Abstract
For cell regulation, E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1) is involved in the transfer of ubiquitin-fold modifier 1 (Ufm1), a ubiquitin like protein which is activated by E1-like enzyme Uba5, to various target proteins. Thereby, Ufc1 participates in the very recently discovered Ufm1-Uba5-Ufc1 ubiquination pathway which is found in metazoan organisms. The structure of human Ufc1 was solved by using both NMR spectroscopy and X-ray crystallography. The complementary insights obtained with the two techniques provided a unique basis for understanding the function of Ufc1 at atomic resolution. The Ufc1 structure consists of the catalytic core domain conserved in all E2-like enzymes and an additional N-terminal helix. The active site Cys116, which forms a thio-ester bond with Ufm1, is located in a flexible loop that is highly solvent accessible. Based on the Ufc1 and Ufm1 NMR structures, a model could be derived for the Ufc1-Ufm1 complex in which the C-terminal Gly83 of Ufm1 may well form the expected thio-ester with Cys116, suggesting that Ufm1-Ufc1 functions as described for other E1-E2-E3 machineries. α-helix 1 of Ufc1 adopts different conformations in the crystal and in solution, suggesting that this helix plays a key role to mediate specificity.
Keywords: Ufc1, Ufm1, Ubiquitin, E2, Ubiquitin Conjugating Enzyme
Introduction
Covalent attachment of a ubiquitin or ubiquitin-like (Ubl) protein molecule to a substrate protein, a process known as ‘ubiquitination’ [1], targets the substrate for a range of possible fates [2-5]. In eukaryotic cells, ubiquitination is critical for the regulation of many cellular processes such as protein degradation [6], DNA repair [7] or cell cycle control [8]. Moreover, dysregulation of ubiquitination is implicated in the etiology of various human diseases [4].
Ubiquitin and Ubls share a common fold [3], and their transfer to the target protein is accomplished through the sequential action of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2 or Ubc), and a ubiquitin-protein ligase (E3) [6, 8, 9]. The mechanisms underlying ubiquitination have been intensely studied and have revealed important differences between the pathways associated with different Ubls [4]. These insights were complemented by atomic resolution structures for several ubiquitination proteins and their protein-protein complexes; the structures contributed to provide a basis for our understanding of ubiquitination at atomic resolution [5].
Recent structural genomics studies have contributed to identifying hitherto unknown ubiquitination pathways. The NMR structure of 11.5 KDa protein ZK652.3 encoded in gene ZK652.3 from C. elegans solved by the Northeast Structural Genomics Consortium (http://www.nesg.org) enabled the identification of this protein as a new Ubl [10]. Since the sequence identity between ZK652.3 and ubiquitin is only 14%, this conclusion could not have been drawn from sequence comparison alone [10]. The human homolog of ZK652.3 was later confirmed to be a Ubl and was named ‘ubiquitin-fold modifier 1’ (Ufm1) [11]. In contrast to many other Ubls (including ubiquitin) which comprise a C-terminal di-glycine segment, ZK652.3 and Ufm1 contain only a single C-terminal Gly. Nonetheless, the first two steps of the ubiquitination cascade associated with Ufm1 are rather similar to those previously described for other Ubls and ubiquitin: the C-terminal carboxylate of Ufm1 is activated by an E1-like enzyme, Uba5 [11]. The thus activated Ufm1 is then transferred to its conjugate E2-like enzyme [protein CGI-126, later named ‘Ufc1’ [11]], and a thio-ester linkage is formed between the C-terminal carboxylate of Ufm1 and the thiol group of the catalytic Cys116 of Ufc1 [11]. In contrast, the E3-type enzymes interacting with Ufc1 remain to be identified.
All members of the family of E2-like enzymes share a conserved ~150-residue catalytic core domain consisting of a 4-stranded β-sheet and four α-helices attached to one side of the sheet [2, 8]. Some members feature N- or C-terminal extensions which are possibly required to convey distinct specificity for protein-protein interactions [4, 8]. The E2-like 167-residue protein Ufc1 belonging to PFam [12] family PF08694 (comprising 27 sequence homologues) is conserved in multicellular organisms, but exhibits low sequence identity with other E2-like enzymes. Hence, Ufc1 was originally annotated as protein CGI-126 until in vitro studies proved its E2-like function [11]. In parallel, this protein was selected as a target of the NESG project (target ID HR41). The NMR structure of HR41 showed that Ufc1 contains the characteristic catalytic core domain of E2 enzymes, and this finding was published in a paper describing a structural genomics technology development project [13].
Insights into protein structures obtained with X-ray in the crystal and with NMR in solution are complementary. In this publication, we present NMR and X-ray crystal structures of this 167-residue protein Ufc1 from H. sapiens that is encoded in gene Ufc1_HUMAN (NCBI ID: 9606; SwissProt ID: Q9Y3C8/Q9BS96/Q9P009; NESGC target ID: HR41; NESGC Rost-cluster ID: 15697).
Materials and Methods
Materials
Uniformly 13C, 15N-enriched human Ufc1 was prepared as was described [13] following standard protocols [14]. SeMet labeled Ufc1 was prepared using the same protocol, except that a culture of MJ9 minimal medium [15] containing SeMet was used.
NMR Structure Determination
The determination of the NMR structure of Ufc1 was accomplished as described previously [13, 16], except that the program CYANA [17, 18] was used.
Crystallization of Ufc1
Starting from results of a crystallization screen conducted by DeTitta and coworkers at the Hauptman-Woodward Medical Research Institute, Ufc1 was crystallized by the hanging drop vapor diffusion method at 21 °C. Two μls of protein solution containing Ufc1 (10 mg/ml), 5 mM Tris (pH 7.5), 100 mM NaCl, and 5 mM DTT were mixed with 2 μls of the reservoir solution consisting of 100 mM sodium acetate (pH 5), 18% PEG 8k, 100 mM ammonium isothionate, and 50 mM lithium isothionate. Crystals were transferred to a solution similar to that of the reservoir to which 25% (v/v) glycerol was added for cryo-protection, and then flash-frozen in liquid propane for data collection at T = 100 K. The crystal has cell parameters of a=41.368 Å, b=64.537 Å, c=66.422 Å, α=90.06°, β=89.95°, and γ=105.18° in spacegroup P1 with 4 Ufc1 protomers in the asymmetric unit (ASU). The crystal exhibits apparent P21 symmetry according to data merging statistics, but, as explained in detail in the next section, the behavior observed during refinement suggests that the lattice packing deviates in subtle way from perfect P21 spacegroup symmetry.
X-ray Crystal Structure Determination
A single-wavelength anomalous diffraction (SAD) [19] data set to 2.54 Å resolution was collected on a single crystal of Ufc1, at the peak absorption wavelength of selenium at the X4A beamline of the National Synchrotron Light Source (NSLS). The diffraction images were processed with the HKL package [20], and the selenium sites were located with the program SnB [21]. The program SOLVE/RESOLVE [22] was used for phasing the reflections and automated model building, which correctly placed 50% of the residues with side chains in both protomers of ASU. The remaining residues were built with the program XtalView [23] and refined by using the program CNS [24]. Very strong non-crystallographic symmetry restraints (300 kcal/Å) were applied during all stages of the refinement, and reflections in the cross-validation set were chosen in thin shells to avoid possible symmetry bias in Rfree values [25-27] (although this procedure only increased the overall Rfree by 0.3%). The crystal structure was initially determined and refined in the spacegroup P21, which gave canonical merging statistics during data processing that were very similar to those obtained in the lower symmetry spacegroup P1. However, the refinement of the structure was better in the latter spacegroup based on a 1.9% reduction in Rfree at 2.54 Å for the same starting model when applying the same strong NCS restraints. This result was interpreted to indicate that there is likely to be some small deviation from P21 symmetry in the lattice and that the spacegroup symmetry of the crystal is more likely to be P1 than P21. The Rfree increased as the strength of NCS restraints was reduced, indicating minimal conformational differences between the four NCS-related molecules in the P1 cell. However, there is a 0.04° inclination between the unit cell axis and the 2-fold screw axis describing the non-crystallographic symmetry operation relating pairs of protomers in the P1 cell. This small deviation from exact P21 symmetry is likely to account for the improved refinement of the crystal in spacegroup P1.
Atomic Coordinates
The atomic coordinates, NMR constraint lists, and structure factors were deposited in the Protein Data Bank [accession codes: 2K07 (NMR) and 3EVX (X-ray)]. NMR resonance assignments have been deposited in the BioMagResDB (accession code: 6546)
Results and Discussion
Ufc1 Structure
The NMR structure reported here (PDB ID 2K07) was further refined when compared with the one (PDB ID 1YWZ) considered for the publication on the structural genomics technology development project [13, 16] and submitted to the PDB in 2005. Specifically, a large number of stereo-specific assignments were added and the program CYANA2.1 was used for structure calculations. Statistics of the refined Ufc1 NMR structure (Table 1) show that a high-quality structure was obtained. After the NMR structure was first made available in 2005, the X-ray crystal structure was determined at a resolution of 2.54 Å (Table 2). Ufc1 exhibits the characteristic E2-like catalytic core domain comprising an anti-parallel β-sheet and five α-helices attached to one side of the sheet, which enabled classification of Ufc1 as an E2-like enzyme [13].
Table 1.
Statistics of Ufcl NMR structures determination
| Conformationally-restricting distance constraints | |
| Intraresidue [i = j] | 707 |
| Sequential [(i - j) = l] | 944 |
| Medium Range [1 < (i - j) ≤ 5] | 808 |
| Long Range [(i - j) > 5] | 1099 |
| Total | 3558 |
| Number of dihedral angle constraints (Φ/Ψ) | 80 / 80 |
| Number of constraints per residue | 23 |
| Number of long-range constraints per residue | 6.5 |
| Completeness of stereospecific assignmentsa (βCH2/ Val and Leu isopropyl groups) |
58% /73% |
| CYANA target function [Å2] | 1.47 ± 0.10 |
| Average r.m.s.d. to the mean CNS NMR coordinates [Å] | |
| regular secondary structure elementsb, backbone heavy | 0.80 ± 0.14 |
| regular secondary structure elements, all heavy atoms | 1.26 ± 0.11 |
| regular secondary structure elementsb, backbone heavy, exclude αl | 0.73 ± 0.15 |
| regular secondary structure elements, all heavy atoms, excluding helix αl | 1.20 ± 0.12 |
| residues 3-159, backbone heavy atoms N, Cα, C' | 0.96 ± 0.13 |
| residues 3-159, all heavy atoms | 1.41 ± 0.12 |
| heavy atoms of best-defined SCc | 0.75 ± 0.11 |
| PROCHECK G-factorsd (raw/z-score) | NMRf |
X-ray |
|---|---|---|
| Φ and Ψ | −0.08/ 0.00 | −0.52/−1.73 |
| all dihedral angles | −0.14/−0.83 | −0.45/−2.66 |
| MOLPROBITY clash raw /Z scoree | 19.93/−1.89 | 34.65/−4.42 |
| R/P/F/DP scores (%)g | 95/95/95/82 |
| Ramachandran plot summary ordered residue ranges: 25-37, 45-65, 70-90 [%] | NMRf | X-ray |
|---|---|---|
| most favored regions | 92.6 | 86.6 |
| additionally allowed regions | 7.1 | 11.9 |
| generously allowed regions | 0.1 | 0.7 |
| disallowed regions |
0.1 |
0.7 |
| Average number of distance constraints violations per CNS conformer [Å] | ||
| 0.2 – 0.5 / > 0.5 | 0.3 / 0 | |
| Average number of dihedral-angle constraint violations per conformer [degrees] | ||
| > 10 | 0 |
Relative to pairs with non-degenerate chemical shifts.
For backbone heavy atoms of regular secondary structure elements: residues 4-12, 25-48, 121-127, 134-140, 142-155 (α-helices), and 54-59, 64-73, 76-85 (β-stands).
Residues: Residues with best defined side-chains, including residues 13, 14, 15, 16, 20, 22, 27, 28, 29, 38, 39, 40, 43, 45, 49, 63, 65, 66, 82, 84, 86, 87, 90, 91, 92, 94, 95, 97, 98, 100, 102, 115, 117, 124, 125, 126, 129, 134, 135, 136, 137, 139, 140, 144, 145, 146, 150, 151, 154.
Scores defined in Ref.[46].
Scores defined in Ref.[47].
NMR structure after CNS refinement in explicit water, and ProCheck scores shown for the NMR structures for only the ordered residues: 4-17, 22-52. 54-73, 76-97, 100-102, 115-159.
Scores defined in Ref.[48].
Table 2.
Refinement statistics of Ufcl X-ray crystal structure
| Maximum resolution (Å) | 2.54 |
| Number of observations | 19,876 (4,903) |
| Rmerge (%)a | 7.4 (19.8) |
| Number of reflections | 19,876 (1,486) |
| Resolution range used in refinement | 2.54 – 19.96 |
| Completeness (%) | 91.0 (76.9) |
| R factor (%)b | 23.3 (27.6) |
| free R factor (%) | 27.9 (36.9) |
| rmsd in bond lengths (Å) | 0.008 |
| rmsd in bond angles (°) | 1.2 |
| Number of small molecules (per 4 protein protomers in asymmetric unit) | 4 isothiocyanate ions 49 waters |
. Numbers in parenthesis are for the highest resolution shell.
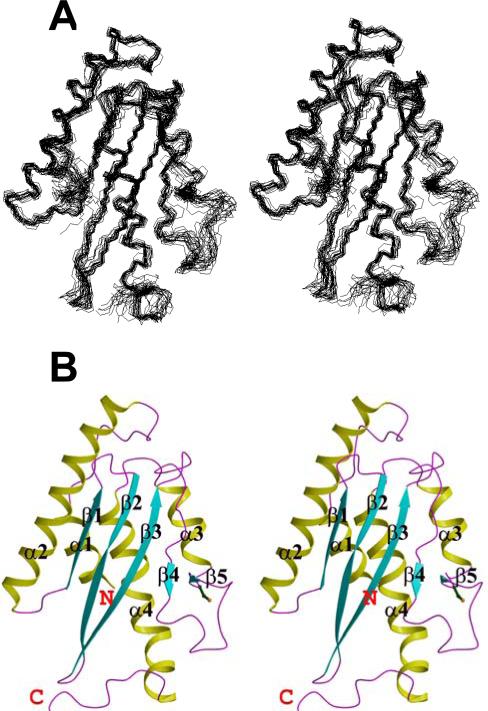
Inspection of the NMR structure (Fig. 1) reveals three β-strands (β1 to β3) comprising residues 54-59, 64-73 and 76-85, and five α-helices (α1 to α5) comprising residues 4-12, 25-48, 121-127, 134-140, and 142-155. These regular secondary structure elements are locally and globally well defined (Table 1). The β-strands form an anti-parallel β-sheet with topology β1(↑), β2(↓) and β3(↑). In some of the 20 conformers representing the NMR solution structure, two additional short β-strands are identified for residues 98-99 (β4) and 114-115 (β5).The five α-helices are attached to one side of the β-sheet while its other side is solvent exposed. The C-terminal deca-peptide segment 158-167 is flexibly disordered in solution. Notably, out of the ten prolyl residues in Ufc1, Pro91 and Pro130 adopt a cis-conformation in solution, as is evidenced by chemical shifts and nuclear Overhauser effect peak patterns.
Fig. 1.
NMR structure of human Ufc1. A, The 20 CYANA conformers with the lowest residual CYANA target function representing the NMR solution structure of Ufc1 are shown after superposition for minimal r.m.s.d. of the backbone heavy atoms N, Cα and C’ of residues of regular secondary structure elements. B, Ribbon drawing of the CYANA conformer with the lowest residual target function value (Table 1). The α-helices (α1-α5) are shown in yellow, the β-strands (β1 to β5) are in cyan, other polypeptide segments are in magenta (see table 1 for the starting and ending residues of regular secondary elements), and the N- and C-terminal ends of the protein are indicated as ‘N’ and ‘C’. The programs MOLMOL [43] and Molscript [44] were used to generate the figure.
The polypeptide segment 86-118 connecting β4 and α3 contains the catalytic Cys116 and is less well defined than the regular secondary structure elements. This suggests the presence of increased local disorder which might well be of importance for an induced-fit interaction with Ufm1 when the Ufc1-Ufm thioester is formed. In agreement with this interpretation, it is observed that (i) the backbone 1H-15N peaks in 2D [15N,1H]-HSQC of Met109 and Arg111 are broadened beyond detection due to slow conformational exchange, and that (ii) only rather weak NOEs are registered for the backbone amide moieties of Lys108 and Arg111.
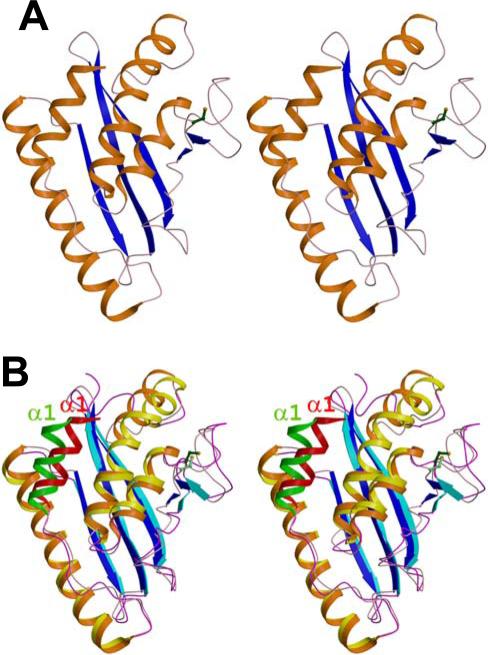
Consistent with the flexibility of the C-terminal segment 158-167 in solution, residues 164-167 are not observed in the X-ray crystal structure (Fig. 2). The ASU contains two Ufc1 molecules, but those are interacting only with a comparably small interaction surface. This is in agreement with the monomeric nature of Ufc1 in solution, as evidenced by (i) a rather short correlation time for overall rotational tumbling [13] and (ii) static light scattering data (data not shown).
Fig. 2.
X-ray crystal structure of human Ufc1. A, Ribbon drawing of the X-ray crystal structure of human Ufc1. The coloring is as in Fig. 1. B, Superposition of X-ray and NMR structure. α-helices, β-strands and loops of the NMR structure are shown in yellow, cyan, and magenta, respectively. The corresponding coloring scheme of the crystal structure involves orange, blue and pink, respectively. Helix α1 of NMR structure is depicted in green in order to highlight its different location with respect to the crystal structure where the helix α1 is shown in red. The side chain of the catalytic Cys116 is shown in dark green.
The comparison of NMR and X-ray crystal structure reveals the expected structural similarity. However, given the fact that the corresponding r.m.s.d. values within the ‘bundle’ of NMR conformers is smaller than 1 Å, the r.m.s.d. values calculated between the X-ray conformer and the mean NMR coordinates indicate some significant differences (Table 3 and Fig. 2B). First, whereas helix α4 in the crystal structure shows a kink at position 140, two smaller helices α4 and α5 connected by a non-helical residue Gly141 (confirmed by NMR chemical shift analysis, data not shown) are observed in the NMR structure (Fig. 2B). The catalytic Cys116 is located in the segment immediately following strand β5 and its side chain has poor electron density with high B-factor values in the X-ray structure, thus confirming the flexibly disordered nature (Fig. 2A) inferred from the NMR data for a larger part of the loop comprising 86-118. Second, and most pronounced, helix α1 is partially solvent exposed in the NMR structure (only few NOEs were observed between residues in helix α1 and residues in helices α3 and α4), whereas in the X-ray structure several hydrophobic interactions are detected with side-chains of helices α2 and α4 (Fig. 2). The conformational variation observed in solution at 298 K and in the crystal at 100 K might indicate that helix α1 can adopt different orientations when protein-protein complexes are formed.
Table 3.
Comparison of NMR (2K07) and X-ray (3E2G and 2Z6Pd) Ufcl structures (r.m.s.d. values in [Å])
| Atoms selected for r.m.s.d. calculationa | 2K07 vs 3EVX | 2K07 vs 2Z6Pd | 3EVX vs 2Z6Pd |
|---|---|---|---|
| Backbone heavy atoms N, Cα, C’ of regular secondary structure elementsb | 1.13 | 1.12 | 1.09 |
| All heavy atoms of regular secondary structure elements | 1.86 | 1.65 | 1.77 |
| Backbone heavy atoms of regular secondary structure elementsb except for α1 | 0.90 | 0.76 | 0.56 |
| All heavy atoms of regular secondary structure elements except for αl | 1.63 | 1.43 | 1.38 |
| Backbone heavy atoms N, Cα, C’ of residues 5-159 | 1.29 | 1.25 | 0.91 |
| All heavy atoms of residues 5-159 | 1.89 | 1.78 | 1.61 |
| All heavy atoms of best-defined side chainsc | 1.09 | 1.03 | 0.83 |
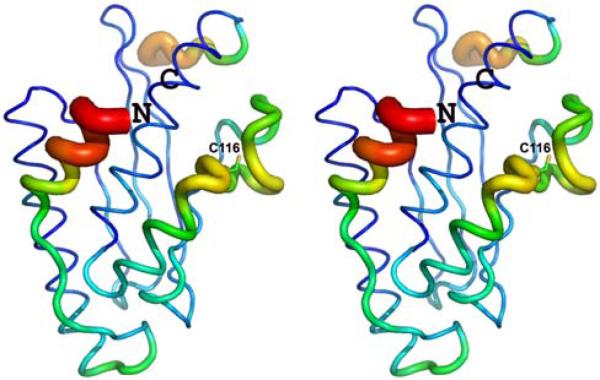
Very recently, another X-ray structure (PDB ID 2Z6P; deposited August 6 2007), solved at 1.6 Å resolution, was reported [28]. Calculation of global r.m.s.d. values (Table 3) reveal that the NMR structure is slightly more similar to the new X-ray structure 2Z6P. However, the major structural difference between the NMR and the new X-ray structure is also due to the different orientation of the N-terminal helix α1. While only weak NOEs are registered between helix α1 and helices α3 and α4, which goes along with its partial solvent exposure, for the NMR structure, helix α1 is oriented along helix α4 in our X-ray structure 3EVX but is in contact with both helices α3 and α4 in the new X-ray structure 2Z6P. Specifically, Thr6 interacts with Pro144 in helix α4 in 3EVX but with Leu124 in helix α3 in 2Z6P. Moreover, although helix α1 adopts a similar conformation in the four molecules in the asymmetric unit in structure 3EVX (even when released from non-crystallographic symmetry restraints), it exhibits strongly elevated backbone B-factors (Fig. 7), indicating some degree of disorder in its packing in the crystal. This observation further supports the view that helix α1 can adopt different orientations (see below).
Fig. 7.
Stereo representation of Cα B-factors along the polypeptide chain of Ufc1 in crystal structure 3EVX: the thickness of the cylindrical rod is proportional to the B-factors. The N- and C-terminal ends of the protein are labeled as ‘N’ and ‘C’. Blue polypeptide segments show the lowest B-factors, followed by green, yellow, orange, and red indicating increasing B-factors.
Identification of Structurally Similar Proteins
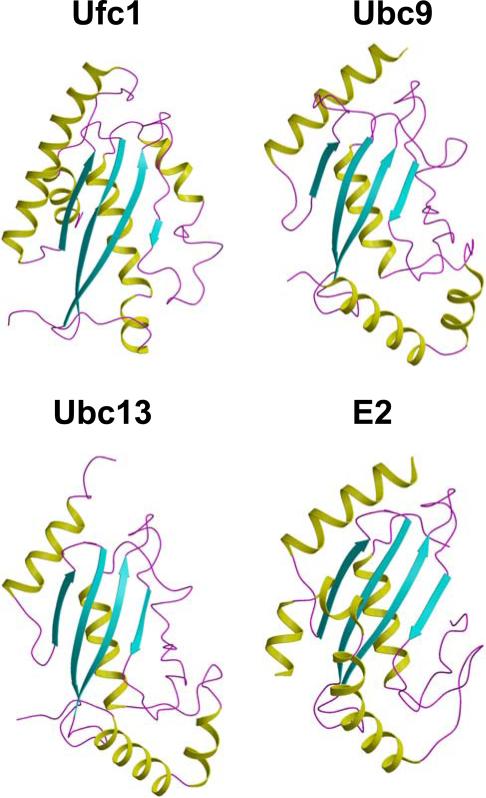
The Dali server [29] was used to search for proteins which are structurally similar. The three structures with the highest score turned out to be (i) human ubiquitin-conjugating enzyme Ubc9 (E2 of SUMO, PDB code: 1U9A, Dali Z-score: 7.7, r.m.s.d between backbone Cα atoms of 72% of the polypeptide chain: 2.8 Å, sequence identity: 16%), (ii) ubiquitin-conjugating enzyme Ubc13 in complex with ubiquitin E2 variant protein mms2 (1JAT:B, 7.4, 3.1 Å, 15%), and (iii) human ubiquitin-conjugating enzyme E2 (2F4W, 7.3, 2.7 Å, 15%) (Fig. 3). These proteins belong to the SCOP [30] family ‘Ubiquitin Conjugation Enzyme’ (Ubc) which comprises a total of 18 members. In comparison to these three most structurally similar E2-type enzymes, Ufc1 possesses the additional N-terminal helix α1. Moreover, helix α5 in Ufc1 is shorter and oriented differently compared to the corresponding helix in Ubc9 and Ubc13.
Fig. 3.
Structurally similar E2-like enzymes. Ribbon drawing of the structures of the E2-like enzymes. Ufc1, Ubc9, Ubc13 and E2 are shown for comparison, and the color code is the same as in Fig.1B.
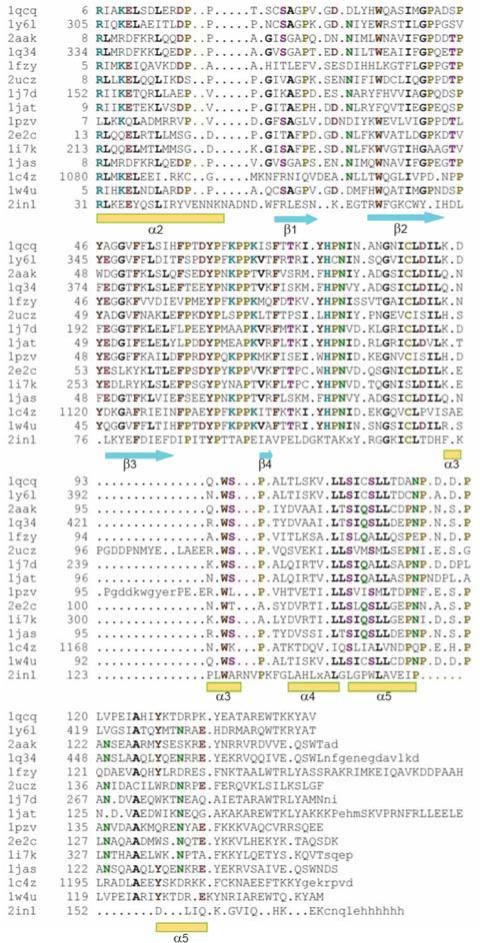
A structure-based sequence alignment of the SCOP Ubc family by using the program STRAP [31] (Fig. 4) reveals that the following residues of Ufc1 are structurally conserved in the other E2-like enzymes: Arg31, Glu35, Phe80, Tyr90, and Trp125. Those residues form a cavity on the surface of the catalytic core domain. Arg31 and Glu35 are located within the putative E1 binding site, but not directly interact with E1 (Fig. 4 and Fig. 6). Notably, Arg31, Glu35, Phe80, Tyr90, and Trp125 are also located close to the putative E3 binding site to E3. Moreover, the two cis-Pro91 cis-Pro130 residues observed in the NMR structure are likewise strongly conserved in this SCOP Ubc family.
Fig. 4.
Structure-based sequence alignment for SCOP classified UBC proteins. The PDB IDs are listed in the first column. 1qcq: from Saccharomyces cerevisiae, ubc4; 1y61: from Homo sapiens, ubch8; 2aak: from Arabidopsis thaliana; 1q34: from Caenorhabditis elegans, E2-21.5 kDa; 1fzy: from Saccharomyces cerevisiae, ubc1; 2ucz: UBC Saccharomyces cerevisiae, ubc7; 1j7d: UBC from Homo sapiens, mms2; 1jat: from Saccharomyces cerevisiae, ubc13; 1pzv: from Caenorhabditis elegans, E2-19 kDa; 2e2c: from Spisula solidissima, E-2C; 1i7k: from Homo sapiens, ubch10; 1jas: from Homo sapiens, ubc2b; 1c4z: from Homo sapiens, ubch7; 1w4u: UBC from Homo sapiens, ubch5b; and 2in1: from Homo sapiens. The alignment and the figures were generated by using the program STRAP [31]. Residues that are identical or highly conserved are highlighted with colors, and regular secondary structure elements are also indicated in the figures.
Fig. 6.
Residues in Ufc1 corresponding to E3 binding sites in other E2-like enzymes. A, Structure based sequence alignment of the segments of Ufc1 corresponding to the E3 binding segments in UbcH7 and Ubc13. Residues interacting with both HECT and RING type E3 in UbcH7 are colored green, residues interacting with HECT E3 in UbcH7 are colored golden, residues interacting with RING E3 in UbcH7 are colored blue, residues interacting with U-box E3 in Ubc13 are colored cyan, residues that are identical to residues in UbcH7 or Ubc13 are highlighted in red. B, View of Tyr90 in Ufc1 which exhibits the unusually slow ring flipping rate [44] (see text). The two cis-prolines Pro91 and Pro130 are labeled. C, Stereo-view of the electron-density for residues 90-92 and 129-131 taken from a simulated annealing omit map contoured at 1 σ. All six of these residues were omitted prior to simulated annealing at 1000° and map calculation.
The sequence of the loop connecting strand β4 and helix α3, which contains the solvent accessible active site Cys residue, is highly conserved among the E2-like enzymes, but shows different conformations (Fig. 3). In contrast to Ubc9 and Ubc13, the active site Cys116 in Ubc1 is not embedded in a largely negatively charged surface patch. This might correlate with different substrate specificity.
Model of the Ufc1-Ufm1 Complex
The interface of the Ubc1-Ub intermediate, which was mapped by using NMR chemical shift perturbation measurements [32, 33], is formed by residues of the C-terminal segment of ubiquitin and residues proximal to the active site Cys in the E2-like enzyme Ubc1. The high structural similarity of Ubc1 and Ufc1 catalytic core domain, and Ufm1 and Ubiquitin suggests that the protein-protein interface in the Ufc1-Ufm1 complex is similar. Hence, a model for the Ufc1-Ufm1 complex was derived using the model of the E2-Ub thio-ester intermediate (PDB accession code 1FXT) [33] as a template (Fig. 5). To generate this model, the backbone of the regular secondary structure elements of Ufc1 were superimposed onto the corresponding polypeptide segments in Ubc1. Since helix α1 could not be accommodated without introducing severe steric clashes, this helix was removed (see Fig. 2). Consistently, helix α1 was found to be dispensable for E1-E2 interaction and thioester linkage formation with Ufm1 in vitro [33]. The steric incompatibility of helix α1 with the Ubc1-Ub model further supports the view that this helix is functionally critical in regulating complex formation, and may serve to convey specificity in the Ufm1-Ufc1 system, as large conformational changes are common features among enzymes involved in Ubl transfer [34-36].
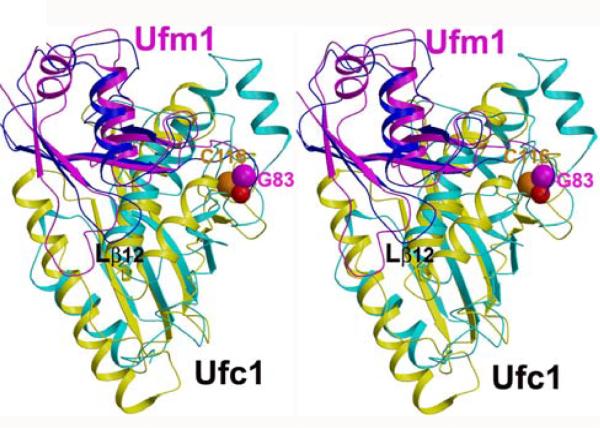
Fig. 5.
Model of the Ufc1-Ufm1 complex. The model was derived on the basis of the structure of the Ubc1-ubiquitin complex (1FXT) [33] by superimposing Ufc1 (in yellow) onto Ubc1 (in cyan) and Ufm1 (in magenta) onto ubiquitin (in blue). Helix α1 of Ufc1 was removed (see text). The thio-ester bond between Gly83 of Ufm1 and Cys116 of Ufc1 is shown with space-filling residues (carbon and oxygen atoms of Gly83 and the sulfur atom of Cys116 are, respectively, shown in magenta, red, and orange). The loop Lβ12 is labeled in black. The figure was produced using the programs Molscript [44] and Raster3D [45].
Subsequently, the NMR structure of Ufm1 [10] was superimposed in a similar manner onto Ubiquitin. Here, the NMR conformer was chosen with the most favorable conformation of the loop connecting strands β1 and β2, which differs in length and adopts two different conformations between Ubiquitin (blue) and Ufm1 (magenta in Fig. 5). The interface of the resulting model (Fig. 5) exhibited several minor steric clashes, which could readily be removed by minor structural rearrangements. In agreement with the expectation that the Ufc1-Ufm1 complex resembles the one of Ufc1-Ubiquitin, the model of the complex places the terminal Gly83 of Ufm1 in close spatial proximity to the catalytic Cys116 in Ufc1. This is evidently required for forming the corresponding thio-ester and confirms the validity of the model.
Interactions of Ufc1 with E1 and E3-like Enzymes
Structures of E1-E2 and E2-E3 complexes revealed that E1- and E3-binding sites on E2-like enzymes overlap partially [5, 34]. Specifically, the E2 binding domain in E1 enzymes resembles Ubl, and E1-like binding to E2-like enzymes occurs by mimicking its interaction with ubiquitin [34]. Residues located in E2 helix α1 (corresponding to helix α2 in Ufc1) and the loop connecting strands β1 and β2 loop are involved in the interface between Ubc12 and NEDD8's E1 [34-36]. Those residues are not conserved in Ufc1 and Uba5, respectively. This reflects the fact that the interface of E1-E2 complexes conveys specificity for any given E1-E2-E3 machinery.
Further specificity is encoded in the E2-E3 interaction [8]. Currently known E3-like enzymes fall into three categories: the HECT domain class, the RING and RING-like domain class, and those not belonging to either the HECT or the RING class [5]. Known UbcH7-E3 complex structures [5, 8, 37-39] suggest that, although there is no structural similarity between the different classes of E3s domains, they bind UbcH7 in a similar way. Future research may identify which E3-like enzymes interact with Ufc1. Notably, some of the E3-interacting residues identified in UbcH7 or Ubc13 are conserved in Ufc1, including Thr92, Pro130 and Lys131. This may indicate that similar contacts mediate interactions between Ufc1 and its E3 enzyme(s).
In fact, Tyr90 of Ufc1 is strongly conserved in other E2-like enzymes (Fig. 4), and is likewise located in the loop connecting strands β3 to β4. Hence, Tyr90 is in spatial proximity to the putative Ufc1-E3 interface. Intriguingly, it has been shown that the aromatic ring of Tyr90 undergoes unusually slow ring flips about the dihedral angle χ1 with a rate constant of about 0.3 s-1 [40]. Ring flipping rate constants primarily reflect the rates of larger amplitude motional modes which allow the ring to flip [41, 42], and the vast majority of buried aromatic rings in proteins flip much faster that Tyr90. Thus, the slowly-flipping ring of Try90 indicates that the sub-structure, in which the ring is embedded, is remarkably rigid [40]. Furthermore, Pro91 and Pro130 of Ufc1, both of which are in cis-conformation in both NMR and crystal structures of Ufc1, are located close to Tyr90. These two prolyl residues are likewise strongly conserved in UbcH7, Ubc13 and other member of the Ubc SCOP family (Fig. 4), and Pro62 in UbcH7 and Pro63 in Ubc13 (both corresponding to Pro91 in Ufc1) also adopt a cis-conformation. Taken together, the rigidity of this conserved substructure (Fig. 6) potentially involved in E3-binding, might well be of significance for functioning of cellular E1-E2-E3 machines.
Conclusions
The structures of human Ufc1 provides a basis for the understanding of the functioning of Ufc1 at atomic resolution. The novel insights presented here will enable one to properly design and reconcile future research aimed at the exploring the Ufc1-Ufm1 ubiquination pathway. Specifically, the comparison of NMR and X-ray crystal structures reveals that helix α1 of Ufc1 can adopt multiple orientations depending on its environment. Modeling suggests that this plasticity might be required to modulate interactions between Ufc1 and various Ub-like modifiers and/or E3 ligases. An immediately apparent future research goal seems to be the identification of E3-like enzyme that interacts with Ufc1.
Acknowledgements
This work was supported by the Protein Structure Initiative of the National Institutes of Health (U54-GM074958) and the National Science Foundation (MCB 0416899 to T.S.). The authors thank Dr. Goldsmith-Fischman for helpful discussions on the structural biology of Ufc1.
Abbreviations
- ASU
asymmetric unit
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin-protein ligase
- NESG
Northeast Structural Genomics Consortium
- NMR
nuclear magnetic resonance
- NSLF
national synchrotron light source
- PDB
protein data bank
- SAD
single-wavelength anomalous diffraction
- UBC
ubiquitin conjugating enzyme
- UBL
ubiquitin-like (Ubl)
- UFC1
ubiquitin-fold modifier conjugating enzyme 1
- UFM1
ubiquitin-fold modifier 1
Contributor Information
Gaohua Liu, Department of Chemistry, Northeast Structural Genomics Consortium, The State University of New York at Buffalo, Buffalo, NY 14260.
Farhad Forouhar, Department of Biological Sciences, Northeast Structural Genomics Consortium, Columbia University, New York, NY 10027.
Alexander Eletsky, Department of Chemistry, Northeast Structural Genomics Consortium, The State University of New York at Buffalo, Buffalo, NY 14260.
Hanudatta S. Atreya, Department of Chemistry, Northeast Structural Genomics Consortium, The State University of New York at Buffalo, Buffalo, NY 14260
James M. Aramini, The Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, Northeast Structural Genomics Consortium, Rutgers University, Piscataway, NJ 08854
Rong Xiao, The Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, Northeast Structural Genomics Consortium, Rutgers University, Piscataway, NJ 08854.
Yuanpeng J. Huang, The Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, Northeast Structural Genomics Consortium, Rutgers University, Piscataway, NJ 08854
Mariam Abashidze, Department of Biological Sciences, Northeast Structural Genomics Consortium, Columbia University, New York, NY 10027.
Jayaraman Seetharaman, Department of Biological Sciences, Northeast Structural Genomics Consortium, Columbia University, New York, NY 10027.
Jinfeng Liu, Department of Biochemistry and Molecular Biophysics, Northeast Structural Genomics Consortium, Columbia University, New York, NY 10032.
Burkhard Rost, Department of Biochemistry and Molecular Biophysics, Northeast Structural Genomics Consortium, Columbia University, New York, NY 10032.
Thomas Acton, The Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, Northeast Structural Genomics Consortium, Rutgers University, Piscataway, NJ 08854.
Gaetano T. Montelione, The Center for Advanced Biotechnology and Medicine, Department of Molecular Biology and Biochemistry, Northeast Structural Genomics Consortium, Rutgers University, Piscataway, NJ 08854
John F. Hunt, Department of Biological Sciences, Northeast Structural Genomics Consortium, Columbia University, New York, NY 10027
Thomas Szyperski, Department of Chemistry, Northeast Structural Genomics Consortium, The State University of New York at Buffalo, Buffalo, NY 14260.
References
- 1.Finley D, Chau V. Annu. Rev. Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- 2.VanDemark AP, Hill CP. Curr. Opin. Struct. Biol. 2002;12:822–830. doi: 10.1016/s0959-440x(02)00389-5. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Eddins MJ. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Kerscher O, Felberbaum R, Hochstrasser M. Annu Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 5.Dye BT, Schulman BA. Annu. Rev. Biophys. Biomol. Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 6.Hershko A, Ciechanover A. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 7.Huang TT, D'Andrea AD. Nat. Rev. Mol. Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 9.VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. Cell. 2001;105:711–720. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 10.Cort JR, Chiang YW, Zheng DY, Montelione GT, Kennedy MA. Proteins. 2002;48:733–736. doi: 10.1002/prot.10197. [DOI] [PubMed] [Google Scholar]
- 11.Komatsu M, Chiba T, Tatsumi K, Iemura S, Tanida I, Okazaki N, Ueno T, Kominami E, Natsume T, Tanaka K. EMBO J. 2004;23:1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu GH, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, Acton TA, Arrowsmith CH, Montelione GT, Szyperski T. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang YW, Climent T, Cooper B, Denissova NG, Douglas SM, Everett JK, Ho CK, Macapagal D, Rajan PK, Shastry R, Shih LY, Swapna GVT, Wilson M, Wu M, Gerstein M, Inouye M, Hunt JF, Montelione GT. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 15.Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. J. Biomol. NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- 16.Liu GH, Aramini J, Atreya HS, Eletsky A, Xiao R, Acton T, Ma LC, Montelione GT, Szyperski T. J. Biomol. NMR. 2005;32:261–261. doi: 10.1007/s10858-005-7941-9. [DOI] [PubMed] [Google Scholar]
- 17.Guntert P, Mumenthaler C, Wuthrich K. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann T, Guntert P, Wuthrich K. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickson WA. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Macromol. Crystallogr. A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Weeks CM, Miller R. J. of Appl. Crystallogr. 1999;32:120–124. [Google Scholar]
- 22.Terwilliger TC. Acta Crystallogr. D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McRee DE. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Kleywegt GJ. Acta Crystallogr. Section D-Biol. Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt GJ, Brunger AT. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt GJ, Jones TA. Acta Crystallogr. Section D-Biol. Crystallogr. 1996;52:826–828. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima T, Tatsumi K, Ozaki Y, Kawakami T, Suzuki A, Ogasahara K, Komatsu M, Kominami E, Tanaka K, Yamane T. Biochem. and Biophys. Res. Commun. 2007;362:1079–1084. doi: 10.1016/j.bbrc.2007.08.129. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Sander C. Nucleic Acids Res. 1996;24:206–209. doi: 10.1093/nar/24.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murzin AG, Brenner SE, Hubbard T, Chothia C. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 31.Gille C, Frommel C. Bioinform. 2001;17:377–378. doi: 10.1093/bioinformatics/17.4.377. [DOI] [PubMed] [Google Scholar]
- 32.Miura T, Klaus W, Gsell B, Miyamoto C, Senn H. J. Mol. Biol. 1999;290:213–228. doi: 10.1006/jmbi.1999.2859. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Struct. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 34.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Mol. Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA. J. Biol. Chem. 2002;277:47938–47945. doi: 10.1074/jbc.M207442200. [DOI] [PubMed] [Google Scholar]
- 36.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang L, Kinnucan E, Wang GL, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 38.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang MH, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Eletsky A, Atreya HS, Liu GH, Szyperski T. J. Am. Chem. Soc. 2005;127:14578–14579. doi: 10.1021/ja054895x. [DOI] [PubMed] [Google Scholar]
- 41.Wagner G. Q. Rev. Biophys. 1983;16:1–57. doi: 10.1017/s0033583500004911. [DOI] [PubMed] [Google Scholar]
- 42.Skalicky JJ, Mills JL, Sharma S, Szyperski T. J. Am. Chem. Soc. 2001;123:388–397. doi: 10.1021/ja003220l. [DOI] [PubMed] [Google Scholar]
- 43.Koradi R, Billeter M, Wuthrich K. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 44.Kraulis PJ. J. Appl.Crystallogr. 1991;24:946–950. [Google Scholar]
- 45.Merritt EA, Bacon DJ. Macromol. Crystallogr. B. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 46.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. J. Appl.Crystallogr. 1993;26:283–291. [Google Scholar]
- 47.Word JM, Bateman RC, Presley BK, Lovell SC, Richardson DC. Protein Sci. 2000;9:2251–2259. doi: 10.1110/ps.9.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang YJ, Powers R, Montelione GT. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]