Abstract
The structure of the 142-residue protein Q8ZP25_SALTY encoded in the genome of Salmonella typhimurium LT2 was determined independently by NMR and X-ray crystallography, and the structure of the 140-residue protein HYAE_ECOLI encoded in the genome of Eschericia coli was determined by NMR. The two proteins belong to Pfam [1] PF07449, which currently comprises 50 members, and belongs itself to the ‘thioredoxin-like clan’. However, protein HYAE_ECOLI and the other proteins of Pfam PF07449 do not contain the canonical Cys-X-X-Cys active site sequence motif of thioredoxin. Protein HYAE_ECOLI was previously classified as a [NiFe] hydrogenase-1 specific chaperone interacting with the twin-arginine translocation (Tat) signal peptide. The structures presented here exhibit the expected thioredoxin-like fold and support the view that members of Pfam family PF07449 specifically interact with Tat signal peptides.
Keywords: Chaperones, GFT NMR, HYAE_ECOLI, Q8ZP25_SALTY, Structural genomics, Thioredoxin
Introduction
Pfam[1] family HyaE, which was named after its functionally characterized member HYAE_ECOLI [2] (accession number PF07449) and currently contains 50 members, was selected by the Protein Structure Initiative-2 (PSI-2) of the United States National Institutes of Health and assigned to the Northeast Structural Genomics consortium (NESG; http://www.nesg.org). Here we describe the structures of two proteins belonging to PF07449, that is, protein Q8ZP25_SALTY encoded by gene STM1790 (UniProtKB/TrEMBL entry Q8ZP25; NESG target ID StR70) of Salmonella typhimurium LT2 [3] and the homologous protein HYAE_ECOLI (73% identity) encoded in gene hyaE (UniProtKB/Swiss-Prot entry P19931; NESG target ID ER415) of Escherichia coli. The NMR structures were determined using the previously described rapid NMR data collection and analysis protocol for high-throughput structure determination [4]. The structure of Q8ZP25_SALTY was also determined independently by X-ray crystallography. The three-dimensional structures of Q8ZP25_SALTY (17 kDa) and HYAE_ECOLI (16.7 kDa) are the first for Pfam family PF07449.
Materials and methods
Protein sample preparation for NMR
Proteins Q8ZP25_SALTY and HYAE_ECOLI were cloned, expressed and purified following standard protocols to produce uniformly U-13C,15N-labeled protein samples.[5] Briefly, the full length STM1790 gene from Salmonella typhimurium LT2 was cloned into a pET21 (Novagen) derivative, yielding the plasmid StR70-21.4. The full length hyaE gene from Escherichia coli was also cloned into a pET21 derivative, yielding the plasmid ER415-21.1. The resulting sequence verified constructs contain the complete native protein coding sequence plus eight nonnative residues at the C-terminus (LEHHHHHH) that facilitate protein purification. Escherichia coli BL21 (DE3) cells containing the pMGK plasmid, which expresses several tRNAs to enhance translation of rare codons, were transformed with StR70-21.4 and separately with ER415-21.1, and cultured in MJ9 minimal medium containing (15NH4)2SO4 and U-13C-glucose as sole nitrogen and carbon sources.[6] U-13C,15N Q8ZP25_SALTY and U-13C,15N HYAE_ECOLI were purified using an AKTAxpress (GE Healthcare) based two step protocol consisting of IMAC (HisTrap HP) and gel filtration (HiLoad 26/60 Superdex 75) chromatography. The final yield of purified U-13C,15N Q8ZP25_SALTY (> 98% homogenous by SDS-PAGE; 17.05 kDa by MALDI-TOF mass spectrometry) was about 26 mg/L. The final sample of U-13C, 15N labeled Q8ZP25_SALTY was prepared at a concentration of ~1.2 mM in 95% H2O/5% D2O solution containing 50 mM MES, 10 mM DTT, and 50 mM arginine, at pH 6.0. The final yield of purified U-13C,15N HYAE_ECOLI (> 98% homogenous by SDS-PAGE; 16.72 kDa by MALDI-TOF mass spectrometry) was about 42 mg/L. The final sample of U-13C, 15N labeled HYAE_ECOLI was prepared at a concentration of ~0.6 mM in 95% H2O/5% D2O solution containing 20 mM MES, 10 mM DTT, 100 mM NaCl, 5mM CaCl2 and 0.02% NaN3 at pH 6.5.
Isotropic overall rotational correlation times of ~8 ns were inferred from 15N spin relaxation times for both proteins, indicating that these proteins are monomeric in solution. This conclusion was further confirmed by gel-filtration and dynamic light scattering (data not shown but available online at www.nesg.org). In order to align protein Q8ZP25_SALTY for measurement of backbone 15N-1H residual dipolar couplings, Pf1 phages [7] were added to the protein solution, yielding a 0.25 mM Q8ZP25_SALTY solution in the presence of 15.5 mg/mL Pf1 phage.
NMR structure determination
All NMR experiments for resonance assignment, identification of slowly exchanging amide protons and measurement of residual dipolar couplings (RDCs) were performed on a Varian INOVA 600 spectrometer equipped with a cryogenic 1H{13C,15N} probe, while NOESY data to derive 1H-1H distance constraints were recorded on a 750 MHz spectrometer equipped with a conventional 1H{13C,15N} probe. For Q8ZP25_SALTY, a standard set of five through-bond G-matrix Fourier transform (GFT) NMR experiments [6; 8; 9] was acquired for resonance assignment (total measurement time: 110 hours), and a simultaneous 3D 15N/13Caliphatic/13Caromatic-resolved NOESY spectrum [9] (measurement time: 48 hours) was acquired to derive distance constraints. For HYAE_ECOLI, the comparably low protein concentration made conventional 3D HNNCACB, CBCA(CO)NHN, and HBHA(CO)NHN [10] recorded in conjunction with (4,3)D HCCH [4] the preferred choice (total measurement time: 216 hours). These spectra were complemented with 3D 15N-resolved (measurement time: 24 hours) and 13C-resolved NOESY spectra (measurement time: 48 hours) to derive distance constraints. All other spectra were processed with the program nmrPipe [11] and analyzed using the program XEASY [12]. Polypeptide backbone (1HN, 15N, 13Cα) and 13Cβ resonance assignments were obtained by using the program AutoAssign [13].
For Q8ZP25_SALTY, polypeptide backbone 15N-1H RDCs were extracted from pairs of 2D [15N,1H]-TROSY and 2D [15N,1H]-HSQC spectra [10] (measurement time 10 hours) acquired for both unaligned and partially aligned protein. A virtually complete set of RDCs was obtained. The spectra were processed using the program NMRPipe [11] and automatically peak picked using the program NMRDraw [11]. To identify slowly exchanging backbone amide protons a 2D [15N,1H]-HSQC spectrum was recorded (measurement time 10 minutes) starting 5 minutes after lyophilizing and re-suspending protein Q8ZP25_SALTY in 99.8% D2O.
The programs CYANA [14; 15] and AUTOSTRUCTURE [16] were used in parallel to automatically assign long-range NOEs. Identical assignments from both programs (“consensus assignments”) were used as the starting point for manual completion of iterative NOE assignment, peak picking and structure calculation [4]. For protein Q8ZP25_SALTY, hydrogen bond constraints were derived for slowly exchanging amide protons whenever NOEs supported hydrogen bond formation [10]. The final structure calculations were performed using version 2.1 of CYANA [15]. For Q8ZP25_SALTY, the alignment tensor was fitted to 15N-1H RDC values using the program REDCAT [17]. Subsequently, orientational constraints derived from RDCs measured for residues in rigid regular secondary structure elements were implemented in the program XPLOR [18; 19] for structure refinement. Finally, additional refinement for the structures of proteins Q8ZP25_SALTY and HYAE_ECOLI was performed in an explicit ‘water bath’ [20] by using the program CNS [21]. Prior to submission to the protein data bank [22], structures were validated using the PSVS server [23].
Protein sample preparation for X-ray crystallography
Selenomethionine labeled Q8ZP25_SALTY was produced as described above for the NMR sample, except that an MJ9 minimal medium containing selenomethionine was used. The yield of purified Q8ZP25_SALTY (> 98% homogenous by SDS-PAGE; 16.19 kDa by MALDI-TOF mass spectrometry) was about 57 mg/L. The final sample of selenomethionine labeled Q8ZP25_SALTY was prepared at a concentration of 10 mg/mL in a solution containing 10 mM Tris, 5 mM DTT and 100 mM NaCl.
Crystallization, X-ray diffraction data collection, and processing
Monoclinic-shaped crystals of Q8ZP25_SALTY grew at 293 K in 1 + 1 μl hanging-drop vapor-diffusion reactions over a reservoir containing 18% PEG 1000, 15 mM NaBr and 100 mM NaOAc at pH 5.0. Crystals were frozen in liquid propane using paratone-N as cryoprotectant. Multi-wavelength anomalous diffraction data were collected from a single crystal at the selenium edge on a Quantum-4 CCD detector (ADSC, San Diego, CA) on beamline X4A at the National Synchrotron Light Source in consecutive 400° sweeps by the inverse beam method at 0.97925 Å (peak), 0.97955 Å (edge) and 0.96782 Å (remote) using 1°, 8 s oscillations. Data were processed using the programs DENZO and SCALEPACK [24]. For each data set, the scaling B factors of the final frames were within 3 Å2 of the initial frames, indicating that there was minimal decay.
Results and Discussion
NMR solution structures of proteins Q8ZP25_SALTY and HYAE_ECOLI
Resonance assignments were obtained for 78% (Q8ZP25_SALTY, which comprises 134 residues, excluding affinity purification tag) and 89% (HYAE_ECOLI, which comprises 132 residues, excluding affinity purification tag) of the assignable backbone (excluding the C-terminal NH3+, the Pro 15N and the 13C′ shifts) and 13Cβ shifts, and for 74% (Q8ZP25_SALTY) and 88% (HYAE_ECOLI) of the side chain shifts (excluding Lys NH3+, Arg NH2, OH, side chain 13C′ and aromatic quaternary 13C shifts; Table I). Stereo-specific assignments were obtained for 28% (Q8ZP25_SALTY) and 25% (HYAE_ECOLI) of the β-methylene groups exhibiting non-degenerate proton chemical shifts using GLOMSA [25]. GLOMSA was also used to stereo-specifically assign 20% of the HYAE_ECOLI Val and Leu isopropyl moieties (Table I). For Q8ZP25_SALTY, a constant-time 2D 13C HSQC was preformed on a 5% fractionally 13C-labeled sample and used to assign 67% of the Val and Leu isopropyl moieties [26]. Upper distance limit constraints for structure calculations were obtained from NOESY, and backbone dihedral angle constraints for residues located within well defined secondary structure were derived from chemical shifts as described [27]. For Q8ZP25_SALTY, 10 hydrogen bond constraints and 69 RDC constraints[28] were used in addition for structure refinement. The RDC “quality factor” Q (i.e., the r.m.s.d. calculated between measured RDC values and RDC values predicted based on the structure) improved from 0.52 to 0.42 as a result of the RDC-based refinement, which also reduced the r.m.s.d. value calculated for heavy atoms N, Cα, and C′ between the NMR structure and the x-ray structure from 1.60 to 1.38 Ǻ(residues 11–25, 34–42, 55–90, 94–101, 106, 107, 113–122). The statistics for the structure determinations of Q8ZP25_SALTY and HYAE_ECOLI (Table I) show that high-quality NMR structures were obtained. Chemical shifts, distance constraints and RDCs were deposited in the BioMagResBank [29] (BMRB accession number 7178 for Q8ZP25_SALTY and 7256 for HYAE_ECOLI) and coordinates were deposited in the Protein Data Bank [22] (PDB 2JZT for Q8ZP25_SALTY and 2HFD for HYAE_ECOLI).
Table 1.
Statistics of Q8ZP25_SALTY and HYAE_ECOLI NMR structures.
| Protein | Q8ZP25_SALTY | HYAE_ECOLI |
|---|---|---|
| Conformationally-restricting distance constraints | ||
| Intraresidue [i = j] | 397 | 533 |
| Sequential [(i − j) = 1] | 428 | 542 |
| Medium Range [1 < (i − j) ≤ 5] | 206 | 200 |
| Long Range [(i − j) > 5] | 510 | 339 |
| Total | 1541 | 1524 |
| Dihedral angle constraints | ||
| Φ | 41 | 56 |
| ψ | 41 | 56 |
| Number of constraints per residue | 15.7 | 12.4 |
| Number of long-range constraints per residue | 5.2 | 2.5 |
| Completeness of stereo-specific assignments a[%] | ||
| βCH2 | 28 (16/58) | 25 (19/77) |
| Val and Leu isopropyl groups | 67 (16/24) | 20 (5/25) |
| CYANA target function [Å2] | 1.1 ± 0.17 | 1.88 ± 0.3 |
| Hydrogen bond constraints | 10 | 0 |
| NH RDC-derived orientational constraints | 69 | 0 |
| Average r.m.s.d. to the mean CYANA coordinates [Å] | ||
| regular secondary structure elements, backbone heavy atoms N, Cα, C′ | 0.72 ± 0.15b | 0.82 ± 0.09c |
| regular secondary structure elements, all heavy atoms | 1.14 ± 0.12 | 1.22 ± 0.09 |
| residues 11–41, 56–127, backbone heavy atoms | 0.95 ± 0.12 | 0.94 ± 0.13 |
| residues 11–41, 56–127, all heavy atoms | 1.49 ± 0.13 | 1.52 ± 0.12 |
| heavy atoms of best-defined side-chains | 0.5 ± 0.07d | 0.58 ± 0.1e |
| PROCHECK G-factorsf (ϕ and Ψ/all dihedral angles) | 0.04/−1.6 | 0.04/−1.48 |
| MOLPROBITY clash scoreg | −1.74 | −2.43 |
| AutoQF R/P/DP scores (%)h | 0.94/0.93/0.76 | 0.97/0.93/0.66 |
| Ramachandran plot summary ordered residues: [%] | ||
| most favored regions | 93.5 | 92.3 |
| additionally allowed regions | 6.4 | 7.2 |
| generously allowed regions | 0.1 | 0.5 |
| disallowed regions | 0 | 0 |
| Average number of distance constraints violations per CYANA conformer [Å] | ||
| 0.2–0.5 | 0.1 | 0.1 |
| > 0.5 | 0 | 0 |
| Average number of dihedral-angle constraint violations per CYANA conformer [degrees] | ||
| > 5 | 0 | 0 |
Relative to pairs with non-degenerate chemical shifts.
Residues: 20–22, 37–40, 71–74, 95–99, 102–107 (β-strands), and 8–16, 26–34, 58–62, 79–86, 115–123 (α-helices).
Residues: 19–22, 35–42, 69–75, 94–99, 102–108 (β-strands), and 5–16, 24–33, 56–63, 77–87, 112–124 (α-helices).
34 residues: 15, 19, 21, 22, 24, 27, 30, 31, 34, 38–41, 58, 59, 68, 70, 72–75, 82, 83, 94–99, 103, 106, 118, 122, 123.
30 residues: 10, 11, 14–16, 19, 21, 38–40, 61, 62, 70, 72–75, 80, 82, 83, 94, 97, 107, 113, 114, 117–120, 123
Defined in Reference [42],
Defined in Reference [43],
Defined in Reference [44]
X-ray crystal structure of protein Q8ZP25_SALTY
Given four molecules of StR70 per asymmetric unit in the P21 lattice, the packing density in the crystal is 2.1 Å3/Da, in the most probable range for proteins [30]. SOLVE [31] identified 7 of the 12 Se atoms in the asymmetric unit, yielding a map that was used for non-crystallographic symmetry (NCS) averaging, density modification (with 41% solvent content) and automated iterative model building in AUTO_RESOLVE [31]. This program identified 50% of the backbone and 35% of the side chains in the final model and produced a map that enabled the structure to be built by hand using O [32]. Completion of the structure required iterative cycles of refinement in CNS [33] and manual rebuilding using standard geometric and van der Waals parameters [34]. The refinement was monitored by a randomly selected Rfree set containing 10% of the reflections. B-factors were refined using standard vicinal restraints (1.5–2.0 Å2 for main-chain atoms and 2.0–2.5 Å2 for side-chain atoms). Strong NCS restraints (1050 kJÅ−2, σB = 1.5) were maintained throughout the refinement. Water-molecule sites were selected automatically using CNS and checked for consistency with 2Fo − Fc electron-density and hydrogen-bonding criteria. The statistics of the 2.8 Å X-ray structure determination are shown in Table II, and the coordinates were deposited in the PDB (id: 2ES7).
Table 2.
Statistics of Q8ZP25_SALTY X-ray crystal structure
| Crystal Parameters: | ||
| Space group | P21 | |
| Unit-cell at 100 K [Å] | 39.5, 71.2, 95.0 | 90.0°, 90.1°, 90.0° |
| Data quality: | ||
| Resolution [Å] | 20–2.8 (2.85–2.80) | |
| No. of measured reflections | 88422 (3992) | |
| No. of unique reflections | 24561 (1174) | |
| Rsym [%] | 4.9 (12.2) | (I ≥ −3σ1 for observations) |
| Mean redundancy | 3.6 (3.4) | |
| Completeness [%] | 98.6 (95.5) | (All measured reflections) |
| 88.6 (83.9) | (I ≥ 2σ1) | |
| Mean I/σI | 35.4 (9.2) | (I ≥ σ1 after merging) |
| Refinement residuals (F≥2σF): | ||
| Rfree [%] | 33.8 | |
| Rwork [%] | 28.5 | |
| Model quality: | ||
| RMSD bond lengths [Å] | 0.017 | |
| RMSD bond angles | 2.4 ° | |
| Ramachandran plot summary [%] | ||
| most favored regions | 86.6 | |
| allowed regions | 12.2 | |
| generously allowed regions | 1.2 | |
| Average B factors (Å2): | ||
| All | 43.3 | |
| Main chain | 33.7 | |
| Side chain | 35.2 | |
| Waters | 45.8 | |
| Model contents: | ||
| Protein residues | 4×103 | |
| Water molecules | 560 |
Structure description and comparisons
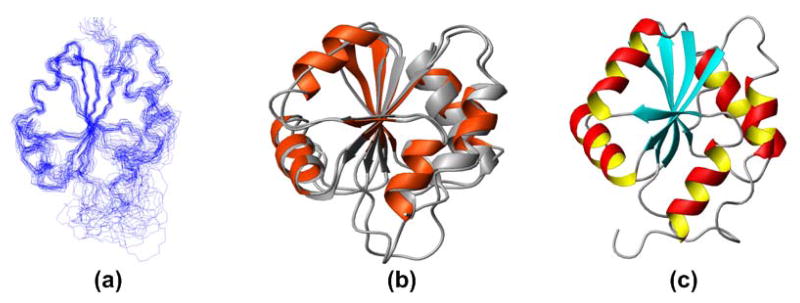
The structures of proteins Q8ZP25_SALTY (NMR and X-ray) and HYAE_ECOLI (NMR) are shown in Figure 1. They exhibit the expected canonical “thioredoxin-like” fold characterized by an internal twisted 5-stranded β-sheet surrounded by 5 α-helices. Starting at the N-terminus, the topology of the regular secondary structure elements can be summarized as α1-βA(↑)-α2-βB(↑)-α3-βC(↑)-α4-βD(↓)-βE(↑)-α5. The close similarity of the structures is evidenced by rather small r.m.s.d. values (Table III) calculated for the backbone heavy atoms N, Cα and C′ of residues for which resonance assignments were obtained for the NMR structures and electron density was observed in the X-ray structure.
Fig 1.
(a) Superposition of the 20 conformers representing the NMR solution structure of Q8ZP25_SALTY, (b) NMR structure of Q8ZP25_SALTY (in orange) superimposed on the X-ray structure of Q8ZP25_SALTY (in grey), (c) ribbon drawing of the NMR structure of HYAE_ECOLI.
Table 3.
R.m.s.d. values calculated for structure comparison
| Backbonea r.m.s.d values | Q8PZ25_SALTY (X-RAY)b | HYAE_ECOLI (NMR)b,c |
|---|---|---|
| Q8PZ25_SALTY (NMR)b,c | 1.23 Å | 1.58 Å |
| Q8PZ25_SALTY (X-RAY)b | - | 1.87 Å |
Heavy atoms N, Cα, and C′ were superimposed for minimal r.m.s.d.
Residues well defined in both structures: 11–25, 34–42, 55–90, 94–101, 106, 107, 113–124
Mean conformer calculated for the 20 best conformers (Table I)
Complementary information is obtained from the NMR and X-ray structures. The polypeptide segments comprising residues 1–10, 43–54, 91–93, 108–112 and 128–142 (in HYAE_ECOLI: 1–2, 43–55, 91–92, and 108–111) are locally not well defined and appear flexibly disordered in the NMR structures, whereas no electron density is observed for residues 1–6, 26–33, 48–51, 102–105, 126–142 in the X-ray structure. Furthermore, α-helix 2 comprising residues 26–35 in the NMR structure (in HYAE_ECOLI: residues 23–34), was not observed in the X-ray structure, suggesting that the structure of this segment is stable in solution but that it may have assumed a range of conformations in the crystal. Conversely, a defined conformation is observed for loop residues 91–92 and 108–111 in the X-ray structure but remained poorly defined in both NMR structures. This suggests that these segments are flexibly disordered in solution but became trapped in a fixed conformation during crystallization.
Given the fact that the level of sequence identity between some members of Pfam family PF07449 is very low (as low as 13%), it is expected that the structures of both proteins Q8ZP25_SALTY and HYAE_ECOLI will be useful as templates in deriving high-quality homology models for most members of Pfam PF07449. In fact, conservation of residues participating in the molecular core of proteins Q8ZP25_SALTY and HYAE_ECOLI within PF07449 is high (Figure 2).
Fig 2.
Multiple sequence alignment of the members of Pfam family PF07449. The sequences of HYAE_ECOLI and Q8ZP25_SALTY are shown in the top two rows. The secondary structure elements of Q8ZP25_SALTY are shown above the alignment, and residues participating in the molecular core are marked with an asterisks (*). Conserved residues with 85% or greater conservation are highlighted in burgundy. Those residues are also seen in the most conserved group resulting from the analysis using the program Consurf analysis (Figure 3a). For comparison, the sequence of thioredoxin (PDB ID 1THX) was aligned with the sequence of protein Q8ZP25_SALTY using a structure based alignment algorithm [40] and is shown at the bottom. The Cys-X-X-Cys motif characteristic of thioredoxins is highlighted in blue (and absent in PF07449).
A search of structurally similar proteins domains using the DALI server [35] identifies 30 structurally similar proteins with Z-scores > 4.0, many of which are thioredoxins containing the catalytic sequence motif Cys-X-X-Cys. The best scoring proteins are (i) thioredoxin from Anabaena sp. Strain PCC1720 (PDB ID 1THX; Z-score 8.8, 102 aligned residues with rmsd. 3.3 Å), (ii) a C73S mutant human thioredoxin (1ERV; Z-score 7.9, 99 aligned residues; 3.2 Å), (iii) an unannotated protein from Haemophilus ducreyi (2DO8; Z-score 7.7, 108 aligned residues; 4.6 Å), and (iv) a theoretical model of a disulfide isomerase from Plasmodium chabuadi (1Y9N; Z-score 7.5, 96 aligned residues; 3.7 Å).
Functional implications of of Q8ZP25_SALTY and HYAE_ECOLI structures
In contrast to the structurally similar thioredoxins identified by the DALI server, proteins Q8ZP25_SALTY and HYAE_ECOLI do not possess the canonical thioredoxin catalytic motif Cys-X-X-Cys and thus cannot participate in the biochemical reactions characteristic for thioredoxins. In contrast, protein HYAE_ECOLI has been shown to interact with the Tat signal peptide-bearing subunit of hydrogenase-1 HyaA, and was thus classified as a hydrogenase-1 β-subunit-specific chaperone [2]. Consistently, the deletion of the gene encoding protein HoxO in Ralstonia eutropha, another member of PF07449, leads to complete loss of the uptake [NiFe] hydrogenase activity, suggesting that hoxO has a critical role in the assembly of the hydrogenase [36].
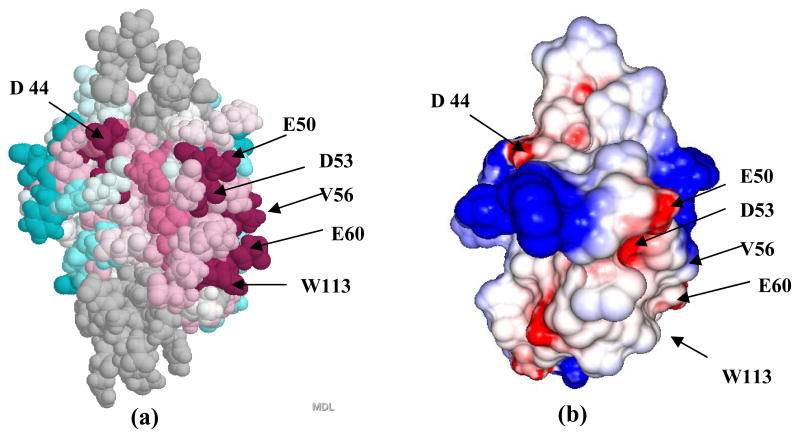
The multiple sequence alignment for proteins of Pfam PF07449 (Figure 2) reveals conservation of surface exposed residues which have no obvious structural role in the molecular core. The program ConSurf [37] was used to map those residues from onto the surface of Q8ZP25_SALTY (Figure 3a). Intriguingly, three of the most highly conserved residues (shown in red) are the negatively charged residues Asp 44, Asp 53 and Glu 50 located in the flexibly disordered loop comprising residues 43–55, which align structurally with the catalytic motif of thioredoxin (Figure 2). Calculation of the electrostatic surface using the program GRASP [38] shows the resulting negatively charged patch on the protein surface (Figure 3b). This structural bioinformatics analysis suggests that the highly conserved, negatively charged surface residues interact with the arginine rich, positively charged, Tat signal peptide.
Fig 3.
(a) Conserved residues from PFAM PF07449 are mapped onto the surface of protein Q8ZP25_SALTY. Colors are based on Bayesian conservation score [41] which characterizes the evolution rate of each residue compared to the average rate for all residues in the protein as follows: burgundy −1.6 to −1.3 (highly conserved), dark pink −1.29 to −0.9, pink −0.89 to −0.5, light pink −0.49 to −0.2, white −0.19 to 0.19, very light blue 0.2 to 0.5, light blue 0.51 to 0.7, blue 0.71 to 1.25, dark blue 1.26 to 2.2 (highly variable). (b) Electrostatic surface potential for Q8ZP25_SALTY. Negative and positive electrostatic potentials are shown, respectively, in red and blue.
Conclusions
The NMR solution and X-ray structures of the protein Q8ZP25_SALTY and the NMR solution structure of homologous protein HYAE_ECOLI were determined and shown by their structure statistics to be of high quality. Both proteins exhibit a “thioredoxin-like” fold likely representative of all members of protein family PF07449 which currently contains 50 proteins. However, none of the members of PF07449 bear the Cys-X-X-Cys active site motif characteristic of thioredoxins. Conserved, negatively charged surface patches on HYAE_ECOLI and Q8ZP25_SALTY could potentially be involved in binding with the arginine rich Tat signal peptide that regulates hydrogenase complex assembly and export.
Acknowledgments
This work was supported by the National Institutes of Health (U54 GM074958-01) and the National Science Foundation (MCB 0416899 to T.S.). We thank Kellie Cunningham, Chi Kent Ho, Haleema Janjua, Li-Chung Ma, and Li Zhao (Rutgers University) for help preparing the protein samples.
Abbreviations
- NESG
Northeast Structural Genomics Consortium
- NMR
nuclear magnetic resonance
- NSLF
national synchrotron light source
- PDB
protein data bank
- SAD
single-wavelength anomalous diffraction
- RDC
residual dipolar coupling
- PSVS
protein structure validation suite
References
- 1.Finn RD, Mistry J, Shuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Nucl Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubini A, Sargent F. FEBS Lett. 2003;549:141–146. doi: 10.1016/s0014-5793(03)00802-0. [DOI] [PubMed] [Google Scholar]
- 3.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterson R, Wilson RK. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 4.Liu GH, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, Acton T, Arrowsmith C, Montelione G, Szyperski T. Proc Natl Acad Sci USA. 2005;102:10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acton TB, SGK, Xiao r, Ma LC, Aramini JM, Baran MC, Chiang YW, Climent T, Cooper B, Denissova N, Douglas SM, Everett JK, Ho CK, Macapagal D, Paranji RK, Shastry R, Shih LJ, Swapna GVT, Wilson M, Wu MJ, Gerstein M, Inouye M, Hunt JF, Montelione GT. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 6.Atreya HS, Szyperski T. Proc Natl Acad Sci USA. 2004;101:9642–9647. doi: 10.1073/pnas.0403529101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen MR, Hanson P, Pardi A. Rna-Ligand Interactions Pt A. ACADEMIC PRESS INC; San Diego: 2000. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions; pp. 220–240. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Szyperski T. J Am Chem Soc. 2003;125:1385–1393. doi: 10.1021/ja028197d. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y, Atreya HS, Liu GH, Szyperski T. J Am Chem Soc. 2005;127:9085–9099. doi: 10.1021/ja0501870. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy. Academic Press; San Diego: 2007. [Google Scholar]
- 11.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 12.Bartels C, Xia T, Billeter M, Guntert P, Wuthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerman DE, Kulikowski CA, Huang YP, Feng WQ, Tashiro M, Shimotakahara S, Chien CY, Powers R, Montelione GT. J Mol Biol. 1997;269:592–610. doi: 10.1006/jmbi.1997.1052. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann T, Güntert P, Wüthrich K. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 15.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 16.Huang YJ, Moseley H, Baran MC, Arrowsmith C, Powers R, Tejero R, Szyperski T, Montelione G. Methods Enzymol. 2005;394:111–141. doi: 10.1016/S0076-6879(05)94005-6. [DOI] [PubMed] [Google Scholar]
- 17.Valafar H, Prestegard JH. J Magn Reson. 2004;167:228–241. doi: 10.1016/j.jmr.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 19.Schwieters CD, Kuszewski J, Clore GM. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. doi: 10.1016/j.pnmrs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 21.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucl Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya A, Tejero R, Montelione G. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Guntert P, Braun W, Wuthrich K. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 26.Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Biochemistry. 1989;28:7510–6. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 27.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 28.Prestegard JH, Bougault CM, Kishore AI. Chem Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 29.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Loannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger KR, Yao H, Markley JL. Nucl Acids Res. 2007 doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews BW. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger TC, Berendzen J. Acta Cryst Sect D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Cryst Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, MN, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Cryst Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 34.Engh R, Huber R. Acta Cryst Sect A. 1991;47:392–400. [Google Scholar]
- 35.Holm L, Sander CR. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 36.Bernhard M, Schwartz E, Rietdorf J, Friedrich B. J Bacteriol. 1996;178:4522–4529. doi: 10.1128/jb.178.15.4522-4529.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glaser F, Pupko T, Paz I, Bell R, Bechor-Shental D, Martz E, Ben-Tal N. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls A, Sharp KA, Honig B. Proteins: Structure, Function, and Genetics. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA. J Mol Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Skolnick J. Nucl Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayrose I, Graur D, Ben-Tal N, Pupko T. Molecular Biology and Evolution. 2004;21:1781–1791. doi: 10.1093/molbev/msh194. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 43.Word JM, Bateman RC, Presley BK, Lovell SC, Richardson DC. Protein Sci. 2000;9:2251–2259. doi: 10.1110/ps.9.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang YJ, Powers R, Montelione G. J Am Chem Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]