Abstract
To assay the preferential binding of eukaryotic type IB topoisomerases to supercoiled DNA, two methods are described that make use of a catalytically inactive mutant form of the enzyme. In the gel shift assay, the preference for binding to supercoiled plasmid DNA is detected in the presence of linear and nicked forms of the same DNA by a reduction in the mobility of the supercoiled plasmid during electrophoresis in agarose. The more quantitative filter binding assay compares the ability of nicked and supercoiled forms of the circular DNA to compete for the binding of a 3H-labeled nicked DNA to the topoisomerase where the enzyme-DNA complexes are quantitated by the retention of the labeled DNA on a nitrocellulose membrane.
Keywords: topoisomerases, supercoiled DNA, plasmid DNA, nicked DNA, gel shift assay, filter binding assay, nitrocellulose filters
1. Introduction
Type I DNA topoisomerases release the torsional strain that builds up in DNA during essential biological processes such as DNA replication and transcription by introducing a transient single-strand break into the DNA (for review, see reference (1)). During the lifetime of the break, the enzyme is covalently attached to one end of the DNA through a phosphodiester bond to a tyrosine side chain. The type I topoisomerases are further classified into two subfamilies based on the polarity of the covalent linkage. For the type IA subfamily, the linkage is to the 5′ end of the DNA at the site of the break; this subfamily includes the prototypical bacterial topoisomerase I. The eukaryotic topoisomerases I belong to the type IB subfamily since the linkage in this case is to the 3′ end of the DNA. In the nicked state for the type IB enzymes, supercoils are relaxed by rotation of the free 5′ end of the broken strand around the intact strand of the DNA (2). The eukaryotic topoisomerases I are able to relax both negative and positive supercoils.
Human topoisomerase I has been shown to preferentially bind supercoiled DNA over relaxed DNA (3-5), providing a likely explanation for how the enzyme is targeted to torsionally-strained regions of the chromosomal DNA in the cell. An early method for detecting this preferential binding relied on measuring the amount of nicked topoisomerase I-DNA covalent complex formed after incubation of supercoiled or relaxed plasmid DNAs with the topoisomerase (3), but this method is complicated by the relaxation of the supercoiled DNA during the course of the analysis. A second method that suffers from the same weakness and is also laborious involves electron microscopic visualization of enzyme bound to DNA nodes in supercoiled DNA (4). Here we describe in detail two methods for measuring the preferential binding of topoisomerase I to supercoiled DNAs that circumvent the problem of DNA relaxation during the assay by utilizing a mutant form of the enzyme in which the active site tyrosine in the human topoisomerase I has been replaced with phenylalanine (Y723F mutation herein referred to as Y/F). In the gel shift assay, topoisomerase I binding to the DNAs in an equimolar mixture of supercoiled circles, nicked circles, and linear molecules with increasing protein concentration is detected by a reduction in the mobility of the DNA during electrophoresis in agarose. A similar assay has been described for the preferential binding of the p53 protein and HMG proteins to supercoiled DNA (6-10). The second more quantitative filter binding assay measures DNA binding by capturing labeled DNA-topoisomerase I complexes on a nitrocellulose filter (11). The relative ability of unlabeled competitor DNAs (supercoiled versus nicked) to reduce the binding of a labeled nicked DNA to the enzyme as measured by retention of the radioactive label on the filter provides a direct measure of the preference for supercoiled DNA.
2. Materials
2.1. Topoisomerase I
An N-terminally-truncated (topo70, missing amino acids 1-174) and catalytically inactive Y/F mutant form of human topoisomerase I designated as topo70 Y/F was purified from recombinant baculovirus-infected insect SF9 cells as described (12) (see Note 1). Topo31 (containing residues 175 to 433 of human topoisomerase I) was purified as described elsewhere (13).
2.2 Gel Shift Assay
2.2.1 Enzymes
EcoR I (20μ/μl) (cat. R0101S) and BamH I (20μ/μl) (cat. R0136S) were purchased from New England Biolabs.
2.2.2 Plasmid DNA
The pKSII+ plasmid DNA (Stratagene) (3.0 kb, see Note 2) was purified from plasmid-bearing Escherichia coli using the Qiagen plasmid purification kit (Qiagen Corp.). The purified DNA is free of contaminating protein and is primarily composed of negatively supercoiled DNA molecules with nicked circles representing no more than 20% of the total DNA.
2.2.3 Buffers
10X TBE: 0.89 M Tris-borate, 20 mM EDTA, pH 8.0.
TE: 10 mM Tris-HCl, pH 7.5, 1 mM EDTA.
3X agarose gel loading buffer: 30 mM EDTA, 1.5% SDS, 15% Ficol-400, 0.03% bromophenol blue.
10X DNA binding buffer: 100 mM Tris-HCl, pH 7.5, 500 mM KCl, 10mM EDTA and 10 mM DTT.
EcoR I reaction buffer (New England Biolabs, cat. B0101S): 50 mM NaCl, 100 mM Tris-HCl, pH 7.5,10 mM MgCl2, 0.025%Triton X-100.
BamH I reaction buffer (New England Biolabs, cat. B0136S): 150 mM NaCl, 10 mM Tris-HCl, pH 7.9, 10 mM MgCl2, 1mM DTT, 100 μg/ml BSA.
2.3 Filter Binding Assay
2.3.1 DNAs
Unlabeled and 3H-labeled (2-4 × 104 cpm/μg) supercoiled SV40 DNAs were isolated from SV40 infected-CV-1 cells using the Hirt procedure (14) and purified by CsCl-ethidium bromide equilibrium centrifugation (5) (see Note 3).
2.3.2 Nitrocellulose Filters and Filter Apparatus
13 mm nitrocellulose filters with a 0.45 μm pore size (mixed cellulose ester, A045A013A) can be purchased from Advantec MFS Inc. (Dublin, CA). Set up a suction filter apparatus equipped for a 13 mm filter and adjust the vacuum for a filtration rate of 3-4 ml/min (see Note 4).
3. Methods
3.1. Preparation of Nicked Circular DNA for Both Assays
Nicked DNA is prepared from supercoiled DNA (pKSII+ plasmid DNA for the gel shift assay and 3H-labeled SV40 DNA for the filter binding assay) as described in Shortle et al. (15).
Digest 500 μg of supercoiled SV40 DNA or pKSII+ DNA with 200 units of EcoR I (see Note 5) in 0.5 ml EcoR I reaction buffer in the presence of ethidium bromide (15 μg/ml) for 5 h at 37°C.
Extract the sample with an equal volume of phenol-chloroform and precipitate the DNA with an equal volume of isopropanol.
Centrifuge the sample at 16,000X g for 15 min and wash the pellet with ice cold 70% ethanol.
Resuspend the DNA in TE buffer and determine the DNA concentration from the absorbance at 260 nm.
Add a one-half volume of 3X agarose gel loading buffer to a small sample of the DNA (~0.6 μg) and subject the sample to electrophoresis in a 1% agarose gel in 1X TBE buffer to verify that the conversion to nicked circles was complete without any substantial production of linear molecules (see Note 6).
3.2. Gel Shift Assay
The gel shift assay for DNA binding by topoisomerase I depends on the observation that bound protein will reduce the mobility of a DNA during gel electrophoresis. Under the conditions described below, when the catalytically inactive form of human topoisomerase I (topo70 Y/F) is combined with a mixture of supercoiled, linear, and nicked circular DNAs, any selectivity of the protein for one or more of the topological forms of the DNA will result in a preferential reduction in the mobility of that DNA in the gel. This gel shift procedure provides the basis for a semiquantitative assay to detect the preferential binding of human topoisomerase I to supercoiled DNA.
3.2.1. Preparation of linear plasmid DNA
Digest 500 μg of pKSII+ DNA with 200 units of BamH I in BamH I buffer for 1 h at 37°C.
Purify and characterize the linearized DNA as described in steps 3-6 of Subheading 3.1.
3.2.2. Sample preparation
Prepare a master mix containing equimolar concentrations of supercoiled, nicked and linear pKSII+ DNAs in 1X DNA binding buffer in which the concentration of each DNA is 8 fmols/μl.
Prepare two-fold serial dilutions of topo70 Y/F (or other protein of interest) in 1X DNA binding buffer where the amount of protein typically ranges from 1.5-0.088 pmol/μl and aliquot 10 μl of each dilution into a reaction tube on ice.
Initiate the reaction by mixing 10 μl of the DNA substrate mix with the serially diluted samples of topo70 Y/F and incubate at room temperature for 20 min.
3.2.3 Agarose Gel Electrophoresis
Add 5 μl of 50% glycerol to the reaction and load the sample onto a 1% agarose gel cast in 0.5X TBE. Include control samples containing no protein to mark the mobilities of the three DNA forms.
Runs the gel in 0.5X TBE at 1.2 volts /cm at 4° C for 20 h
Stain the gel in 0.15 μg/ml ethidium bromide for 30 min and visualize the DNA bands photographically.
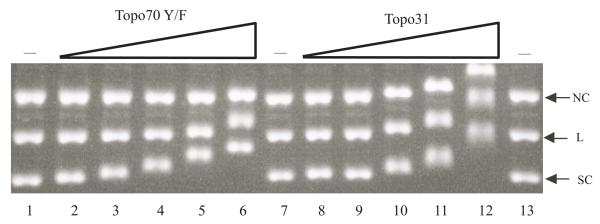
A typical analysis is shown in Fig. 2 comparing the binding properties of topo70 Y/F with topo31, which is a C-terminally truncated form of topo70 that displays no preference for supercoils (Z. Yang and J.J. Champoux, unpublished). As can be seen in lanes 2-4 of Fig. 2, the binding of topo 70 at the lower protein concentrations retards the mobility of the supercoiled DNA (SC) without noticeably affecting the mobility of either the linears (L) or the nicked circles (NC). With higher protein concentrations, the mobilities of all three forms of the DNA are affected. This observation is in sharp contrast to the results with topo31, where the protein binds and shifts the mobility of all three forms of the DNA equally at all ratios of protein to DNA (Fig. 2, lanes 10-12, note that higher protein concentrations were required for topo31, owing to its lower affinity for DNA).
Fig. 2.
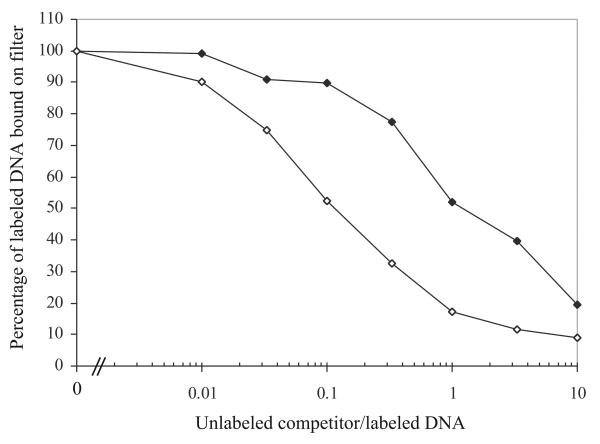
Filter binding assay. 3H-labeled nicked SV40 DNA (0.2 pmol) was incubated with topo70 Y/F and then challenged with increasing amounts of unlabeled nicked competitor (filled diamonds) or unlabeled supercoiled competitor (open diamonds) as described in the text. The percentage of 3H-labeled nicked SV40 DNA retained on the filter is plotted against the ratio of the unlabeled competitor to labeled DNA.
3.3. Filter Binding Assay
In the filter binding assay, 3H-labeled nicked DNA is first incubated with the catalytically inactive form of human topoisomerase I (topo70 Y/F) (or mutant forms of the protein), followed by the addition of an unlabeled competitor DNA (either nicked or supercoiled). After an additional incubation, the fraction of labeled DNA containing bound protein is assessed by collecting the complexes on a nitrocellulose membrane. By varying the molar ratios of the unlabeled competitors to the labeled DNA, competition profiles are generated that permit one to compare the binding properties of the supercoiled and nicked DNAs for the protein of interest.
3.3.1. Sample Preparation
Incubate 3H-labeled nicked SV40 DNA (0.2 pmol per reaction) with an equimolar amount of topo70 Y/F in 10 μl of 1X DNA binding buffer at room temperature for 20 min.
Prepare a series of 10 μl samples containing the following amounts of the unlabeled nicked DNA in 1X DNA binding buffer to serve as the “like” competitor: 2, 0.67, 0.2, 0.067, 0.02 0.0067, and 0.002 pmols. These amounts cover the range from 10 times to 0.01 times the amount of labeled nicked DNA in the reactions. Likewise prepare a series of 10 μl samples containing the same amounts of the supercoiled competitor DNA.
Initiate the competition reactions by mixing the 10 μl samples containing the unlabeled competitor DNAs with the 10 μl reactions that contain the 3H- labeled nicked SV40 DNA with bound topo 70 Y/F, and incubate the reactions at 23° C for 30 min.
3.3.2 Filtration and Determination of Bound Radioactivity
During the incubation, soak the 13 mm, 0.45 μm pore size nitrocellulose membranes in distilled water (see Note 7).
After the incubation, position a pre-wetted 13 mm nitrocellulose filter on a suction apparatus set for a filtration rate of 3-4 ml/min, and apply the 20 μl reaction directly to the filter.
Immediately wash the filter with 0.8 ml of 1X DNA binding buffer (see Note 8).
Air dry the filters and count the samples in 5 ml of toluene/Ominifluor (4 g/l) scintillation fluid (see Note 9) in a scintillation counter.
A typical experiment is shown in Fig. 2 where the percentage of labeled DNA bound on the filter is plotted as a function of the ratio of unlabeled competitor to labeled DNA. With the “like” nicked competitor, half-maximal binding of the labeled DNA occurs at a ratio of 1.0 as expected (closed symbols). However, with the unlabeled supercoiled competitor, it can be seen that approximately 10-fold less competitor DNA is required to compete the labeled DNA to the level of half-maximal binding (open symbols). We have previously suggested that this difference reflects the existence of ~10 times more binding sites for the enzyme on supercoiled DNA as compared with nicked or relaxed DNA (5).
4. Notes
Although we describe the methods and show data for the N-terminally truncated form of human topoisomerase I (topo70), similar results are obtained for the full length protein.
Any circular plasmid DNA in the size range from 2.5 to 3.5 kb is suitable for the analysis. However, since the magnitude of the gel shift is inversely related to the size of the DNA, it is recommended that larger plasmids DNA not be used.
We find it convenient to prepare 3H-labeled SV40 DNA as described, but the nature and source of the supercoiled circular DNA are not important as long as the specific activity is at least 2 × 104 cpm/μg.
A suction filter holder appropriate for this use is available from Millipore Corp. (Catalogue number XF3001200).
Other single-cut restriction enzymes can be used for this purpose, but the reaction conditions must be adjusted as described by Shortle et al. (15).
If the nicked DNA preparation contains an unsatisfactory level of linear molecules, the ethidium bromide concentration in the restriction enzyme reaction should be empirically titrated upwards to the point where nicking is complete without the generation of linears.
If complete wetting of the filter does not occur as judged by the presence of a uniform gray color, the filter should be discarded.
Care should be taken to avoid drying the filter prior to the wash step. For this reason, we recommend carrying out the filtrations one at a time.
Any scintillation fluid suitable for non-aqueous samples is satisfactory for this purpose.
Fig. 1.
Gel shift assay with topo70 Y/F and topo31. Equal amounts of supercoiled circles (SC), linears (L) and nicked circles (NC) of pKSII+ plasmid DNA (0.08 pmol each) were incubated with increasing amounts of the indicated proteins in 20 μl of DNA binding buffer and analyzed by agarose gel electrophoresis as described in the text. Lanes 1, 7 and 13 contained no protein to mark the mobilities of the unbound DNAs. Lanes 2-6 contained 0.88, 1.75, 3.5, 7 and 15 pmol of topo70 Y/F protein, respectively. Lanes 8-12 contained 1.75, 3.5, 7, 15, and 30 pmol of topo31, respectively.
Acknowledgments
This work was supported by National Institutes of Health Grant GM49156.
References
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 3.Muller MT. Quantitation of eukaryotic topoisomerase I reactivity with DNA. Preferential cleavage of supercoiled DNA. Biochim. Biophys. Acta. 1985;824:263–267. doi: 10.1016/0167-4781(85)90057-0. [DOI] [PubMed] [Google Scholar]
- 4.Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madden KR, Stewart L, Champoux JJ. Preferential binding of human topoisomerase I to superhelical DNA. EMBO J. 1995;14:5399–5409. doi: 10.1002/j.1460-2075.1995.tb00224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stros M, Muselikova E. A role of basic residues and the putative intercalating phenylalanine of the HMG-1 box B in DNA supercoiling and binding to four-way DNA junctions. J. Biol. Chem. 2000;275:35699–35707. doi: 10.1074/jbc.M007167200. [DOI] [PubMed] [Google Scholar]
- 7.Grasser KD, Teo SH, Lee KB, Broadhurst RW, Rees C, Hardman CH, Thomas JO. DNA-binding properties of the tandem HMG boxes of high-mobility-group protein 1 (HMG1) Eur. J. Biochem. 1998;253:787–795. doi: 10.1046/j.1432-1327.1998.2530787.x. [DOI] [PubMed] [Google Scholar]
- 8.Payet D, Travers A. The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J. Mol. Biol. 1997;266:66–75. doi: 10.1006/jmbi.1996.0782. [DOI] [PubMed] [Google Scholar]
- 9.Palecek E, Brazdova M, Brazda V, Palecek J, Billova S, Subramaniam V, Jovin TM. Binding of p53 and its core domain to supercoiled DNA. Eur. J. Biochem. 2001;268:573–581. doi: 10.1046/j.1432-1327.2001.01898.x. [DOI] [PubMed] [Google Scholar]
- 10.Mazur SJ, Sakaguchi K, Appella E, Wang XW, Harris CC, Bohr VA. Preferential binding of tumor suppressor p53 to positively or negatively supercoiled DNA involves the C-terminal domain. J. Mol. Biol. 1999;292:241–249. doi: 10.1006/jmbi.1999.3064. [DOI] [PubMed] [Google Scholar]
- 11.Hinkle DC, Chamberlin MJ. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J. Mol. Biol. 1972;70:157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- 12.Stewart L, Champoux JJ. Purification of baculovirus expressed human DNA topoisomerase I. In: Bjornsti M-A, Osheroff N, editors. DNA Topoisomerase Protocols, Volume 1: DNA topology and enzymes. Vol. 94. Humana Press; Totowa, New Jersey: 1996. pp. 223–234. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Champoux JJ. Reconstitution of enzymatic activity by the association of the cap and catalytic domains of human topoisomerase I. J. Biol. Chem. 2002;277:30815–30823. doi: 10.1074/jbc.M205302200. [DOI] [PubMed] [Google Scholar]
- 14.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 15.Shortle D, Grisafi P, Benkovic SJ, Botstein D. Gap misrepair mutagenesis: efficient site-directed induction of transition, transversion, and frameshift mutations in vitro. Proc. Natl. Acad. Sci. U.S.A. 1982;79:1588–1592. doi: 10.1073/pnas.79.5.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]