitro and in vivo (7–12). Among these, a number of various salt forms of chitosan including chitosan acetate (CS-Ac), chitosan hydrochloride (CS-HCl), and chitosan glutamate (CS-Glu) were employed. Different acids possess different physicochemical properties such as pKa, solubility, charge, and size, and these properties might affect the physicochemical properties of respective chitosan, resulting in different transfection efficiencies. No previous systematic comparison study of the effect of salt form on CS/siRNA complex formation and gene-silencing efficiency has been reported. In this study, CS/siRNA complexes formulated with different CS including CS-HCl, CS-Asp, CS-Glu, and CS-Ac were compared for their ability to form complexes with siRNA, and for their transfection efficiencies in HeLa cells, a human cervical carcinoma cell line, stably expressing enhanced green fluorescent protein (EGFP). The effect of CS salt form and other formulation parameters including molecular weight (MW) of CS and weight ratio on the physicochemical properties, the gene-silencing efficiency and cytotoxicity of the CS/siRNA complex were also investigated.
MATERIALS AND METHODS
Materials
Chitosan was purchased from Seafresh Chitosan Lab., Thailand with MW of 20, 45, 200, and 460 kDa and 85% degree of deacetylation. Hydrochloric acid, acetic acid, and dimethylsulfoxide (DMSO) were purchased from Fisher Scientific (Fairlawn, NJ, USA). Aspartic acid and glutamic acid were purchased from ACROS Organics (Geel, Belgium). Polyethylenimine (PEI, branched, MW 25 kDa) was purchased from Aldrich (Milwaukee, WI, USA). Lipofectamine2000™ was purchased from Invitrogen (NY, USA). Agarose, diethylpyrocarbonate (DEPC) and 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were purchased from Sigma (St Louis, MO, USA). Modified Eagle’s medium (MEM), fetal bovine serum (FBS), Trypsin–EDTA and penicillin–streptomycin were purchased from Gibco BRL (Rockville, MD, USA). siRNA-EGFP(+) and siRNA-EGFP(−) were synthesized using Ambion’s Silencer™ siRNA Construction Kit (Ambion, USA). HeLa cells, a human cervical carcinoma cell line, were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). All other chemicals were cell culture and molecular biology grade.
Preparation of Chitosan Salts
Chitosan of different MW was dissolved in distilled water containing aspartic acid, glutamic acid, hydrochloric acid, and acetic acid in 1:0.850, 1:0.825, 1:0.800, and 1:2 molar ratios, respectively. The solution was adjusted with distilled water to make a 1% w/w solution and stirred for 12 h. This solution was spray-dried using a spray dryer (Minispray Dryer, Büchi 190, Switzerland). The physicochemical properties of chitosan salts were determined by Fourier-transform infrared (FTIR) spectroscopy, Differential scanning calorimetry (DSC), Powder X-ray diffraction (XRPD), and solid-state 13C NMR spectra.
Preparation of siRNA
The EGFP targeted siRNA, siRNA-EGFP(+), and the mismatch siRNA, siRNA-EGFP(−) were synthesized using Ambion’s Silencer™ siRNA Construction Kit (Ambion, USA). The duplex siRNA-EGFP(+) contains sense 5′-gcu gac ccu gaa guu cau cuu-3′ and antisense 5′-gau gaa cuu cag ggu cag cuu-3′. The siRNA-EGFP(−), contains sense 5′-gca ccg cuu acg uga uac uuu-3′, and antisense 5′-agu auc acg uaa gcg gug cuu-3′. The siRNA-EGFP(+) targets to position 124–144 of EGFP open-reading frame (13) and siRNA design suggested by the company. The mismatch siRNA-EGFP(−) were designed by scrambling the nucleotide sequence of siRNA-EGFP(+) and performing blast analysis to identify the sequences that lack of significant homology to human genes.
Complex Formation
CS salt was dissolved in DEPC-treated water to a final concentration of 0.88 mg/ml (stock solution). Purified siRNA from the construction reaction was diluted to 2 μM in DEPC-treated water (pH 4.6). The siRNA solution was added to CS salt solution to obtain the weight ratios of 4, 8, 16, and 32. The mixture was gently pipetted and vortexed for 3–5 s to initiate CS/siRNA complex formation and left for 30 min at room temperature. Complex formation was confirmed by electrophoresis. Briefly, the mixture of 10 μl of the complex containing 140 ng of siRNA and 2 μl of 50% glycerol in water was loaded in the well of 2% agarose gel. The electrophoresis was carried out for 20 min at 100 V in Tris-borate-EDTA (TBE) running buffer pH 8.3. The siRNA in the complexes were visualized under a UV transilluminator.
Size and Zeta Potential Measurement
The particle sizes and surface charge of the CS/siRNA complexes were measured by photon correlation spectroscopy (PCS) using the Zetasizer Nano ZS (Malvern Instruments Ltd., Malvern, UK) at room temperature. The complexes were diluted with phosphate buffer saline (PBS) pH 7.2 which was passed through 0.22 µm membrane filter prior used. All samples were measured in triplicate.
Morphology
The morphology of the CS/siRNA complexes was determined by an atomic force microscope (AFM) using tapping-mode AFM in air. The complexes were diluted with distilled water which was passed through 0.22 µm membrane filter prior used. These samples were dropped immediately onto freshly cleaved mica and air-dried.
HeLa Cells Stably Expressing Green Fluorescent Protein
HeLa cells with stable constitutive expression of EGFP were generated. Briefly, HeLa cells were transfected with pEGFP-C2 plasmids (Clontech, USA) using lipofectamine2000™ (Invitrogen, USA). The cells were trypsinized, diluted, and transferred to a 10-cm diameter plate to get a few hundred cells. Several single cells were marked under the plate and treated with 0.5 mg/ml G418 to generate clones of stable cell lines. Individual clones were isolated and expanded in modified Eagle’s medium (MEM), supplemented with 10% fetal bovine serum (FBS) and treated with 0.1 mg/ml G418 every 3–4 weeks to maintain the EGFP gene expression.
In Vitro Transfection
In vitro transfection was carried out as previous report (14). Briefly, the HeLa stable cells were seeded at the density of 8,000 cells/well in black clear-bottom, 96-well plates (Corning, USA) with MEM, supplemented with 10% FBS in a humidified atmosphere (5% CO2, 95% air, 37°C) 24 h prior to transfection. Measurement of EGFP was performed using a fluorescence microplate reader (Universal Microplate Analyzer, Model AOPUS01 and AI53601, Packard BioScience, CT, USA) with excitation/emission at 485/530 nm. The seeding variation as measured by the fluorescence intensity of each well before transfection was less than 10%. The CS/siRNA-EGFP(+) and the CS/siRNA-EGFP(−) complexes with various weight ratios were formed as described in the complex formation. Just prior to transfection, the cells were washed with PBS and added with the mixtures of 100 μl culture medium and 25 μl of complexes, containing 350 ng of siRNA. These cells were incubated with the transfection medium for 24 h before changing culture medium. Plates were incubated for a further 5 days at 37°C and 5% CO2, culture medium changed every other day, and the fluorescence intensity measured daily for 5 days.
To take into account the variation of fluorescent intensity due to unequal initial number of cells in each well, we first calculated % seeding variation (Eq. 1) and then used the values to adjust the fluorescent intensity obtained from each well by Eq. 2. The percentage of EGFP gene silencing was calculated by Eq. 3.
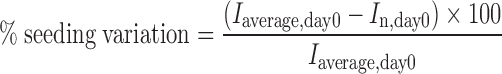 |
1 |
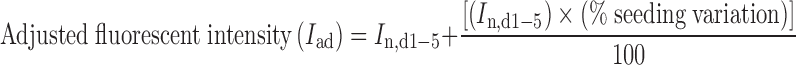 |
2 |
 |
3 |
where Iaverage,day0 is the average fluorescent intensity of all wells prior to transfection (day 0), In,day0 is the fluorescent intensity of individual well at day 0, In,d1−5 is the fluorescent intensity at days one through five of each well, Iad is adjusted fluorescent intensity, Iad,siRNA(+) is adjusted fluorescent intensity of individual well with CS/siRNA(+) complexes, and Iad,siRNA(−) is the average value of adjusted fluorescent intensity of 3 wells with CS/siRNA(−) complexes.
Cytotoxicity
Evaluation of CS/siRNA complexes cytotoxicity was performed by MTT assay (15). EGFP-HeLa cells were seeded at a cell density of 8,000 cells/well in 96-well plates and incubated for 24 h prior to transfection with CS/siRNA complexes in the same concentrations as in vitro transfection experiment. After 24 h of transfection, the medium was removed and 20 μl of MTT (5 mg/ml) was added to each well and the incubation was continued for 4 h. The medium was then removed, the cells were rinsed with PBS pH 7.4, and formazan crystals formed in the living cells were dissolved in 100 μl DMSO per well. Relative viability (%) was calculated based on absorbance at 550 nm using a microplate reader (Universal Microplate Analyzer, Model AOPUS01 and AI53601, Packard BioScience, CT, USA). The viability of non-treated control cells was arbitrarily defined as 100%.
Statistical Analysis
Data are presented as the means ± standard deviations. The statistical significance of gene-silencing efficiency and cell viability were determined using one-way analysis of variance (ANOVA) followed by an LSD post hoc test. P values of <0.05 were considered significant.
RESULTS
Characterization of Chitosan Salts
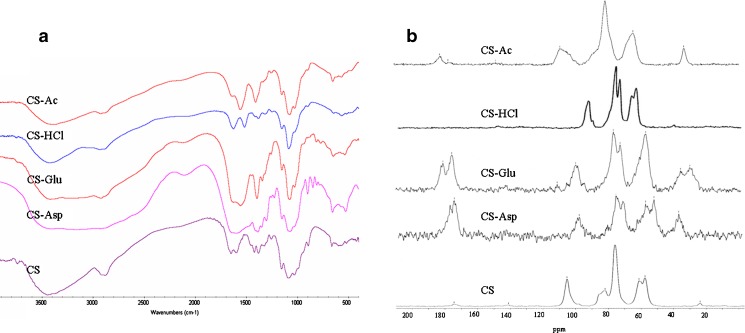
The results from FTIR and 13C NMR spectra of all spray-dried salts indicated the presence of salt groups in the chitosan structure (Fig. 1). CS is in a semi-crystalline while CS salts showed a diffraction pattern of an amorphous state with a broad endothermic peak at onset temperature around 110–140°C as investigated by XRPD and DSC, respectively.
Fig. 1.
a FTIR and b 13C NMR spectra of spray-dried chitosan salts
Characterization of CS/siRNA Complexes
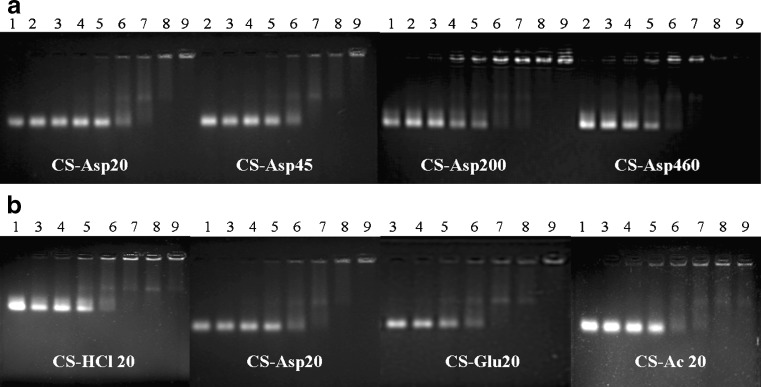
In order to determine the optimal complexation conditions, it was necessary to evaluate the degree of binding between siRNA and CS with different MW, salt forms, and the weight ratio of CS/siRNA. The 2% gel electrophoresis of CS/siRNA complexes formulated with CS-Asp MW 20, 45, 200 and 460 kDa and four salt forms including CS-HCl, CS-Asp, CS-Glu, and CS-Ac with MW of 20 kDa formed at various weight ratios were examined as shown in Fig. 2a, b respectively. As compared to the control naked siRNA, CS binding caused retarded mobility of siRNA with heterogeneous size and charge of the CS/siRNA complexes appeared as an upper smear band with bright band of complete complexes carrying net zero or positive charge retaining within the well. The results showed that siRNA binding to CS was dependent on both the MW of CS and the weight ratio. At a weight ratio of less than 4 (Fig. 2, Lanes 2–5), the migration behavior was almost the same as the naked siRNA (Fig. 2, Lane 1). The gradually increasing retardation effect on the migration of siRNA was observed when the weight ratio increased. When the MW of CS increased (Fig. 2a), a greater retardation effect on the migration of siRNA was observed. The complexes completely retained within the well at a weight ratio above 8 (Lane 7) for high MW of 200 and 460 kDa and above 16 (Lane 8) for low-MW CS of 20 and 45 kDa. Figure 2b showed the effect of salt form on complex formation. CS-HCl and CS-Ac formed complexes with siRNA slightly better than CS-Asp and CS-Glu. The initial gel retardation was observed at a weight ratio of 4 (Lane 6) for CS-HCl and CS-Ac and 8 (Lane7) for CS-Asp and CS-Glu. The complexes completely retained within the well were observed at a weight ratio above 16 (Lane 8) for all CS salts.
Fig. 2.
The 2% agarose gel electrophoresis of CS/siRNA complexes (a) formulated with CS-Asp of different MWs (20, 45, 200, and 460 kDa) and b CS/siRNA complexes containing CS MW 20 kDa of different salt (CS-HCl, CS-Asp, CS-Glu, and CS-Ac). Lane 1 naked siRNA; lanes 2–9: CS/siRNA at weight ratio of 0.25, 0.5, 1, 2, 4, 8, 16, and 32, respectively
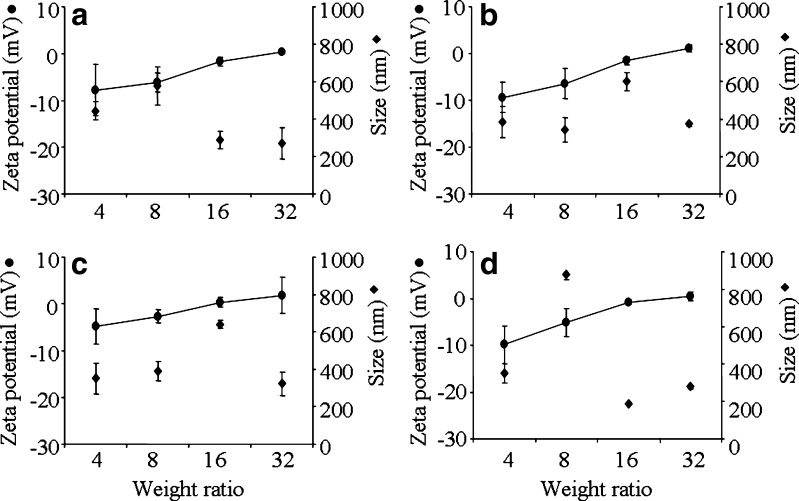
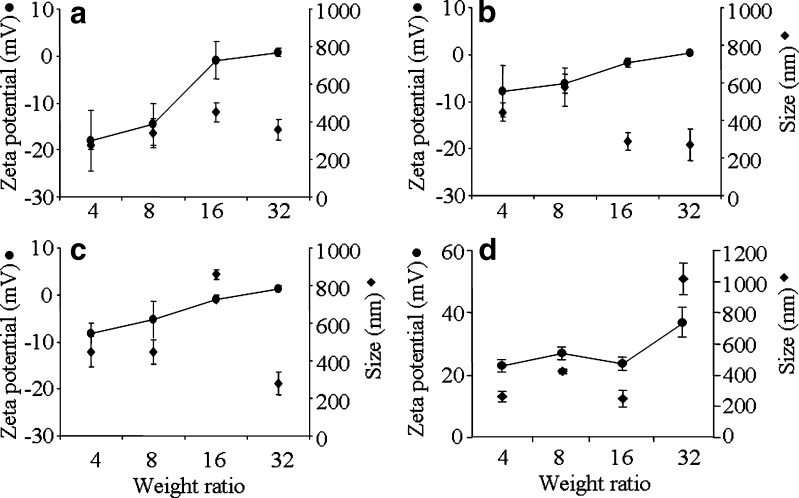
The complexes were formed at acidic pH of 4.6 in DEPC-treated water and the physical properties including size and surface charge that influence cellular interactions and nanoparticles distribution were measured at transfection pH of 7.2. Particle size and zeta potential were plotted against weight ratio of CS/siRNA complexes formulated with CS-Asp MW 20, 45, 200, and 460 kDa (Fig. 3) and with 4 chitosan salts: HCl, Asp, Glu, and Ac (Fig. 4). Zeta potential value increased when increasing weight ratio. MW of chitosan showed no effect on surface charge and size of complexes. For complexes of all MW and all salt except CS-Ac, initial negative charge of around 10 were observed at low weight ratio of 4 and slightly positive charge was observed at weight ratio 32. The complexes of CS-Ac with weight ratio of 4 to 32 showed positive surface charge in the range of 23.3 ± 1.2 to 36.9 ± 4.5. The particle sizes of CS/siRNA complexes of all tested CS were in nano-range. In general, the particle size of the complexes increased with increasing weight ratio from 4 to 8 or 16 and decreased after a weight ratio of 16 or 32 except the complex of CS-Ac/siRNA at weight ratio of 32 had the largest size.
Fig. 3.
Zeta potential (filled circle) and particle size (filled diamond) at varying weight ratios of CS/siRNA complexes formulated with CS-Asp of different MW (20, 45, 200, and 460 kDa). a CS-Asp 20 kDa; b CS-Asp 45 kDa; c CS-Asp 200 kDa; d CS-Asp 460 kDa. Each value represents the mean ± SD of three measurements
Fig. 4.
Zeta potential (filled circle) and particle size (filled diamond) at varying weight ratios of CS/siRNA complexes formulated with different salt forms of 20 kDa CS a CS-HCl; b CS-Asp; c CS-Glu; d CS-Ac. Each value represents the mean ± SD of three measurements
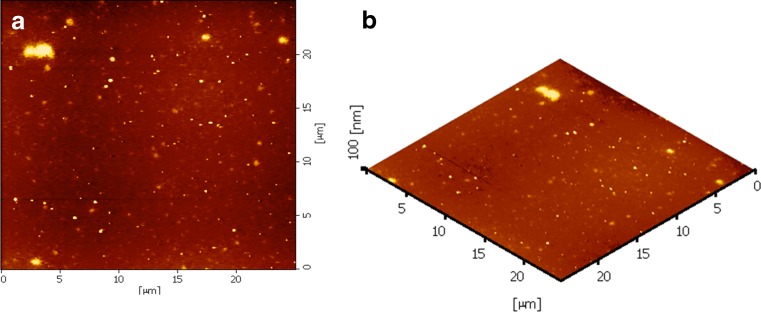
The morphological examination of the CS/siRNA complexes formulated with CS-HCl MW 20 kDa at a weight ratio of 16 was performed by AFM (Fig. 5). The AFM images revealed that the complexes were spherical with a wide range of size distribution in nanoscale which was consistent with the effective diameter measured by PCS (Fig. 4) and explain the detectable upper smear band on gel analysis which partly was the result of size heterogeneity of complexes (Fig. 2).
Fig. 5.
The AFM images of CS-HCl/siRNA complexes at a weight ratio of 16 and a CS MW of 20 kDa; a planar image, and b three-dimensional image
In Vitro siRNA Transfection
The HeLa cells that stably expresses EGFP was used to investigate the gene-silencing efficiency of various CS/siRNA complexes. Polyethylenimine (PEI, 25 kDa) was used as a positive control. siRNA concentration was kept constant at 350 ng/well and the weight ratio was varied from 4 to 32. Transfection of EGFP stable cells with CS/siRNA(−) complexes containing EGFP-mismatch siRNA and that with CS/siRNA(+) complexes were performed in parallel. The low nonspecific EGFP inhibition was observed in well receiving mismatch siRNA. The mean fluorescent intensity of wells treated with CS/siRNA(+) complexes was subtracted from that of wells treated with CS/siRNA(−) complexes of the same weight ratio to gain specific EGFP gene silencing. The naked siRNA showed non-significant gene-silencing effect. The positive control of PEI/siRNA complexes at an N/P ratio of 16 showed the maximum gene-silencing efficiency of 55.3 ± 1.1%.
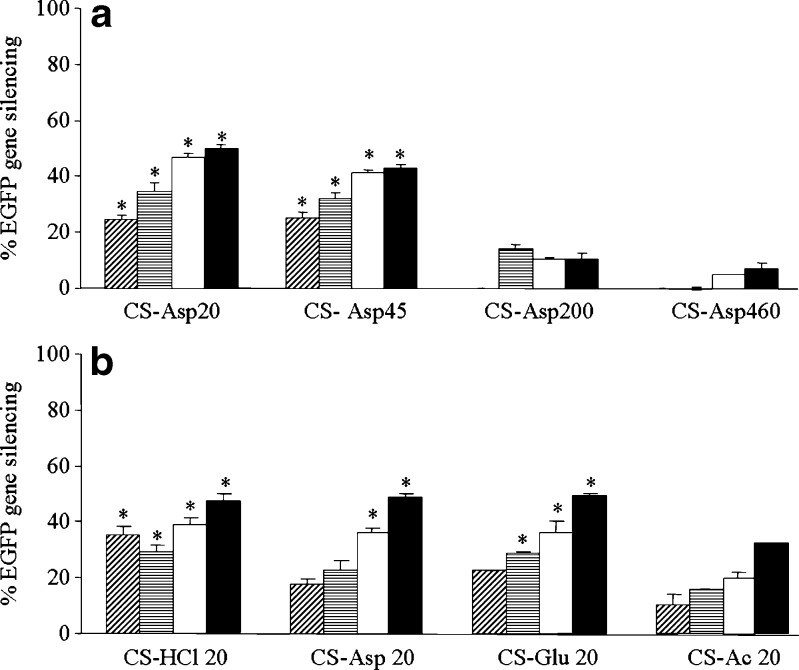
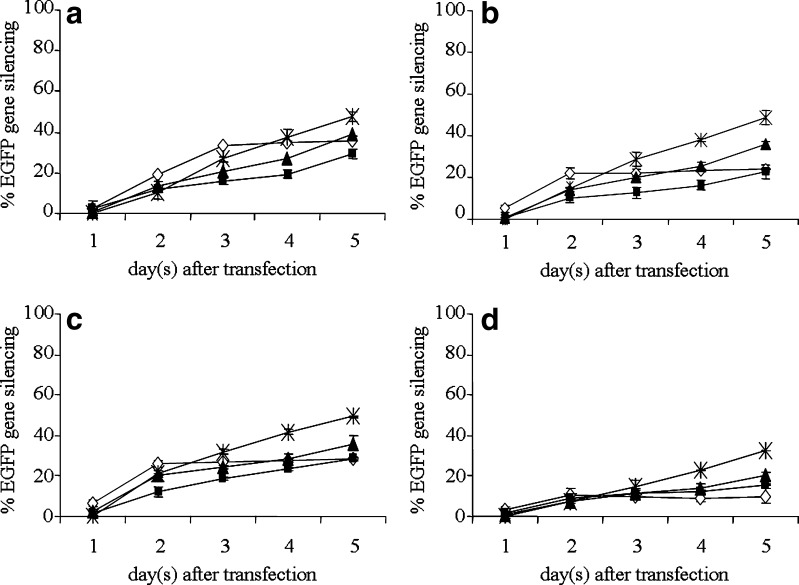
Figure 6 shows the reduction of EGFP expression in stable constitutive EGFP-HeLa cells treated with CS/siRNA complexes at day 5 post-transfection. The MW of CS has great effect on gene-silencing efficiency. The low MW of CS-Asp20 and CS-Asp45 showed much greater gene-silencing efficiency than the high MW of CS-Asp200 and CS-Asp460 (Fig. 6a). Salt form showed less influence on gene silencing than MW. Complexes formulated with HCl, Asp and Glu salt of CS gave no different level of gene silencing at the same weight ratio whereas the complexes of CS-Ac showed significantly less efficiency than other salt at the same weight ratio (Fig. 6b). The level of gene silencing gradually increased when increasing weight ratio as observed in all CS salt (Fig. 6b). The maximum gene silencing was gained at weight ratio of 32 (Fig. 6). The kinetics of gene silencing of CS MW20 kDa with four chitosan salts: HCl, Asp, Glu, and Ac were shown in Fig. 7. Gene-silencing effect of complexes formed at weight ratio of 4 greatly enhanced from day 1 to days 2–3 post-transfection and reached the plateau with low gene-silencing level whereas the gene-silencing effect of complexes formed at weight ratio of 8, 16, and 32 gradually increased from day 1 to day 5.
Fig. 6.
Percentage of EGFP gene silencing at day 5 of transfection by CS/siRNA complexes formulated with a CS-Asp of different MWs (20, 45, 200, and 460 kDa) and b CS MW 20 kDa of different salt (CS-HCl, CS-Asp, CS-Glu, and CS-Ac) at various weight ratio of 4 ( ), 8 (
), 8 ( ), 16 (□) and 32 (■) in HeLa cells stably expressing EGFP. Each value represents the mean ± SD of three wells. Differences values * were statistically significant (P < 0.05)
), 16 (□) and 32 (■) in HeLa cells stably expressing EGFP. Each value represents the mean ± SD of three wells. Differences values * were statistically significant (P < 0.05)
Fig. 7.
The kinetic of EGFP gene silencing induced by CS/siRNA complexes formulated with CS MW of 20 kDa a CS-HCl; b CS-Asp; c CS-Glu; d CS-Ac at various weight ratios of 4 (empty diamond), 8 (filled square), 16 (filled upright triangle) and 32 (exmark) in HeLa cells stably expressing EGFP. Each value represents the mean ± SD of three wells
Cytotoxicity
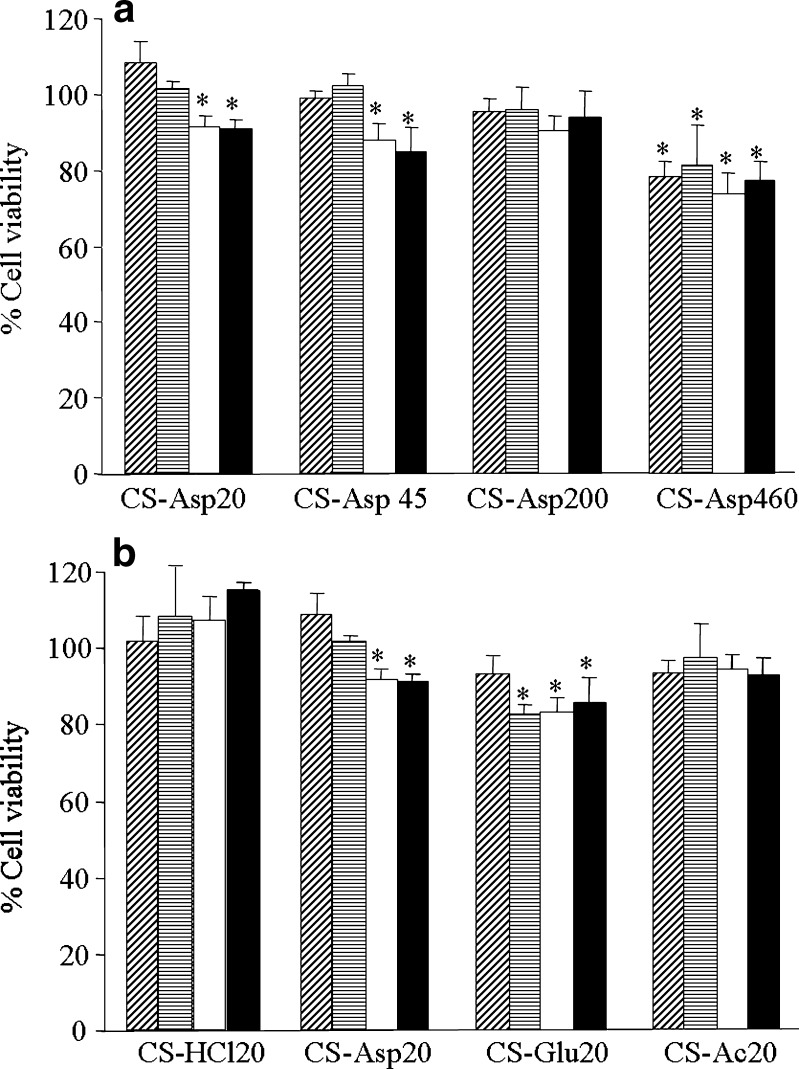
A required characteristic of a gene/siRNA delivery system is that it is not cytotoxic. The cytotoxicity of chitosan/DNA complexes was found to be low compared to other cationic complexes (2,16–18). To investigate the cytotoxicity of CS/siRNA complexes, the cell viability was determined by MTT assay after 24 h incubation with CS/siRNA complexes. Cells without treatment of the CS/siRNA complexes were considered as a control with a cell viability of 100%. Figure 8 showed the effect of MW (Fig. 8a) or salt form of CS (Fig. 8b) on cell viability. The results showed that CS/siRNA complexes formulated with different salts and MW of CS at various weight ratios of 4, 8, 16, and 32 did not affect the viability of EGFP-HeLa cells. The observed relative cytotoxicities of CS/siRNA complexes are comparable to the results reported by the others (7,9,14,19), over 80% average cell viability was observed for CS/siRNA complexes in comparison to untreated cells. It was clear that all CS/siRNA formulation were safe.
Fig. 8.
Cell viability of CS/siRNA complexes formulated with a CS-Asp of different MWs (20, 45, 200, and 460 kDa) and b CS/siRNA complexes containing CS MW 20 kDa of different salt (CS-HCl, CS-Asp, CS-Glu, and CS-Ac). at various weight ratio of 4 ( ), 8 (
), 8 ( ), 16 (□), and 32 (■) in HeLa cells stably expressing EGFP. Each value represents the mean ± SD of three wells. Differences values * were statistically significant (P < 0.05)
), 16 (□), and 32 (■) in HeLa cells stably expressing EGFP. Each value represents the mean ± SD of three wells. Differences values * were statistically significant (P < 0.05)
DISCUSSION
Chitosan salts including CS-HCl, CS-Ac, and CS-Glu has been used to formulate CS/siRNA complexes for gene silencing in vitro and in vivo (7–12). However, the effect of different salt forms of chitosan on gene-silencing efficiency has not been elucidated in the same experiment. In this study, CS/siRNA complexes formulated with various chitosan salts including CS-HCl, CS-Ac, CS-Asp, and CS-Glu were investigated for their gene-silencing abilities. The EGFP-siRNA was used to monitor the gene-silencing efficiency in HeLa cells stably expressing EGFP. It was found that CS/siRNA complexes could sufficiently transfect stable cells with low cytotoxicity.
Agarose gel electrophoresis was performed to confirm the polyionic complex formation of CS with siRNA. The rate of migration of CS/siRNA complexes in agarose gel is dependent on the size and surface charge of complexes. CS binding to siRNA generated complexes varying in size and charge, which were observed as an upper smear band. The complexes with larger size and less negative charge run slower than the complexes with smaller size and higher negative charge. The noncomplex-forming siRNA remained in the reaction migrated at the same level as the control siRNA. Analysis by 2% agarose gel electrophoresis indicated that the high-MW CS bound siRNA slightly better than the low-MW CS (Fig. 2a). The influence of MW in complex formation, as explained previously, could be attributed to a chain entanglement effect since longer CS chains in high-MW CS more easily entangle and trap-free siRNA than shorter chain abundantly found in low-MW CS (7,14). Study of the effect of salt form on siRNA binding showed that CS-HCl and CS-Ac bound siRNA slightly better than others. In comparison of gel analysis of CS/siRNA complex in this study and CS/pDNA complexes in previous study of our group (16), the result indicates that pDNA easily completely condensed to the CS even at a very low N/P ratio of 2 (weight ratio of 1) and needed as low as weight ratio of 0.25 to initiate complex formation whereas siRNA required much higher excess amount of CS up to 32 of weight ratio (Fig. 2a, lane 9) to form complex completely and required at least a weight ratio above 2 to initiate complex formation (Fig. 2a, lane 5). The possible explanation could be the size difference. siRNA is a few thousand times smaller than pDNA, therefore, it is not surprising that it would need much higher density of CS to catch the protonated amine in the CS backbone and initiate complex formation.
The other different characteristic between the CS/pDNA and the CS/siRNA nanoparticles is the net surface charge of nanoparticles. For CS/pDNA nanoparticles at the N/P above the level that complete complexes were formed (N/P ratios higher than 2 or weight ratio higher than 1) the zeta potential reached plateau of +15 to +28 mV whereas the zeta potential of complete complexes of CS/siRNA formed at a very high weight ratio of 32 was less than +2 mV. This suggests that siRNA bind to the chitosan in a different manner from that observed with pDNA. The shorter length of siRNA as well as its rigid linearity can be expected to contribute to the weak interaction with CS (9). It is most likely that the supercoiled pDNA are fully wrapped up and their negative charges are obscured by the positive charge of the protonated amine group of CS whereas part of the short and rigid siRNA molecules protrude and partially neutralize positive charge of CS resulting in low net positive surface charge of the CS/siRNA particles.
Previous study on pDNA has shown that the CS/pDNA particles formulated with high-MW CS has higher particle size and zeta potential than those formulated with low MW CS (16). In this study, we do not observe any effect of MW on particle size and zeta potential of CS/siRNA. MW of CS had less influence on surface charge and particle size than salt form and weight ratio. The comparative positive value of surface charge of the CS/siRNA complexes increased with the increasing weight ratio due to the increase in the number of positive charge of the CS. The result of zeta potential was closely consistent with the result of gel analysis in that the increasing number of complexes remaining in the gel loading well was observed when increasing the weight ratio. The complete immobility of complexes formed at the highest weight ratio (Fig. 2, lane 9) was due to the positive surface charge since the complex size was relatively smallest at this weight ratio (Figs. 3 and 4).
Liu et al. (7) proposed that CS with high MW of 114 and 170 and deacetylation over 80% are highly suitable for formulation (N/P of 50 or weight ratio of 25) of siRNA/CS complex. In this study, we have shown that CS with 85% deacetylation and low MW of 20 and 45 kDa can initially form stable complex with siRNA at weight ratio of 4 (Fig. 2a, lane 6) and achieve a significant gene-silencing effect when increasing weight ratio to adequately high level (Fig. 6). Moreover, we have found that increasing MW of CS up to 200 and 460 kDa reduces the gene knockdown level tremendously although stable complexes are formed as observed in gel analysis. These might be due to the complexes formed with a large number of short-chain CS, abundantly found in low-MW CS, released siRNA better than the complexes formed with a long chain of high-MW CS.
At the same pH (pH 7.2), the gene-silencing efficiency was affected by salt form, MW of CS and weight ratio. The result of gene silencing are consistent with the physicochemical properties of CS/siRNA complexes in that the complete complexes formed at highest weight ratio of 32, carried positive surface charge with relatively smallest size and induces highest gene-silencing effect. In addition, increasing weight ratio resulted in increasing the number of complete complexes and high gene-silencing efficiency. At a low weight ratio (4 and 8), the gene-silencing efficiency of all CS/siRNA complexes was lower than that of high weight ratio (16 and 32). This might be because the amount of positively charged amines in chitosan in the CS/siRNA complexes at low weight ratios was not sufficient to transfect cells. The CS/siRNA complexes achieved sufficient transfection efficiencies at higher weight ratios. This might be because an increase in the concentration of chitosan at higher weight ratios could yield a higher amount of complete complexes and have positively charged complexes to successfully transfect cells. The increase in transfection ability with an increase in chitosan concentration in CS/siRNA complexes was also demonstrated by Liu et al. (7). The complexes derived from CS-HCl, CS-Asp, and CS-Glu at the same weight ratio showed comparative level of gene knockdown except CS-Ac showed the lowest gene-silencing efficiency. This result indicated that the MW of CS and weight ratio rather than the salt form greatly influences the gene-silencing efficiency. The low gene-silencing efficiency of CS-Ac/siRNA complex was possibly due to the large particle size of the complete complex at weight ratio 32.
The kinetics of gene-silencing effect of CS/siRNA complexes formulated from CS MW20 kDa found that the highest gene silencing of all chitosan salts were at the post-transfection time of 2 to 3 days for CS/siRNA complexes formed at weight ratio of 4, and 5 days for CS/siRNA complexes formed at weight ratio of 8, 16, and 32 (Fig. 7). The degree of specific gene silencing is dependent on both the number of complexes that enter cells and the level of siRNA released from the complex which occurs in cell cytoplasm. The point that the complexes formed with low MW CS (20 and 45 kDa) at low weight ratio of 4 induces the lowest gene silencing and achieved the maximum level faster than the complexes formed at higher weight ratios, could be explained by the quick release of siRNA from the complexes rather than the better cell entry. The complexes formed at this low weight ratio are unstable with negative surface charge therefore gain less entry and quickly dissociate in the cytoplasm.
The system used to examine the gene-silencing effect of CS/siRNA complexes in this study is a constitutive gene expression which generally observed that the inhibition effect would be less and slower than the transient expression system (9,20). However, this system provides a more realistic model for clinical application in which a chronic disease gene such as viral hepatitis genes or tumor genes is the target for the delivered therapeutic siRNA. Taking the advantage of green fluorescence protein, cells in the transfected wells were observed daily under a fluorescence microscope. Several groups of expanding number of fade green cells derived from the original transfected cells carrying CS/siRNA complexes were found indicating that the gene-silencing effect was transferred to the daughter cells after each cycle of cell division. The gene-silencing effect remained for at least 7 days.
CONCLUSION
Successful siRNA delivery was observed in stable HeLa cells expressing green fluorescent protein (EGFP). The ability of chitosan to transfer functionally active siRNA into cell culture is greatly dependent on weight ratios and molecular weight of chitosan and less dependent on the salt forms of chitosan. The highest gene-silencing effect of CS/siRNA complex achieved at MW of 20 kDa and weight ratio of 32 with no apparent cell cytotoxicity.
Acknowledgments
The authors wish to thank the National Center for Genetic Engineering and Biotechnology (BIOTEC) Thailand, Commission of Higher Education (Thailand), The Thailand Research Funds, and National Research Council of Thailand for financial support.
References
- 1.Sharp PA. RNA interference. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 2.Lee M, Nah JW, Kwon Y, Koh JJ, Ko KS, Kim SW. Water-soluble and low molecular weight chitosan-based plasmid DNA delivery. Pharm Res. 2001;18:427–431. doi: 10.1023/A:1011037807261. [DOI] [PubMed] [Google Scholar]
- 3.Ishii T, Okahata Y, Sato T. Mechanism of cell transfection with plasmid/chitosan complexes. Biochim Biophys Acta. 2001;1514:51–64. doi: 10.1016/S0005-2736(01)00362-5. [DOI] [PubMed] [Google Scholar]
- 4.Weecharangsan W, Opanasopit P, Ngawhirunpat T, Rojanarata T, Apirakaramwong A. Chitosan lactate as a nonviral gene delivery vector in COS-1 cells. AAPS PharmSciTech. 2006;7:E1–E6. doi: 10.1208/pt070366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Yu SB, Wu FL, Mao ZB, Yu CL. Transfection of primary chondrocytes using chitosan-pEEGFP nanoparticles. J Control Release. 2006;112:223–228. doi: 10.1016/j.jconrel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 6.MacLaughlin FC, Mumper RJ, Wang J, Tagliaferri JM, Gill I, HinCHyiffe M, et al. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release. 1998;56:259–272. doi: 10.1016/S0168-3659(98)00097-2. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Howard KA, Dong M, Andersen MO, Rahbek UL, Johnsen MG, et al. The influence of polymeric properties on chitosan/siRNA nanoparticle formulation and gene silencing. Biomaterials. 2007;28:1280–1288. doi: 10.1016/j.biomaterials.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Pille JY, Li H, Blot E, Bertrand JR, Pritchard LL, Opolon P, et al. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Hum Gene Ther. 2006;17:1019–1026. doi: 10.1089/hum.2006.17.1019. [DOI] [PubMed] [Google Scholar]
- 9.Katas H, Alpar HO. Development and characterisation of chitosan nanoparticles for siRNA delivery. J Control Release. 2006;115:216–225. doi: 10.1016/j.jconrel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 10.Howard KA, Rahbek UL, Liu X, Damgaard CK, Glud SZ, Andersen MO, et al. RNA interference in vitro and in vivo using a novel chitosan/siRNA nanoparticle system. Mol Ther. 2006;14:476–484. doi: 10.1016/j.ymthe.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Andersen MO, Howard KA, Paludan SR, Besenbacher F, Kjems J. Delivery of siRNA from lyophilized polymeric surfaces. Biomaterials. 2007;29:506–512. doi: 10.1016/j.biomaterials.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Duan Y, Guan X, Ge J, Quan D, Zhou Y, Ye H, et al. Cationic nano-copolymers mediated IKKbeta targeting siRNA inhibit the proliferation of human Tenon’s capsule fibroblasts in vitro. Mol Vis. 2008;14:2616–2628. [PMC free article] [PubMed] [Google Scholar]
- 13.Caplen NJ, Parrish S, Iman F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. PNAS. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rojanarata T, Opanasopit P, Techaapornkul S, Ngawhirunpat T, Ruktanonchai U. Chitosan–thiamine pyrophosphate as a novel carrier for siRNA delivery. Pharm Res. 2008;25:2807–2814. doi: 10.1007/s11095-008-9648-6. [DOI] [PubMed] [Google Scholar]
- 15.Smith MD, Barbenel JC, Courtney JM, Grant MH. Novel quantitative methods for the determination of biomaterial cytotoxicity. Int J Artif Organs. 1992;15:191–194. [PubMed] [Google Scholar]
- 16.Weecharangsan W, Opanasopit P, Ngawhirunpat T, Apirakaramwong A, Rojanarata T, Ruktanonchai U, et al. Evaluation of chitosan salts as non-viral gene vectors in CHO-K1 cells. Int J Pharm. 2008;348:161–168. doi: 10.1016/j.ijpharm.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Thanou M, Florea BI, Geldof M, Junginger HE, Borchard G. Quaternized chitosan oligomers as novel gene delivery vectors in epithelial cell lines. Biomaterials. 2002;23:153–159. doi: 10.1016/S0142-9612(01)00090-4. [DOI] [PubMed] [Google Scholar]
- 18.Corsi K, Chellat F, Yahia L, Fernandes JC. MesenCHymal stem cells, MG 63 and HEK293 transfection using chitosan-DNA nanoparticles. Biomaterials. 2003;24:1255–1264. doi: 10.1016/S0142-9612(02)00507-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang SL, Yao HH, Guo LL, Dong L, Li SG, Gu YP, et al. Selection of optimal sites for TGFB1 gene silencing by chitosan-TPP nanoparticle-mediated delivery of shRNA. Cancer Genet Cytogenet. 2009;190:8–14. doi: 10.1016/j.cancergencyto.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Koping-Hoggard M, Varum KM, Issa M, Danielsen S, Christensen BE, Stokke BT, et al. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11:1441–1452. doi: 10.1038/sj.gt.3302312. [DOI] [PubMed] [Google Scholar]