Abstract
Hydrogen peroxide (H2O2) is a potent small-molecule oxidant that can exert a diverse array of physiological and/or pathological effects within living systems depending on the timing and location of its production, accumulation, trafficking, and consumption. To help study the chemistry and biology of this reactive oxygen species (ROS) in its native cellular context, we now present a new method for monitoring local, subcellular changes in H2O2 levels by fluorescence imaging. Specifically, we have exploited the versatility of the SNAP-tag technology for site-specific protein labeling with small molecules on the surface or interior of living cells with the use of boronate-capped dyes to selectively visualize H2O2. The resulting SNAP-Peroxy-Green (SNAP-PG) probes consist of appropriately derivatized boronates bioconjugated to SNAP-tag fusion proteins. Spectroscopic measurements on the SNAP-PG constructs confirm their ability to detect H2O2 with specificity over other biologically relevant ROS. Moreover, these hybrid small-molecule/protein reporters can be used in live mammalian cells expressing SNAP-tag fusion proteins directed to the plasma membrane, nucleus, mitochondria, and endoplasmic reticulum. Imaging experiments using scanning confocal microscopy establish organelle-specific localization of the SNAP-tag probes and their fluorescence turn-on in response to changes in local H2O2 levels. This work provides a general molecular imaging platform for assaying H2O2 chemistry in living cells with subcellular resolution.
Introduction
Reactive oxygen species (ROS) molecules derived from the metabolism of oxygen by living organisms are classically known as indicators for oxidative stress and potential contributors to aging1–6 and diseases ranging from cancer7–12 to diabetes13–15 to neurodegeneration.16–18 However, organisms that live in aerobic environments can also harness and incorporate ROS chemistry into their normal physiology by means of regulated production, release, and compartmentalization of these oxygen metabolites.19–26 A prime example of ROS chemistry used for beneficial purposes is the oxidative burst within phagosomes of macrophages and neutrophils that serves as a defense against invading pathogens.27–29 In this context, a major ROS produced in living systems is hydrogen peroxide (H2O2), which is a newly recognized messenger in cellular signal transduction but also a participant in oxidative stress and damage. Whereas high concentrations of H2O2 can trigger apoptotic or necrotic cell death, controlled bursts of H2O2 produced in response to stimulation by various growth factors, cytokines, and neurotransmitters can act as secondary messengers to signal cell growth, proliferation, and differentiation.19–26, 30–39
Because variations in the spatial and temporal production of H2O2 lead to disparate downstream biological effects, new methods that allow detection of this ROS in living systems with subcellular resolution can help decipher its complex cellular chemistry. To this end, an elegant example of a fluorescent protein sensor based on a circularly permuted YFP with an OxyR insert has been reported for cellular H2O2 detection.40 We envisioned developing an alternative chemical biology approach that combines the versatility of small-molecule reporters with the targetability of genetically encodable protein scaffolds for subcellular localization. Currently, selective protein labeling in living cells with synthetic compounds is feasible in either a covalent and non-covalent manner.41–45 Non-covalent labeling methods utilizing the specific binding of ligands to short peptide or protein domains include the binding of biarsensic compounds to tetracysteine haiprins (FlAsH, ReAsH),46 the binding of metal complex to oligo-His47, 48 or oligo-Asp motifs,49 the recognition of trimethoprim by dihydrofolate reductase,50, 51 and a high affinity binding interaction of protein FKBP12(F36V) with its ligand FK-SLF.52 Examples of covalent self-labeling mediated by fusion enzymes include attachment of alkyl chlorides to dehalogenase enzymes (HaloTag),53–55 formation of covalent adduct between cutinase and p-nitrophenyl phosphonate (pNPP) derivatives,56 labeling of AGT (O6-alkyguanine-DNA alkyltransferase) fusion proteins with SNAP/CLIP substrates,57–59 and covalent labeling by enzymes involved in post-translation modifications such as phosphopantetheine transferase,60, 61 transglutaminase,62, 63 biotin ligase BirA,64, 65 lipoic acid ligase,66 sortaseA,67, 68 protein farnesyltransferase,69 and formylglycine generating enzyme.70, 71 In particular, we were attracted to the SNAP tag technology, which within short time after its invention by Johnsson and coworkers, has found broad utility in a variety of biological applications. Use of the SNAP tag allows precise targeting of bioactive reagents or probes to specific subcellular locales; recent examples include photosensitizers for chomophore-assisted laser inactivation (CALI) of fusion proteins72 and fluorescent sensors for intracellular metal ions including zinc(II)73 and calcium(II).74 Simultaneous labeling of two different targets can also be achieved using tandem SNAP and CLIP tags.59 The SNAP tag has been used for protein tracking with spatial and temporal resolution, where a fusion protein at a given time point is initially marked by a fluorescent or non-fluorescent SNAP tag substrate label, and subsequent newly synthesized proteins can be differentiated by a second label.59, 75 Other applications of the SNAP tag technology include its use in combination with fluorescent proteins as FRET-based reporters for studying protein-protein interactions76 and the development of semisynthetic fluorescent sensors or photoactivable switches.77, 78
In this report, we present the design, synthesis, properties, and cellular applications of a new class of organelle-targetable fluorescent probes for H2O2 based on SNAP-AGT bioconjugation chemistry. We have prepared and characterized a pair of boronate-caged, Peroxy Green-type fluorescent peroxide indicators bearing SNAP substrates, where one is a membrane-permeable version for intracellular use and another is a membrane-impermeable version for cell surface labeling, and coupled them to AGT proteins expressed in various cellular compartments. The resulting SNAP-Peroxy-Green (SNAP-PG) hybrid small-molecule/protein conjugates are capable of detecting H2O2 over a variety of ROS and can be targeted to various subcellular locales, including the plasma membrane, nucleus, mitochondrial inner membrane, and endoplasmic reticulum, to sense changes in H2O2 levels within or on the surface of living cells. This method provides a general platform for studying the chemistry of H2O2 by molecular imaging in living biological specimens with subcellular resolution.
Results and Discussion
Design and Synthesis of SNAP-Peroxy-Green Bioconjugates as Organelle-Targetable Fluorescent Reporters for Cellular Hydrogen Peroxide
Figure 1 depicts our overall strategy for creating subcellular-targetable fluorescent H2O2 probes by combining the SNAP-tag methodology for site-specific labeling79, 80 with the chemoselective H2O2-mediated deprotection of boronate esters to phenols for reaction-based detection of H2O2.81–86 Fusion of AGT with proteins containing a signaling sequence allows expression of AGT in defined subcellular compartments. These localized AGT scaffolds can then be tagged by covalent labeling at Cys145 with a boronate Peroxy Green (PG) fluorescent probe bearing an appended SNAP substrate. The resulting SNAP-PG conjugates are hybrid small-molecule/protein reporters that give a fluorescent turn-on response upon H2O2-mediated boronate cleavage.
Figure 1.
Design strategy for organelle-specific hydrogen peroxide reporters using the SNAP tag methodology.
To this end, we previously reported the synthesis and application of Peroxy Green 1 (PG1),87 a cell-permeable and selective H2O2 probe that can be activated by a single boronate deprotection. Along these lines, we sought to create PG1 analogs for organelle-specific labeling by replacing its 4-methoxy moiety with benzylguanine or benzyl-2-chloro-6-aminopyrimidine SNAP tag substrates. Scheme 1 depicts the synthesis of SNAP tag derivatizes for coupling to a PG1 scaffold. Benzylguanine 4 was synthesized according to literature protocols.58 In a similar manner, benzyl-2-chloro-6-aminopyrimide 3 was synthesized by nucleophilic substitution of 2,6-dichloropyrimidin-4-amine with trifluoro-N-(4-hydroxymethyl-benzyl)-acetamide 1. The trifluoroacetamide protecting group was subsequently removed with 33% methylamine in ethanol.
Scheme 1.
Synthesis of SNAP-tag substrates
Scheme 2 details the synthesis of SNAP-PeroxyGreen-1 (SPG1) and SNAP-PeroxyGreen-2 (SPG2) probes by coupling the aforementioned SNAP tag substrates with a carboxylate-derivatized PG1; the latter is prepared in five steps from the fluorescein-like dye Tokyo Green.88 Treatment of Tokyo Green 5 with boron tribromide at −78 °C in dry CH2Cl2 furnishes phenolic Tokyo Green 6. O-alkylation of 6 in anhydrous DMF with excess cesium carbonate and a slow addition of tert-butylbromoacetate gives tert-butyl ester Tokyo Green 7. Reaction of 7 with bis(trifluoromethane)sulfonamide affords triflate 8. Palladium-mediated borylation of triflate 8 with bis(pinacolato)diboron, cyclohexyl JohnPhos, and diisopropylethylamine in anhydrous 1,4-dioxane provides boronic ester 9. Subsequent treatment with trifluoroacetic acid in anhydrous CH2Cl2 delivers carboxyl PG 10. Amide bond formation between carboxyl PG 10 and SNAP substrate 4 or 3 mediated by N,N,N′,N′-tetramethyl-O-(7-azabenzotriazol-1-yl)uroniumhexafluorophosphate (HATU) affords SPG1 (11) or SPG2 (12), respectively.
Scheme 2.
Synthesis of SNAP-Peroxy-Green-1 (SPG1) and SNAP-Peroxy-Green-2 (SPG2)
Finally, we also synthesized Tokyo Green (TG) analogs bearing SNAP tags 3 or 4 as control compounds for cell labeling and imaging as shown in Scheme 3. Treatment of tert-butyl ester Tokyo Green 7 with TFA furnishes carboxyl Tokyo Green 13. The HATU-mediated amide coupling of 13 with either 4 or 3 provides SNAP-TokyoGreen-1 (STG1, 14) and SNAP-TokyoGreen-2 (STG2, 15), respectively.
Scheme 3.
Synthesis of SNAP-Tokyo Green-1 (STG1) and SNAP-Tokyo Green-2 (STG2)
Spectroscopic Properties, In Vitro Protein AGT labeling, and Peroxide Responses of SNAP-PG and SNAP-TG Fluorophores
Table 1 summarizes the spectroscopic properties of the SNAP-PG and SNAP-TG fluorophores in aqueous buffer at neutral pH (20 mM HEPES, pH 7). SPG1 and SPG2 both show visible absorption bands centered at 465 nm (SPG1: ε = 10,200 M−1 cm−1, SPG2: ε = 9,800 M−1 cm−1) and weak fluorescence with an emission maximum at 515 nm. In contrast, the STG1 and STG2 fluorophores exhibit a prominent absorption at 495 nm (STG1: ε = 36,000 M−1 cm−1, STG2: ε = 37,000 M−1 cm−1) and fluorescent emission maximum at 513 nm. The quantum yield of STG1 (Φ = 0.12) is much lower than that of STG2 (Φ = 0.87), presumably due to the greater quenching effect of guanidine moiety; a similar effect has been observed in other SNAP fluorophores.73 We thus proceeded to test the H2O2 reactivity of these free dyes with the SPG2 analog. Reaction of SPG2 with 1 mM H2O2 triggers a ca. 32-fold fluorescence turn-on for the dye (Figure S11) confirming the peroxide response of these SNAP tag reporters. Kinetics measurements of the SPG2 boronate deprotection performed under pseudo-first order conditions (1 μM dye, 1 mM H2O2) give an observed rate constant of kobs = 1.0(1) × 10−3 s−1.
Table 1.
Spectroscopic Properties of SNAP Dyes and Their AGT Bioconjugates.
| Compound | λexc (nm) | (M−1 cm−1) | λem (nm) | Φ |
|---|---|---|---|---|
| STG1 | 495 | 36000 | 513 | 0.12 |
| STG2 | 495 | 37000 | 513 | 0.87 |
| AGT-TG | 500 | 32000 | 515 | 0.57 |
|
| ||||
| SPG1 | 465 | 10200 | 515 | 0.10 |
| SPG2 | 465 | 9800 | 515 | 0.09 |
| AGT-PG | 465 | 11500 | 515 | 0.07 |
We next proceeded to test the ability of the SPG and STG dyes to label purified AGT protein samples and to evaluate the spectroscopic properties and peroxide responses of the isolated fluorophore-protein conjugates. For these in vitro protein labeling assays, hexahistidine-tagged AGT (His-AGT) was overexpressed in E. coli and purified with a standard Ni-NTA affinity column. After incubation of His-tagged AGT with SPG1, SPG2, STG1, or STG2 dyes in 20 mM HEPES at pH 7 for 30 min at 37 °C, we confirmed the formation of the desired covalently labeled products, AGT-PG and AGT-TG, by both LC-MS and 488 nm excitation in-gel fluorescence measurements (Supporting Information, Figures S1 to S7). Specifically, AGT-TG and AGT-PG show mass increases of 478 and 506 Da, respectively, from His-tagged AGT; these values are consistent with the calculated mass of the protein tagged with TG or the boronic acid form of PG, respectively. We purified the AGT-PG and AGT-TG bioconjugates for further spectroscopic evaluation using MWCO spin columns and size-exclusive gel chromatography to remove the free organic fluorophores. This dual purification method is essential to obtain clean dye-protein conjugates without interference from small-molecule impurities. In addition, the labeling proceeds well under oxidizing conditions, as we observe covalent adduct formation in the presence up to 1 mM H2O2 (Figure S2). AGT-TG possesses a dominant absorption maximum in the visible region centered at 500 nm, which is slightly red-shifted relative the free TG dye. The AGT-TG complex shows prominent absorption at 280 nm (standard protein absorption) and 500 nm (TG dye), consistent with labeling of the AGT protein with the small-molecule TG payload (Figure S8). Furthermore, formation of AGT-TG conjugate is also reflected in the observed fluorescence quantum yield, as binding of STG1 to AGT ejects guanidine from the dye and eliminates its quenching effect, resulting in a 6-fold fluorescence increase (Figure S9). The absorption and emission spectra of AGT-PG are similar to SPG1 and SPG2, with an additional 280 nm absorption due to the AGT protein.
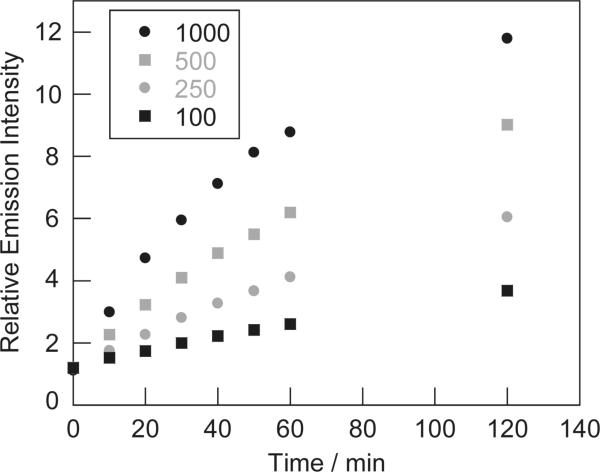
Importantly, the SPG dyes are able to retain their fluorescence response to H2O2 upon protein labeling, as purified AGT-PG conjugates exhibit a marked turn-on emission response to added H2O2 (Figure 2). The H2O2-induced fluorescence increases occur with concomitant growth of the characteristic TG absorption at 500 nm, consistent with boronate cleavage of PG to produce the phenol TG product (Figure 3). Finally, we observe a linear correlation between various H2O2 concentrations added and observed fluorescence emission responses (Figure 4).
Figure 2.
Fluorescence responses of 1 μM AGT-PG, a conjugate formed from the reaction of AGT with SPG2, to 100, 250, 500, and 1000 μM H2O2 in 20 mM HEPES, pH 7 at 25 °C. The plot shows emission responses at 0, 10, 20, 30, 40, 50, 60, and 120 min after H2O2 addition. The reactions are not complete at these early time points. The collected emission was integrated between 500–650 nm (λexc = 488 nm).
Figure 3.
Absorption spectra of 1 μM AGT-PG in 20 mM HEPES, pH 7 upon addition of 1 mM H2O2. Spectra were acquired at one hour intervals for 8 hours. An isosbestic point was observed at 460 nm.
Figure 4.
Fluorescence responses of 1 μM AGT-PG to various concentrations of added H2O2. Spectra were acquired in 20 mM HEPES, pH 7 at 25 °C after incubation of the probe with H2O2 for 30 min. Reactions are not complete at this early time point. The collected emission was integrated between 500–650 nm (λexc = 488 nm).
Fluorescence Detection of H2O2 in Various Compartments of Living Cells with SNAP-AGT Peroxy Green Reporters
With spectroscopic data establishing the formation of AGT-PG conjugates and their fluorescence turn-on response to H2O2, we next sought to apply SPG1 and SPG2 for optical detection of H2O2 in living biological samples. Specifically, we focused on directing these probes to a broad range of subcellular compartments that are either capable of locally generating H2O2 in response to various stimuli or to sites where their function and regulation are sensitive to H2O2 flux. In this study, we targeted SPG reporters to a broad spectrum of locations. Included are the plasma membrane, mitochondria, endoplasmic reticulum, and nucleus. To this end, plasmids for expression of the SNAP-tag in live mammalian cells were obtained from Covalys and then subjected to further modification as necessary. The parent SNAP-tag without a signaling sequence (pSNAP), encoding a genetically optimized AGT protein, gives a uniform intracellular expression of SNAP-tag in the cytoplasm and the nucleus. Localization of the SNAP-tag to nucleus is achieved by fusion of SNAP-tag to the C-terminus of the histone H2B protein (SNAP-H2B). Fusion of the SNAP-tag to the C-terminus of cytochrome c oxidase subunit 8 (Cox8A) enables its localization to the mitochondrial inner membrane (SNAP-Cox8A). For plasma membrane targeting, SNAP-tag is expressed as a fusion to the N-terminus of neurokinin-1 receptor (NK1R) and the C-terminus of 5HT3A serotonin receptor signaling sequence, resulting in a SNAP-tag exposed to the extracellular space (SNAP-NK1R). Addition of KDEL, a signaling peptide for retention of protein in endoplasmic reticulum, to the C-terminus of SNAP results in an ER localized SNAP-tag (SNAP-KDEL).
We first tested the site-specific labeling of various SNAP fusion proteins with control STG1 and STG2 dyes in live HEK293T cells. To our surprise, we found that STG1 and STG2 exhibit marked differences in cell permeability that can be exploited to obtain more selective conjugation to extracellular and intracellular spaces. Specifically, whereas both STG1 and STG2 gave excellent membrane labeling with SNAP-NK1R, intracellular tagging of pSNAP and SNAP-H2B was observed only with STG2 (Figure 5). The same trend applies to SPG1 and SPG2, where only the latter is membrane permeable. Our findings are consistent with previous work that reports that benzylguanine can hamper the cell permeability of its conjugated partners in a fluorophore-dependent manner.58, 89 STG2 shows desirable properties for intracellular site-specific labeling applications, as the dye is cell permeable and unbound probe can be readily washed out. This latter property is crucial to minimizing background fluorescence from non-specific or off-target labeling. Indeed, in HEK293T cells transiently expressing SNAP-KDEL and SNAP-Cox8A to target the endoplasmic reticulum and mitochondria, respectively, we were able to successfully tag these organelles with STG2. The subcellular localization of this dye to these compartments was confirmed by fluorescence overlay of green emissive STG2 with red emission from mCherry-KDEL and mCherry-Cox8A constructs (Figure 6).
Figure 5.
Targeted localization of STG1 and STG2 in living HEK293T cells by conjugation to SNAP-AGT fusion proteins. Cells were incubated with 5 μM STG1 or STG2 for 30 min, and washed with fresh DMEM + 10% FBS for 30 min (2 × 1 mL) before image acquisition. Rows (a) and (b) show HEK 293T cells transiently expressing SNAP-NK1R. Rows (c) and (d) display HEK 293T cells transiently expressing pSNAP for non-specific intracellular tagging, and row (e) presents HEK 293T cells transiently expressing SNAP-H2B. For each series: (1) emission from labeling with STG1 or STG2, (2) nuclear staining with Hoechst 33342, and (3) DIC image. Scale bar = 20 μm.
Figure 6.
Targeted labeling of endoplasmic reticulum and mitochondria organelles with STG2. Row (a) shows HEK 293T cells expressing SNAP-KDEL in the ER lumen, and row (b) depicts HEK 293T cells expressing SNAP-Cox8A for mitochondrial tagging. For each series: (1) emission from labeling with STG2, (2) emission from (a) mCherry-KDEL or (b) mCherry-Cox8A, (3) overlay of STG2 and mCherrry, (4) nuclear staining with Hoechst 33342, and (5) DIC image. Scale bar = 20 μm.
Recognizing these SNAP-tag-dependent differences in membrane permeability, we then proceeded to evaluate the boronate SPG1 and SPG2 reporters for site-specific subcellular H2O2 detection, employing SPG1 for extracellular plasma membrane targeting and SPG2 for intracellular labeling. We utilized real-time imaging of the same cells before and after oxidative stimulation or in untreated samples. This method allows us to perform an important control for varying levels of protein expression as well as a control to show that photodynamic oxidation is not significant with these systems. For extracellular measurements, HEK293T cells transiently expressing SNAP-NK1R display weak but detectable green fluorescent emission localized to the plasma membrane after SPG1 conjugation. The relatively poor membrane permeability of this probe allows for rapid and selective labeling of cell surfaces with washing. Treatment of SPG1-labeled cells with 100 μM H2O2 triggers a marked increase in fluorescence from the plasma membrane region as shown by time lapse image acquisitions (Figure 7a). Importantly, negligible emission comes from non-transfected cells in the same image frame even after treatment with H2O2, establishing that specific SPG1 binding to the SNAP-NK1R fusion protein scaffold is required for imaging changes in H2O2 levels localized to the plasma membrane. As a further control, we do not observe fluorescence turn-on responses for SPG1-loaded cells in the absence of H2O2, and a combination of nuclear staining and brightfield transmission measurements confirm that the cells are viable throughout the imaging experiments.
Figure 7.
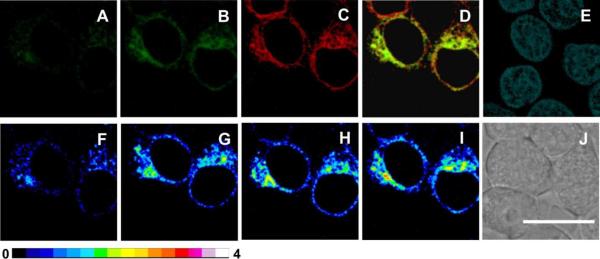
Fluorescence detection of H2O2 on the surface of or within living HEK 293T cells. Row (a) shows cells transiently expressing SNAP-NK1R tagged with SPG1, row (b) displays cells transiently expressing pSNAP tagged with SPG2, and row (c) presents cells transiently expressing SNAP-H2B tagged with SPG2. H2O2 was added from 50 mM solution in Mili-Q water. Time-lapse image acquisition was assisted by a motorized stage equipped with incubator and humidifier maintaining 37 °C and 5% CO2 atmosphere. For each series: (1–4) pseudocolor mode of fluorescent emission at 0, 10, 20, and 30 min after addition of 100 μM H2O2, (5, 6) fluorescent emission before (5) and after (6) treatment of cells with 100 μM H2O2 for 30 min (λexc = 488 nm, λem = 500–550 nm), (7) nuclear staining with Hoechst 33342, (8) DIC image. Scale bar = 20 μm.
To label intracellular compartments, we subjected cells to 5 μM SPG2 for 30 min followed by a 30 min incubation in fresh complete medium (DMEM +10% FBS) to wash out any unbound probe. HEK293T cells transiently expressing pSNAP display weak intracellular fluorescent emission after labeling with SPG2. Addition of 100 μM H2O2 to the cells results in a patent increase in overall intracellular fluorescence from this protein/small-molecule reporter (Figure 7b). Likewise, SPG2-loaded HEK293T cells transiently expressing SNAP-H2B show nuclear-localized fluorescence that becomes more intense after stimulation with H2O2 (Figure 7c). Moreover, SPG2 can be precisely immobilized onto the mitochondrial inner membrane and ER regions of HEK293T cells transiently expressing SNAP-Cox8A (Figure 8) and SNAP-KDEL (Figure 9), respectively. In both cases, the turn-on green fluorescence emission from the SNAP-PG probes after H2O2 treatment display good co-localization with the red mCherry emission of the corresponding SNAP protein target, and the staining pattern can persist for many hours. Because the mitochondrial and ER structures are relatively small compared to the overall volume of the cell, fluorescence emission from these organelles could be easily overwhelmed by noise from any non-specific staining in cytoplasm. These experiments clearly demonstrate that non-specific binding is not an issue with the SPG2 probe, as unbound dye can be effectively removed from cells with a simple washing step. Taken together, these data show that SPG1 and SPG2 can be used to label and sense changes in H2O2 levels in specific subcellular compartments via selective bioconjugation with a variety of localizable SNAP-tag fusion constructs (Figure 10).
Figure 8.
Fluorescence detection of H2O2 in living HEK 293T cells transiently expressing SNAP-KDEL and mCherry-KDEL. Panels (a) and (b) show fluorescent emission from cells labeled with SPG2 before (a) and after (b) treatment with 100 μM H2O2 for 30 min (λexc = 488 nm, λem = 500–550 nm), panel (c) presents emission from mCherry-KDEL, panel (d) displays the overlay of (b) and (c), panel (e) shows nuclear staining with Hoechst 33342, and panels (f–i) display pseudocolor fluorescent emission at 0, 10, 20, and 30 min after addition of 100 μM H2O2. Panel (j) presents the DIC image of cells in (i). Scale bar = 20 μm.
Figure 9.
Fluorescence detection of H2O2 in living HEK 293T cells transiently expressing SNAP-Cox8A and mCherry-Cox8A. Panels (a) and (b) show fluorescent emission from cells labeled with SPG2 before (a) and after (b) treatment with 100 μM H2O2 for 30 min (λexc = 488 nm, λem = 500–550 nm), panel (c) presents emission from mCherry-Cox8A, panel (d) displays the overlay of (b) and (c), panel (e) shows nuclear staining with Hoechst 33342, and panels (f-i) display pseudocolor fluorescent emission at 0, 10, 20, and 30 min after addition of 100 μM H2O2. Panel (j) presents the DIC image of cells in (i). Scale bar = 20 μm.
Figure 10.
Relative fluorescence emission from time-lapse confocal images of SPG1 or SPG2 labeled HEK 293T cells in response to added 100 μM H2O2 at 0, 10, 20, 30 min. Error bars represent standard error measurement (s.e.m).
Concluding Remarks
In summary, we have described the synthesis, spectroscopic properties, H2O2 responses, and live-cell imaging applications of a new class of organelle-targeted reporters for cellular H2O2. SPG1 and SPG2 combine boronate-caged Tokyo Green derivatives for H2O2 detection with SNAP tags for site-specific protein labeling at extracellular and intracellular locations, respectively. When SPG1 and SPG2 are exposed to AGT or an AGT fusion protein, these dyes form covalent AGT-PG adducts that respond to H2O2 by a turn-on fluorescence increase upon H2O2-mediated boronate deprotection. These hybrid small-molecule/protein reporters can be used to label a variety of subcellular locations, including the plasma membrane, nucleus, mitochondria, and endoplasmic reticulum, and detect changes in local H2O2 fluxes in living cells by confocal microscopy. The methodology presented here combines the precision of site-specific labeling using genetically encodable tags with the chemical versatility of small-molecule probes. Ongoing efforts are focused on expanding the expanding color palette of targetable H2O2 probes, optimizing their sensitivities and dynamic turn-on responses to H2O2, creating targetable reporters with ratiometric readouts, and developing additional labeling methodologies for multichannel imaging. We anticipate that simultaneous measurements of H2O2 and other ROS fluxes in multiple locales using hybrid small-molecule/protein conjugates will contribute to a better understanding of oxidation biology in states of health, aging, and disease, with particular interest in elucidating cellular redox regulation and signaling in multiple cellular compartments.
Experimental Section
Materials and Methods
All reactions were carried out under a dry nitrogen atmosphere. Silica gel 60 (230–400 mesh, Fisher) was used for column chromatography. Palladium(II) acetate [Pd(OAc)2], Dichloro[1,1′-bis(diphenylphosphino)ferrocene]palladium (II) [Pd(dppf)Cl2], and 2-(Dicyclohexylphosphino)biphenyl [Cyclohexyl JohnPhos] were purchased from Strem Chemicals (Newburyport, MA). Bis(pinacolato)diboron was purchased from Boron Molecular (Research Triangle Park, NC). 2,2,2-Trifluoro-N-(4hydroxylmethyl-benzyl)-acetamide (1) and O6-(4-Amino-methyl-benzyl)guanine (4) were synthesized following literature procedures.58 All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and were used as received. 1H NMR and 13C NMRspectra were collected in CDCl3, DMSO-d6, or CD3OD (Cambridge Isotope Laboratories, Cambridge, MA) at 25 °C using a Bruker AVQ-400 spectrometer at the College of Chemistry NMR Facility at the University of California, Berkeley. All chemical shifts are reported in the standard δ notation of parts per million. Low-resolution mass spectral analyses were carried out using GC-MS (Agilent Technology 5975C, inert MSD with triple axis detector) or LC-MS (Agilent Technology 6130, Quadrupole LC/MS). High-resolution mass spectral analyses (ESI-MS, FAB-MS) were carried out at the College of Chemistry Mass Spectrometry Facility at the University of California, Berkeley.
Restiction endonuclease and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Hoechst 33342, monoclonal ANTI-FLAG M2 antibody and Alexa Fluor 488 Goat anti-mouse lgG antibody were obtained from Invitrogen (Carlbad, CA). pCEMS-CLIP10m-NK1R, pSEMS-SNAP26m-ADBR, pCEMS1-H2B-CLIP10m, and pSEMS1-Cox8A-SNAP26m plasmids were purchased from Covalys Biosciences (Witterswil, Switzerland). pmCherry and DsRed Golgi were obtained from Clontech Laboratories (Mountain View, CA). pET28a(+) was obtained from Novagen (EMD Chemicals, Gibbstown, NJ). Primers for PCR amplification were synthesized by Intergrated DNA Technologies (San Diego, CA).
N-(4-((2-Amino-6-chloropyrimidin-4-yloxy)methyl)benzyl)-2,2,2-trifluoroacetamide (2)
Compound 1 (1.0 g, 4.3 mmol) in anhydrous DMF (7 mL) was added to a dry 50 mL Schlenk flask under a nitrogen atmosphere. NaH (60% in mineral oil, 515 mg, 12.8 mmol) was then added slowly to the reaction flask. After stirring at room temperature for 15 min, the solution appeared blue-green in color. A solution of 4,6-dichloropyrimidin-4-amine (700 mg, 4.3 mmol) in anhydrous DMF (5 mL) was added dropwise to the reaction flask over a 5 min period, giving a brown-colored solution. The reaction was then heated to 90 °C for 6 h. The reaction was cooled to room temperature, quenched with slow addition of cold water (20 mL), extracted with EtOAc (3 × 30 mL), and dried over Na2SO4. Purification by column chromatography (silica gel, EtOAc:hexanes 2:1) gave 2 as a clear viscous oil (300 mg, 19% yield). 1H NMR (DMSO-d6, 400 MHz): δ 9.99 (1H, br-s), 7.39 (2H, d, J = 8.4 Hz), 7.26 (2H, d, J = 8.4 Hz), 7.09 (2H, s), 6.10 (1H, s), 5.26 (2H, s), 4.36 (2H, s). GC-MS: calculated for [M+] 360, found 360.
4(4-(Aminomethyl)benzyloxy)-6-chloropyrimidin-2-amine (3)
Methylamine (33% in EtOH, 2.5 mL) was added to the solution of 2 (250 mg, 0.69 mmol) in MeOH (5 mL). The mixture was stirred at room temperature for 24 h. The solvent was removed in vacuo, providing product 3 as a white powder (175 mg, 95% yield) 1H NMR (DMSO-d6, 400 MHz): δ 7.33 (2H, d, J = 8.0 Hz), 7.31 (2H, d, J = 8.0 Hz), 7.10 (2H, br-s), 6.10 (1H, s), 5.25 (2H, s), 3.68 (2H, s) LC-MS: calculated for [M+] 265.1, found 265.1.
6-Hydroxy-9-(4-hydroxy-2-methylphenyl)-3H-xanthen-3-one (6)
Tokyo Green 5 (6.3 g, 18.9 mmol) was added to a dried 500 mL round bottom flask. Anhydrous CH2Cl2 (300 mL) was added by cannula and the resulting insoluble mixture stirred at room temperature for 5 min. The flask was cooled to −78 °C in a CO2(s)/acetone bath and boron tribromide (14.2 g, 5.4 mL, 56.7 mmol) was added dropwise over 15 min. The reaction was allowed to warm to room temperature overnight and added slowly to 500 mL of vigorously stirred ice/H2O slurry, causing precipitation of a reddish orange solid. The mixture was stirred for an additional 24 h and the solid collected by vacuum filtration. Purification by flash column chromatography (silica gel, MeOH:CH2Cl 1:10) afforded 4 as a brick red solid (5.3 g, 87% yield). 1H NMR (DMSO-d6, 400 MHz): δ 10.00 (1H, s), 7.35 (2H, d, J = 9.2 Hz), 7.08 (2H, d, J = 8.0 Hz), 7.07 (1H, s), 7.03 (2H, dd, J = 9.2, 2.4), 6.87 (1H, d, J = 2.4 Hz), 6.83 (2H, dd, J = 8.0, 2.4), 1.90 (3H, s). LC-MS: calculated for [MH+] 319, found 319.
tert-Butyl 2-(4-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-3-methylphenoxy)acetate (7)
Phenolic Tokyo Green 6 (3.13 g, 10.0 mmol) was added to a dried 500 mL Schlenk flask. Anhydrous DMF (300 mL) and cesium carbonate (22.2 g, 68.1 mmol) were added, and the mixture stirred at room temperature for 60 min. tert-Butylbromoacetate (1.95 g, 1.46 mL, 10.0 mmol) was added by micropipettor and the resulting reaction mixture stirred at room temperature overnight. The reaction was filtered and solvent concentrated under reduced pressure, giving a dark brown mass. H2O (100 mL) was added and the solution neutralized with 1M HCl, giving a red orange precipitate. The solid was collected by vacuum filtration, washed with ddH2O (100 mL), and dried in air. Purification by flash column chromatography (silica gel, MeOH:CH2Cl2 1:20) furnished 5 as a brick red solid (1.54 g, 36% yield). 1H NMR (CDCl3, 400 MHz): δ 7.10 (2H, d, J = 9.2 Hz), 7.09 (1H, d, J = 8.4 Hz), 6.93 (1H, d, J = 2.4 Hz), 6.89 (1H, s), 6.88 (2H, s), 6.83 (2H, dd, J = 9.2, 2.4 Hz), 4.61 (2H, s), 2.03 (3H, s), 1.54 (9H, s). LC-MS: calculated for [MH+] 433, found 433.
tert-Butyl-2-(3-methyl-4-(3-oxo-6-(trifluoromethylsulfonyloxy)-3H-xanthen-9-yl)phenoxy)acetate (8)
tert-Butyl Tokyo Green 7 (1.06 g, 2.44 mmol) was added to a dried 25 mL Schlenk flask. Anhydrous DMF (5 mL) and DIPEA (1.57 g, 2.1 mL, 12.2 mmol) were added, and the resulting mixture stirred at room temperature for 60 min. N-Phenyl bistrifluoromethane sulfonimide (1.37 g, 3.84 mmol) was dissolved in anhydrous DMF (3 mL) and added dropwise to the stirring basic solution over 5 min. The mixture was stirred at room temperature overnight, diluted in toluene (200 mL), washed with H2O (3 × 100 mL), and dried over MgSO4. Purification by flash column chromatography (silica gel, EtOAc:hexanes 1:4 to 1:2 gradient) gave 8 as a puffy, orange crystalline solid (930 mg, 67% yield). 1H NMR (CDCl3, 400 MHz): δ 7.39 (1H, d, J = 2.4 Hz), 7.18 (1H, d, J = 8.8 Hz), 7.09 (2H, d, J = 8.8, 2.4 Hz), 7.01 (1H, d, J = 10.0 Hz), 6.95 (1H, d, J = 2.4 Hz), 6.91 (1H, dd, J = 10, 2.4 Hz), 6.60 (1H, dd, J = 10.0 2.0 Hz), 6.44 (1H, d, J = 2.0 Hz), 4.61 (2H, s), 2.05 (3H, s), 1.54 (9H, s). LC-MS: calculated for [MH+] 565, found 565.
tert-Butyl-2-(3-methyl-4-(3-oxo-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-3H-xanthen-9-yl)phenoxy)acetate (9)
Compound 8 (641 mg, 1.14 mmol), Pd(OAc)2 (84 mg, 0.12 mmol), Cyclohexyl JohnPhos (201 mg, 0.57 mmol), and bis(pinacolato)diboron (446 mg, 1.76 mmol) were added to a dried 25 mL scintillation vial in a glove box. Anhydrous 1,4-dioxane (10 mL) was added and the solution stirred at room temperature for 5 min. Anhydrous DIPEA (742 mg, 1.00 mL, 5.74 mmol) was added dropwise by syringe and the reaction mixture was stirred at room temperature overnight. The reaction vial was removed from the glove box, poured into saturated aqueous NH4Cl (30 mL), extracted with EtOAc (3 × 30 mL), and dried over MgSO4. Purification by flash column chromatography (silica gel, EtOAc:hexanes 1:4) delivered 9 as an orange crystalline solid (615 mg, 99% yield). 1H NMR (CDCl3, 400 MHz): δ 7.90 (1H, s), 7.57 (1H, d, J = 8.0 Hz), 7.08 (1H, J = 8.4 Hz), 7.07 (1H, d, J = 8.0 Hz), 7.01 (1H, d, J =9.6 Hz), 6.94 (1H, d, J = 2.4 Hz), 6.89 (1H, dd, J = 8.4, 2.4 Hz), 6.59 (1H, dd, J = 9.6, 2.0 Hz) 6.43 (1H, d, J = 2.0 Hz), 4.61 (2H, s), 2.03 (3H, s), 1.54 (9H, s), 1.37 (12H, s). LC-MS: calculated for [MH+] 543, found 543.
2-(3-Methyl-4-(3-oxo-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-3H-xanthen-9-yl)phenoxy)acetic acid (10)
tert-Butyl Peroxy Green 9 (542 mg, 1.0 mmol) was dissolved in CH2Cl2 (3 mL) and added dropwise by Pasteur pipette to a stirring mixture of CH2Cl2:TFA (1.0:1.0, 25 mL). The solution was allowed to stir for 4 h and the solvent removed under reduced pressure. Purification by flash column chromatography (silica gel, MeOH:CH2Cl2 1:10) gave 10 as an orange film (419 mg, 86% yield). 1H NMR (CDCl3, 400 MHz): δ 8.23 (1H, s), 7.82 (1H, d, J = 8.0 Hz), 7.41 (1H, d, J = 8.0 Hz), 7.40 (1H, d, J = 8.8 Hz) 7.15 (3H, m), 7.04 (1H, s), 7.00 (1H, d, J = 8.8 Hz), 4.83 (2H, s), 2.01 (3H, s), 1.39 (12H, s). LC-MS: calculated for [MH+] 487, found 487.
N-(4-((2-Amino-9H-purin-6-yloxy)methyl)benzyl)-2-(3-methyl-4-(3-oxo-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-3H-xanthen-9-yl)phenoxy)acetamide, SNAP-Peroxy-Green-1, SPG1 (11)
Compound 10 (99 mg, 0.20 mmol) was added to a dried 25 mL Schlenk flask. Anhydrous DMF (3 mL) was added by syringe and the solution was stirred at room temperature for 5 min. HATU (89 mg, 0.23 mmol), 6-(4-(aminomethyl)benzyloxy)-9H-purin-2-amine 4 (63 mg, 0.23 mmol), and DIPEA (266 mg, 0.359 mL, 2.06 mmol) were added, and the reaction stirred at room temperature overnight. The reaction was poured into an ice/H2O slurry (30 mL) and stirred to precipitate an orange solid. The solid was collected by vacuum filtration and dried in vacuo. Purification by flash column chromatography (silica gel, MeOH:CH2Cl2 1:10) provided SPG1 11 as an orange film (17.1 mg, 11% yield). 1H NMR (DMSO-d6, 400 MHz): δ 12.41 (1H, s), 8.75 (1H, s), 7.78 (1H, s), 7.71 (1H, s), 7.55 (1H, d, J = 7.6 Hz), 7.45 (2H, d, J = 7.2 Hz) 7.31 (2H, d, J = 7.6 Hz), 7.24 (1H, d, J = 8.4 Hz), 7.11 (1H, s), 7.04 (2H, d, J = 8.4 Hz), 6.94 (1H, d, J = 9.6 Hz), 6.50 (1H, d, J = 9.6 Hz), 6.29 (2H, s), 6.25 (1H, s), 5.44 (2H, s), 4.67 (2H, s), 4.39 (2H, d, J = 6.4 Hz), 1.98 (3H, s), 1.31 (12H, s). HRFAB-MS: calculated for [MH+] 739.3042, found 739.3046.
N-(4-((2-Amino-6-chloropyrimidin-4-yloxy)methyl)benzyl)-2-(3-methyl-4-(3-oxo-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)-3H-xanthen-9-yl)phenoxy)acetamide, SNAP-Peroxy-Green-2, SPG2 (12)
Compound 10 (25 mg, 0.051 mmol) dissolved in anhydrous DMF (2 mL) was added to a dried 5 mL round bottom flask and the solution was stirred at room temperature for 5 min. HATU (25 mg, 0.066 mmol), compound 3 (18 mg, 0.068 mmol), and DIPEA (25 uL) were added, and the reaction stirred at room temperature overnight. DMF was removed under vacuo and the orange oil was redissolved in 10% MeOH in CH2Cl2. Purification by flash column chromatography (silica gel, MeOH:CH2Cl2 1:10) afforded SPG2 12 as an orange film (20 mg, 0.027 mmol, 53% yield). 1H NMR (20% MeOD in CDCl3) δ 7.92 (1H, s), 7.55 (1H, d, J = 6.4 Hz), 7.33 (2H, d, J = 6.4 Hz), 7.28 (2H, d, J = 6.4 Hz), 7.07 (1H, d, J = 6.8 Hz), 7.02 (1H, d, J = 6.4 Hz), 6.9–7.0 (3H, m), 6.58 (1H, d, J = 7.6 Hz), 6.43 (1H, s), 6.06 (1H, s), 5.26 (2H, s), 4.61 (2H, s), 4.53 (2H, d, J = 3.6 Hz), 1.98 (3H, s), 1.32 (12H, s). ESI-MS: calculated for [MH+] 733.25, found 733.26.
2-(4-(6-Hydroxy-3-oxo-3H-xanthen-9-yl)-3-methylphenoxy)acetic acid (13)
Compound 7 (200 mg, 0.46 mmol) was stirred in a mixture of CH2Cl2: TFA (1:2, 20 mL) for 1 h. Removal of the solvent under vacuum gave a viscous orange oil. EtOAc (10 mL) was added, and the mixture was sonicated for 30 min. Slow addition of the EtOAc mixture into a beaker of vigorously stirring hexane (30 mL) gave carboxyl Tokyo Green 13 as orange solid precipitate (120 mg, 0.32 mmol, 69% yield). 1H NMR (CD3OD, 400MHz) δ 7.32 (2H, d, J = 8.6 Hz), 7.19 (1H, d, J = 8.4 Hz), 7.08 (1H, d, J = 2.0 Hz), 7.03 (1H, dd, J = 8.4, 2.0 Hz), 6.96 (2H, s), 6.92 (2H, d, J = 8.6 Hz), 4.80 (2H, s), 1.97 (3H, s). LC-MS: calculated for [MH+] 377.1, found 377.0
N-(4-((2-Amino-9H-purin-6-yloxy)methyl)benzyl)-2-(4-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-3-methylphenoxy)acetamide, SNAP-Tokyo Green 1, SNAP-TG1 (14)
Compound 13 (235 mg, 0.67 mmol) was added to a dried 25 mL Schlenk flask. Anhydrous DMF (5 mL) was added by syringe and the solution was stirred at room temperature for 5 min. HATU (256 mg, 0.67 mmol), 6-(4-(aminomethyl)benzyloxy)-9H-purin-2-amine 4 (182 mg, 0.67 mmol), and DIPEA (0.87 mL) were added, and the reaction stirred at room temperature overnight. The reaction was poured into an ice/ddH2O slurry (30 mL) and stirred to precipitate an orange solid. The solid was collected by vacuum filtration and dried in vacuo. Purification by flash column chromatography (silica gel, MeOH:CH2Cl2 1:10) delivered STG1 14 as an orange solid (197 mg, 0.31 mmol, 46% yield). 1H NMR (DMSO-d6, 400 MHz) δ 12.42 (1H, br-s), 8.74 (1H, s), 7.82 (1H, s), 7.45 (2H, d, J = 7.2 Hz), 7.29 (2H, d, J = 7.2 Hz), 7.20 (1H, d, J =8.4 Hz), 7.09 (1H, s), 7.02 (1H, d, J = 8.0 Hz), 6.87 (2H, d, J =8.8 Hz), 6.50–6.80 (4H, m), 6.29 (2H, br s), 5.44 (2H, s), 4.66 (2H, s), 4.38 (2H, d, J = 5.2 Hz), 3.16 (1H, s), 1.98 (3H, s). ESI-MS: calculated for [MH+] 629.21, found 629.21.
N-(3-((2-Amino-6-chloropyrimidin-4-yloxy)methyl)benzyl)-2-(4-(6-hydroxy-3-oxo-3H-xanthen-9-yl)-3-methylphenoxy)acetamide, SNAP-Tokyo Green 2, STG2 (15)
Compound 13 (30 mg, 0.079 mmol) in anhydrous DMF (2 mL) and HATU (30 mg, 0.078 mmol) were added to a dried 5 mL round bottom flask and the solution was stirred at room temperature for 5 min. Compound 3 (21 mg, 0.079 mmol), and DIPEA (100 μL) were added, and the reaction was stirred at room temperature overnight. DMF was removed in vacuo and the orange oil was redissolved in 10% MeOH in CH2Cl2. Purification by flash column chromatography (silica gel, MeOH:CH2Cl2 1:10) afforded STG2 15 as an orange film (45 mg, 0.072 mmol, 91% yield). 1H NMR (20% CD3OD in CDCl3) δ 7.35 (1H, t, J = 6.0 Hz), 7.25 (2H, d, J = 8.2), 7.21 (2H, J = 8.2 Hz), 6.98 (1H, d, J =8.0), 6.91 (2H, d, J = 8.8 Hz), 6.85 (1H, s), 6.82 (1H, d, J = 8.0), 6.41 (2H, d, J = 2.0 Hz), 6.60 (2H, dd, J = 8.8, 2.0 Hz), 5.97 (1H, s), 5.16 (2H, s), 4.52 (2H, s), 4.44 (2H, d, J = 6.0 Hz), 1.97 (3H, s) ESI-MS: calculated for [MH+] 623.16, found 623.17.
Expression and Purification of His-AGT for SNAP Labeling
The sequence encoding SNAP was PCR amplifed from pSEMS-SNAP-ADBR (Covalys) and subcloned into PET28a(+) (Novagen) using NheI/XhoI restriction sites. The resulting plasmid, after verified by DNA sequencing, was transformed into E.Coli strain BL-21(DE3). A bacterial culture was grown at 37 °C in TB medium (50 mL) with 50 μg/mL Kanamycin for 6–8 h until an optical density (OD600nm) of 0.8 was reached. Expression of His-AGT was induced by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). The culture was grown at 16 °C for 12 h and was harvested by centrifugation at 8000 rpm for 10 min at 4 °C. Cell lysates were prepared by resuspending the pellet in buffer (PBS with 0.5 mM phenylmethylsulfonyl fluoride) followed by FRENCH® press (Thermo Scientific). Insoluble protein and cell debris were removed by centrifugation at 10000 rpm for 30 min. For purification of His-AGT, Ni-NTA (Qiagen) was used according to the supplier's protocol. The fractions with His-AGT were combined and concentrated using an Amicon Ultra-15 Ultracel®-10K centrifugal unit (Millipore). The concentrated His-AGT solution was then desalted using G25 Sephadex (Sigma Aldrich) with the following elution buffer: 150 mM NaCl, 25 mM Tris-Cl, 1mM DTT pH 7.5. The purified His-AGT protein was concentrated using an Amicon Ultra-4 Ultracel®-10K and stored at −20 °C. The protein concentration was determined using the BCA assay, yielding 8.6 mg of purified His-AGT (1.5 mM in 250 μL elution buffer).
In vitro SNAP Labeling and Analysis by ESI-MS and In-Gel Fluorescence Scanning
Purified His-AGT (0.5 uM) was incubated in reaction buffer (PBS pH 7.4, 1 mM DTT) at 37 °C with 0.5 μM of either SPG1, SPG2, STG1, or STG2 for 30 min at 37 °C. Binding of the fluorescent dyes to His-AGT was analyzed by SDS-PAGE, followed by in-gel fluorescence scanning (488 nm Argon excitation, 520 BP 40 emission filter, Typhoon 9410 imaging system, GE Healthcare) and Coomassie protein staining. For mass spectrometric analysis and spectroscopic measurements, His-AGT (1 μM) was incubated in 20 mM HEPES buffer pH 7 (1 mL) with 5 μM of either SPG1, SPG2, STG1, or STG2 for 30 min at 37 °C. The reaction was concentrated to 50 μM with an Amicon© 4 10K column (Milipore) and subjected to size-exclusive gel filtration with Bio-Rad Micro Bio-Spin 6 chromatography column to remove the excess dye. The deconvoluted MS data were collected by Dr. Anthony T. Iavarone at UC Berkeley QB3 Mass Spectrometry Facility.
Spectroscopic Materials and Methods
Millipore water was used to prepare all aqueous solutions. All spectroscopic measurements were performed in 20 mM HEPES buffer, pH 7.0, 25 °C. Absorption spectra were recorded using a Varian Cary 50 spectrophotometer (Walnut Creek, CA). Fluorescence spectra were recorded using a Photon Technology International Quanta Master 4 L-format scanning spectrofluorometer (Lawrenceville, NJ) equipped with an LPS-220B 75-W xenon lamp and power supply, A-1010B lamp housing with integrated igniter, switchable 814 photon-counting/analog photomultiplier detection unit, and MD5020 motor driver. Samples for absorption and fluorescence measurements were contained in 1-cm × 1-cm quartz cuvettes (1.4 mL volume, Starna, Atascadero, CA). Fluorescein in 0.1 M NaOH (Φ = 0.85)90 was used as standard for quantum yield measurements.
Cell Culture and Labeling Procedures
HEK 293T cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Hyclone) and glutamine (2 mM), One day before transfection, cells were passaged and plated on 4-wells Lab-Tek borosilicate chambered coverglass (Nunc). Transient expression of SNAP fusion protein or mCherry was performed by following the standard protocol of Lipofectamine 2000. SNAP-tag labeling was achieved by incubating cells in DPBS containing 5 μM dye and 1 μM Hoechst 33342 for 30 min at 37 °C. For membrane labeling with cell-impermeable SPG1 or STG1, cells were washed with fresh DPBS (2 × 1 mL) before image acquisition. Labeling of intracellular targets with STG2 or SPG2 required incubation of cells in fresh DMEM with 10% FBS (2 × 1 mL) for 30 min after dye loading to remove any unbound fluorophore. All confocal fluorescence images were acquired in DPBS media.
Fluorescence Imaging Experiments
Confocal fluorescence imaging studies were performed with a Zeiss LSM510 NLO Axiovert 200 Laser scanning microscope and a 40× oil-immersion objective lens. The motorized stage on the microscope was equipped with an incubator, maintaining the sample at 37 °C in a 5% CO2 humidified atmosphere. Excitation of the SNAP tag probe at 488 nm was carried out with an Argon laser and emission was collected using a 500–550 nm filter. Excitation of mCherry was carried out with a Helium-Neon 543 nm laser and emission was collected using a LP560 filter. Excitation of Hoechst 33342 was carried out using a MaiTai two-photon laser at 770 nm pulses (mode-locked Ti:sapphire laser, Tsunami Spectra Physics) and emission was collected through 385–425 nm filter. Image analysis was performed in ImageJ (National Institute of Health).
Supplementary Material
Acknowledgements
We thank the Packard and Sloan Foundations, the Hellman Faculty Fund (UC Berkeley), Amgen, Astra Zeneca, and the National Institute of General Medical Sciences (NIH GM 79465) for funding this work. C.J.C. is an Investigator with the Howard Hughes Medical Institute. D.S. was supported by a scholarship from the Ministry of Science, Thailand. A.E.A. thanks the NIH Chemical Biology Graduate Program (T32 GM066698) for support, as well as the ACS Organic Division for an Emmanuil Troyansky Graduate Fellowship and UC Berkeley for a Chancellor's Opportunity Fellowship. We thank Holly Aaron (UCB Molecular Imaging Center), Ann Fischer (UCB Tissue Culture Facility), and Prof. Michelle Chang (UCB Department of Chemistry) for expert technical assistance and helpful discussions.
Footnotes
Supporting Information Available Mass spectrometric and in-gel fluorescence analysis of intact AGT as well as AGT-PG and AGT-TG adducts, details of construction of plasmids, and complete Ref 54. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Harman D. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- (2).Finkel T, Holbrook NJ. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- (3).Stadtman ER. Free Radical Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- (4).Droge W, Schipper HM. Aging Cell. 2007;6:361–370. doi: 10.1111/j.1474-9726.2007.00294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Free Radical Biol. Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- (6).Sharpless NE, Depinho RA. Nat. Rev. Mol. Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- (7).Brandon M, Baldi P, Wallace DC. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- (8).Finkel T, Serrano M, Blasco MA. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- (9).Fruehauf JP, Meyskens FL. Clin. Cancer Res. 2007;13:789–794. doi: 10.1158/1078-0432.CCR-06-2082. [DOI] [PubMed] [Google Scholar]
- (10).Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- (11).Goetz ME, Luch A. Cancer Lett. 2008;266:73–83. doi: 10.1016/j.canlet.2008.02.035. [DOI] [PubMed] [Google Scholar]
- (12).Rossi DJ, Jamieson CHM, Weissman IL. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- (13).Houstis N, Rosen ED, Lander ES. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- (14).Jay D, Hitomi H, Griendling KK. Free Radical Biol. Med. 2006;40:183–192. doi: 10.1016/j.freeradbiomed.2005.06.018. [DOI] [PubMed] [Google Scholar]
- (15).Pop-Busui R, Sima A, Stevens M. Diabetes-Metab. Res. Rev. 2006;22:257–273. doi: 10.1002/dmrr.625. [DOI] [PubMed] [Google Scholar]
- (16).Barnham KJ, Masters CL, Bush AI. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- (17).Lin MT, Beal MF. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- (18).DiMauro S, Schon EA. Annu. Rev. Neurosci. 2008;31:91–123. doi: 10.1146/annurev.neuro.30.051606.094302. [DOI] [PubMed] [Google Scholar]
- (19).Rhee SG. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- (20).Stone JR, Yang S. Antioxid. Redox Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- (21).D'AutrEaux B, Toledano MB. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- (22).Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Nat. Rev. Mol. Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- (23).Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- (24).Miller EW, Chang CJ. Curr. Opin. Chem. Biol. 2007;11:620–625. doi: 10.1016/j.cbpa.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Veal EA, Day AM, Morgan BA. Mol.Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- (26).Winterbourn CC. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- (27).Segal AW, Abo A. Trends Biochem.Sci. 1993;18:43–47. doi: 10.1016/0968-0004(93)90051-n. [DOI] [PubMed] [Google Scholar]
- (28).Lambeth JD. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- (29).Bedard K, Krause KH. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- (30).Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- (31).Bae YS, Kang SW, Seo MS, Baines IC, Tekle E, Chock PB, Rhee SG. J.Biol.Chem. 1997;272:217–221. [PubMed] [Google Scholar]
- (32).Avshalumov MV, Chen BT, Marshall SP, Pena DM, Rice ME. J. Neurosci. 2003;23:2744–2750. doi: 10.1523/JNEUROSCI.23-07-02744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tonks NK. Cell. 2005;121:667–670. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- (34).Poole LB, Nelson KJ. Curr. Opin. Chem. Biol. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bao L, Avshalumov MV, Patel JC, Lee CR, Miller EW, Chang CJ, Rice ME. J. Neurosci. 2009;29:9002–9010. doi: 10.1523/JNEUROSCI.1706-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Eligini S, Arenaz I, Barbieri SS, Faleri ML, Crisci M, Tremoli E, Colli S. Free Radical Biol. Med. 2009;46:1428–1436. doi: 10.1016/j.freeradbiomed.2009.02.026. [DOI] [PubMed] [Google Scholar]
- (37).Kim JS, Huang TY, Bokoch GM. Mol. Biol. Cell. 2009;20:2650–2660. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Leonard SE, Reddie KG, Carroll KS. ACS Chem. Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- (39).Niethammer P, Grabher C, Look AT, Mitchison TJ. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- (41).Chen I, Ting AY. Curr. Opin. Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- (42).Gronemeyer T, Godin G, Johnsson K. Curr. Opin. Biotechnol. 2005;16:453–458. doi: 10.1016/j.copbio.2005.06.001. [DOI] [PubMed] [Google Scholar]
- (43).Miller LW, Cornish VW. Curr. Opin. Chem. Biol. 2005;9:56–61. doi: 10.1016/j.cbpa.2004.12.007. [DOI] [PubMed] [Google Scholar]
- (44).Chattopadhaya S, Abu Bakar FB, Yao SQ. Curr. Med. Chem. 2009;16:4527–4543. doi: 10.2174/092986709789760706. [DOI] [PubMed] [Google Scholar]
- (45).Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- (47).Guignet EG, Hovius R, Vogel H. Nat. Biotechnol. 2004;22:440–444. doi: 10.1038/nbt954. [DOI] [PubMed] [Google Scholar]
- (48).Hauser CT, Tsien RY. Proc. Nat. Acad. Sci. U. S. A. 2007;104:3693–3697. doi: 10.1073/pnas.0611713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Ojida A, Honda K, Shinmi D, Kiyonaka S, Mori Y, Hamachi I. J. Am. Chem. Soc. 2006;128:10452–10459. doi: 10.1021/ja0618604. [DOI] [PubMed] [Google Scholar]
- (50).Calloway NT, Choob M, Sanz A, Sheetz MP, Miller LW, Cornish VW. ChemBioChem. 2007;8:767–774. doi: 10.1002/cbic.200600414. [DOI] [PubMed] [Google Scholar]
- (51).Gallagher SS, Sable JE, Sheetz MP, Cornish VW. ACS Chem. Biol. 2009;4:547–556. doi: 10.1021/cb900062k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Marks KM, Braun PD, Nolan GP. Proc. Nat. Acad. Sci. U. S. A. 2004;101:9982–9987. doi: 10.1073/pnas.0401609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zhang Y, So MK, Loening AM, Yao HQ, Gambhir SS, Rao JH. Angew. Chem. Int. Ed. 2006;45:4936–4940. doi: 10.1002/anie.200601197. [DOI] [PubMed] [Google Scholar]
- (54).Los GV, et al. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- (55).Watkins RW, Lavis LD, Kung VM, Los GV, Raines RT. Org. Biomol. Chem. 2009;7:3969–3975. doi: 10.1039/b907664f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Bonasio R, Carman CV, Kim E, Sage PT, Love KR, Mempel TR, Springer TA, von Andrian UH. Proc. Nat. Acad. Sci. U. S. A. 2007;104:14753–14758. doi: 10.1073/pnas.0705201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. Chem. Biol. 2003;10:313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- (58).Keppler A, Kindermann M, Gendreizig S, Pick H, Vogel H, Johnsson K. Methods. 2004;32:437–444. doi: 10.1016/j.ymeth.2003.10.007. [DOI] [PubMed] [Google Scholar]
- (59).Gautier A, Juillerat A, Heinis C, Correa IR, Kindermann M, Beaufils F, Johnsson K. Chem. Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- (60).Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Proc. Nat. Acad. Sci. U. S. A. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Zhou Z, Cironi P, Lin AJ, Xu YQ, Hrvatin S, Golan DE, Silver PA, Walsh CT, Yin J. ACS Chem. Biol. 2007;2:337–346. doi: 10.1021/cb700054k. [DOI] [PubMed] [Google Scholar]
- (62).Lorand L, Graham RM. Nat. Rev. Mol. Cell Biol. 2003;4:140–156. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- (63).Lin CW, Ting AY. J. Am. Chem. Soc. 2006;128:4542–4543. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Chen I, Howarth M, Lin WY, Ting AY. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- (65).Howarth M, Takao K, Hayashi Y, Ting AY. Proc. Nat. Acad. Sci. U. S. A. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Fernandez-Suarez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, Bertozzi CR, Ting AY. Nat. Biotechnol. 2007;25:1483–1487. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Nat. Chem. Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- (68).Tanaka T, Yamamoto T, Tsukiji S, Nagamune T. ChemBioChem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- (69).Duckworth BP, Zhang ZY, Hosokawa A, Distefano MD. ChemBioChem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- (70).Carrico IS, Carlson BL, Bertozzi CR. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- (71).Wu P, Shui WQ, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Proc. Nat. Acad. Sci. U. S. A. 2009;106:3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Keppler A, Ellenberg J. ACS Chem. Biol. 2009;4:127–138. doi: 10.1021/cb800298u. [DOI] [PubMed] [Google Scholar]
- (73).Tomat E, Nolan EM, Jaworski J, Lippard SJ. J. Am. Chem. Soc. 2008;130:15776–15777. doi: 10.1021/ja806634e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Bannwarth M, Correa IR, Sztretye M, Pouvreau S, Fellay C, Aebischer A, Royer L, Rios E, Johnsson K. ACS Chem. Biol. 2009;4:179–190. doi: 10.1021/cb800258g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Farr GA, Hull M, Mellman I, Caplan MJ. J. Cell. Biol. 2009;186:269–282. doi: 10.1083/jcb.200901021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Maurel D, Comps-Agrar L, Brock C, Rives ML, Bourrier E, Ayoub MA, Bazin H, Tinel N, Durroux T, Prezeau L, Trinquet E, Pin JP. Nat. Methods. 2008;5:561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Mao S, Benninger RKP, Yan YL, Petchprayoon C, Jackson D, Easley CJ, Piston DW, Marriott G. Biophys. J. 2008;94:4515–4524. doi: 10.1529/biophysj.107.124859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Brun MA, Tan KT, Nakata E, Hinner MJ, Johnsson K. J. Am. Chem. Soc. 2009;131:5873–5884. doi: 10.1021/ja900149e. [DOI] [PubMed] [Google Scholar]
- (79).Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- (80).Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc. Nat. Acad. Sci. U. S. A. 2004;101:9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Chang MCY, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2004;126:15392–15393. doi: 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Miller EW, Albers AE, Pralle A, Isacoff EY, Chang CJ. J. Am. Chem. Soc. 2005;127:16652–16659. doi: 10.1021/ja054474f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Albers AE, Okreglak VS, Chang CJ. J. Am. Chem. Soc. 2006;128:9640–9641. doi: 10.1021/ja063308k. [DOI] [PubMed] [Google Scholar]
- (84).Albers AE, Dickinson BC, Miller EW, Chang CJ. Bioorg. Med. Chem. Lett. 2008;18:5948–5950. doi: 10.1016/j.bmcl.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Srikun D, Miller EW, Dornaille DW, Chang CJ. J. Am. Chem. Soc. 2008;130:4596–4597. doi: 10.1021/ja711480f. [DOI] [PubMed] [Google Scholar]
- (86).Dickinson BC, Chang CJ. J. Am. Chem. Soc. 2008;130:11561–11562. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Nat. Chem. Biol. 2007;3:263–267. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- (88).Urano Y, Kamiya M, Kanda K, Ueno T, Hirose K, Nagano T. J. Am. Chem. Soc. 2005;127:4888–4894. doi: 10.1021/ja043919h. [DOI] [PubMed] [Google Scholar]
- (89).Keppler A, Arrivoli C, Sironi L, Ellenberg J. Biotechniques. 2006;41:167–175. doi: 10.2144/000112216. [DOI] [PubMed] [Google Scholar]
- (90).Parker CA, Rees WT. Analyst. 1960;85:587–600. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.