Abstract
Recent studies identified MYH9 as a major susceptibility gene for common forms of non-diabetic end-stage kidney disease (ESKD). A set of African ancestry DNA sequence variants comprising the E-1 haplotype, was significantly associated with ESKD. In order to determine whether African ancestry variants are also associated with disease susceptibility in admixed populations with differing genomic backgrounds, we genotyped a total of 1425 African and Hispanic American subjects comprising dialysis patients with diabetic and non-diabetic ESKD and controls, using 42 single nucleotide polymorphisms (SNPs) within the MYH9 gene and 40 genome-wide and 38 chromosome 22 ancestry informative markers. Following ancestry correction, logistic regression demonstrated that three of the E-1 SNPs are also associated with non-diabetic ESKD in the new sample sets of both African and Hispanic Americans, with a stronger association in Hispanic Americans. We also identified MYH9 SNPs that are even more powerfully associated with the disease phenotype than the E-1 SNPs. These newly associated SNPs, could be divided into those comprising a haplotype termed S-1 whose association was significant under a recessive or additive inheritance mode (rs5750248, OR 4.21, P < 0.01, Hispanic Americans, recessive), and those comprising a haplotype termed F-1 whose association was significant under a dominant or additive inheritance mode (rs11912763, OR 4.59, P < 0.01, Hispanic Americans, dominant). These findings strengthen the contention that a sequence variant of MYH9, common in populations with varying degrees of African ancestry admixture, and in strong linkage disequilibrium with the associated SNPs and haplotypes reported herein, strongly predisposes to non-diabetic ESKD.
INTRODUCTION
Epidemiologic data from the USA consistently demonstrate a significantly greater risk for end-stage kidney disease (ESKD) among African Americans (incidence rate of 1010 per million population) compared with European Americans (279 per million population) (1). This discrepancy is evident for many major etiologies of ESKD, including sporadic cases of focal segmental glomerulosclerosis (FSGS), non-diabetic ESKD associated with hypertension and HIV-1 associated FSGS (African American risk ratio compared with European Americans of 4-, 5.3- and 18-fold, respectively) (1–3). Importantly, these discrepancies were shown to persist even after socioeconomic differences were considered, leading many to hypothesize that a population-based genetic predisposition, likely related to African ancestry, is responsible for the observed elevated risk for ESKD among African Americans. A similar discrepancy in ESKD incidence rates of lesser magnitude is also well documented for Hispanic Americans (incidence rate of 520 per million population) (1).
Concordant with this epidemiological data and the suggested hypothesis, two recently published studies demonstrated a strong association of genetic variants in the MYH9 gene, located on chromosome 22, encoding the molecular motor protein non-muscle myosin heavy chain IIA, with ESKD in African Americans (4,5). These two studies used mapping by admixture linkage disequilibrium (LD) in African Americans to identify the MYH9 gene, as a single major disease susceptibility locus for ESKD. The association in these studies was strongest with a set of four single nucleotide polymorphisms (SNPs) which defined a risk haplotype, termed the E-1 haplotype (5). In particular, the risk conferred by African ancestry at this genetic locus was only evident for non-diabetic ESKD etiologies, and was strongest for FSGS, human immunodeficiency virus-associated nephropathy (HIVAN) and hypertension associated ESKD. A subsequent study confirmed the strong association of the MYH9 gene with hypertension associated ESKD in African Americans (6). More recently, an association with ESKD among diabetic African Americans was attributed to the undiagnosed subset of non-diabetic glomerulopathy, well-documented among patients with coincident diabetes and ESKD (7).
The epidemiological association between individuals of African ancestry and higher incidence of ESKD due to idiopathic FSGS or HIVAN has also been reported outside of the USA (3,8–10) and in the case of HIVAN was further suggested to be specifically related to West African Ancestry (11) but has not been studied at the genetic level in these populations. Recently, Pattaro et al. (12) replicated the genetic association of serum creatinine levels with the MYH9 gene polymorphisms in European non-diabetic individuals, indicating that this gene may influence kidney function in non-Africans as well. Interestingly, two of the SNPs reported by Kopp et al. (5) to comprise part of the E-1 risk haplotype observed among non-diabetic African Americans, were also tested by Pattaro et al. (12), but were not associated with elevated creatinine levels among non-diabetic ESKD Europeans. Another recent study showed an association of the E-1 haplotype with albuminuria levels in hypertensive African Americans (13). Cumulatively, these lines of evidence suggest a marked effect of MYH9 polymorphisms on kidney function or disease susceptibility. However, the question as to whether the same protective or risk polymorphisms similarly affect populations of various distinct geographic origins remains open. In this regard, the potential to study MYH9 risk polymorphisms reported among African Americans in an additional population known to have both African ancestry and a higher incidence of ESKD is of particular interest, and might lend further insight into our understanding of the role of genetic patterns in ESKD vulnerability, particularly with respect to the role of African ancestry risk variants at the MYH9 locus.
To this end, the population of Hispanic Americans presents an important opportunity. It is well established that the various Hispanic American communities have varying degrees of admixture of Native American, African and European ancestries (14,15), and experience a higher risk for ESKD as noted above. Thus, for example it can be postulated that a major component of the increased susceptibility to ESKD among Hispanic Americans as compared with European Americans may be attributed to the African ancestral genetic component per se among Hispanic Americans, and that conversely the reduced risk in Hispanic Americans compared with African Americans can be attributed to a lower level of African genetic ancestry component in the former compared with the latter. Consequently, we have studied the association between MYH9 polymorphisms and ESKD in a study cohort of 531 Hispanic and 894 African Americans ESKD cases and controls. First, we characterized the global differences in the proportions of African ancestry between the two population samples using a set of 40 ancestry informative markers (AIMs) and highlighted the local differences in the proportions of African ancestry using 38 additional SNPs on chromosome 22. Second, following correction for global and local ancestries we found that Hispanic Americans share a higher frequency compared with Europeans of three of the four most closely spaced E-1 risk haplotype SNPs located together on intron 23 (rs4821480, rs2032487, rs4821481) previously described among African Americans. Third, to allow a finer mapping for association within the MYH9 gene, we have extended the genotyping to include 42 SNPs and studied them in the combined dataset of African and Hispanic Americans, yielding novel polymorphisms aggregating in two haplotypes, termed S-1 and F-1. The S-1 polymorphisms show an even stronger association with ESKD susceptibility compared with previously reported polymorphisms in both recessive and additive inheritance modes, although association for the F-1 polymorphisms is evident in the current study in dominant and additive inheritance modes (4–6). We conclude that the common susceptibility to ESKD described among African and Hispanic Americans relates to their shared partial African ancestry.
RESULTS
Clinical characteristics
Following the inclusion and exclusion criteria outlined in ‘Materials and Methods’, a total of 1425 affected and control samples were available for analysis. Clinical characteristics of the 894 and 531 African and Hispanic American samples are presented in Table 1. The average age of ESKD patients was significantly lower (61.7 ± 13.5) when compared with the controls (77.3 ± 10.8; P-value <10−15, both Kolmogorov–Smirnov test and t-test). The average age among ESKD patients from the African American group (62.1 ± 13.6) was not significantly different than that of the Hispanic American group (60.3 ± 13.0; Table 1). There was also no significant difference observed between the two groups in time from initiation of renal replacement therapy, although the gender distribution varied slightly between the African American group (47% female) and the Hispanic American group (38% female, P-value 0.023 for a χ2 test of the difference between the two populations). Of the 977 ESKD cases, the recorded etiology was diabetic nephropathy in 541 (55.4%). The 436 non-diabetic ESKD cases, comprised 366 (83.9%) hypertension associated ESKD, 60 (13.8%) HIVAN and 10 (2.3%) idiopathic FSGS cases. In 7.1% of the hypertension associated ESKD, a diagnosis of hypertensive nephrosclerosis was available based on kidney biopsy and 21.7% of the HIVAN cases were also diabetic. Among the 448 controls, 168 (37.5%) were diabetic, 363 (81%) were hypertensive and 149 (33.3%) were both diabetic and hypertensive. In 60 of the 977 ESKD cases, pathological diagnosis based on kidney biopsy was available, and was defined as non-specific nephrosclerosis in 40, FSGS in 10, HIVAN in 5 and as hypertensive nephrosclerosis in the remaining 5.
Table 1.
Descriptive parameters of samples included in the analysis
| African Americans |
Hispanic Americans |
|||||
|---|---|---|---|---|---|---|
| Controls | Non-diabetic ESKD cases | Diabetic ESKD Cases | Controls | Non-diabetic ESKD cases | Diabetic ESKD Cases | |
| Sample size | 140 | 346 | 408 | 308 | 89 | 134 |
| Age (average ± SD) | 79.6 ± 9.5 | 58.7 ± 14.6 | 64.9 ± 12.1 | 76.3 ± 11.2 | 54.9 ± 13.0 | 63.8 ± 11.7 |
| Gender, Females (%) | 62 | 41 | 52 | 53 | 38 | 38 |
| BMI (average ± SD) | 27.2 ± 6.1 | 26.1 ± 6.3 | 28.5 ± 6.7 | 26.7 ± 5.6 | 25.6 ± 5.5 | 26.6 ± 6.1 |
| Hypertension (HTN) (%) | 81 | 100 | 99 | 81 | 98 | 97 |
| Age at dialysis onset (average ± SD) | – | 52.2 ± 15.1 | 59.5 ± 12.2 | – | 48.1 ± 13.8 | 58.1 ± 11.9 |
Global and local ancestry estimates and association with disease status
The reported cohort comprised 894 and 531 African and Hispanic Americans, respectively. It is important to note that no a priori criteria to identify African or Hispanic American ancestry were set and the sole criterion was the participant's self identification as belonging to one of these two groups.
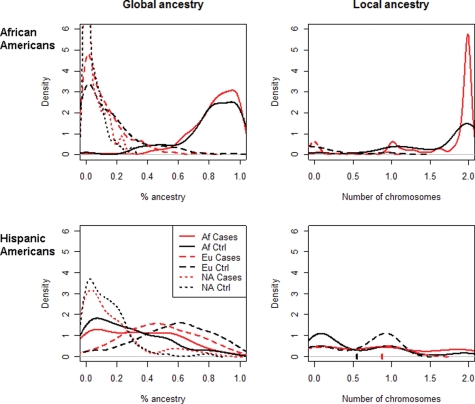
Density graphs of African ancestry in African and Hispanic Americans (cases versus controls) are presented in Figure 1, and show sharp differences between the two population groups. Among self-identified African Americans the global African, European and Native American genetic ancestries (mean ± SD) were 83.4 ± 16.3, 12.2 ± 14.5 and 4.3 ± 6.5% respectively, although in Hispanic Americans as defined and collected in the current study, the proportions were 31.2 ± 24.3, 54.3 ± 23.5 and 14.4 ± 16.1%, respectively. The African American group has a significantly larger African component compared with the Hispanic American group (P-value <10−15 for both Kolmogorov–Smirnov and t-test). As previously reported on the basis of analysis of uniparental genomic regions (16), we find a very low percent of Native American ancestry in self-identified African Americans. Interestingly, the Hispanic American group also showed a relatively small Native American component in our analysis, indicating that the ancestry of the Hispanic American subjects in the current New York City based sample set, is comprised of mostly a European–African admixture. Therefore, it is evident that in both populations admixture is mostly of African and European ancestry, with a significantly larger European component in the Hispanic population. Importantly, a comparable proportion of global African ancestry was found in controls and non-diabetic ESKD cases for both the African American and Hispanic American cohorts, and global African ancestry estimates showed little association with ESKD status in either the Hispanic or African American populations (both P-values >0.2, Kolmogorov–Smirnov test). No significant difference was observed in the percent of Native American ancestry in non-diabetic Hispanic American cases and controls (15.4 ± 18 and 13.9 ± 14.4%, respectively).
Figure 1.
Ancestry distribution Graphs. Density of ancestry proportions among African and Hispanic Americans as discerned from a panel of 40 AIMs for estimating global ancestry (left column) and 38 chromosome 22 SNPs for estimating local ancestry (right column), separately in the African American cohort (top row) and the Hispanic American cohort (bottom row). In the left graphs the ancestry is shown as a percentage (on the x-axis), although in the right graphs the local MYH9 ancestry is shown as the estimated number of chromosomes (0–2) on the x-axis. The average ancestry reported in the text is the mean of the distribution described by the corresponding curve. For example, the significant difference in average local African ancestry in the Hispanic American cohort mentioned in the text (0.87 for non-diabetic ESKD cases, 0.54 for controls) is the difference between the red and black checkmarks on the x-axis in the bottom right plot. ‘Af’ designates African ancestry, ‘Eu’ designates European ancestry and ‘NA’ designates Native American ancestry. Red lines represent non-diabetic ESKD cases, and black lines represent controls.
Local ancestry estimates were calculated using the ANCESTRYMAP program, using a total of 38 widely spread chromosome 22 SNPs. It should be noted that ANCESTRYMAP does not estimate a Native American component, as it is limited to inference on two ancestral populations. The inferred number of African chromosomes in MYH9 was 0.87 ± 0.72 for Hispanic cases compared with 0.54 ± 0.59 for controls, and 1.76 ± 0.42 for African cases compared with 1.65 ± 0.53 in controls. In contrast to the global ancestry estimates noted above, these local ancestry estimates at the MYH9 locus, did show a significant association with non-diabetic ESKD status in the Hispanic American population (P = 0.0002 for Welch's two-sample t-test, P = 0.0009 for Kolmogorov–Smirnov test) and marginally significant association in the African American population (P = 0.02 for Welch's two-sample t-test, P = 0.06 for Kolmogorov–Smirnov test).
Confirmation of the ESKD E-1 allele risk in Hispanic Americans
To examine the E-1 haplotype association with ESKD in Hispanic Americans we have restricted our analysis to the 14 previously reported MYH9 SNPs (5,6). We considered these 14 SNPs in a recessive risk inheritance model, corrected for local and global ancestry, and applied a false discovery rate (FDR) multiple comparison correction to their P-values (17). Table 2 shows the results of single SNP association analyses limited to these 14 SNPs and our set of Hispanic patients and controls. We conclude that three of the four E-1 polymorphisms are significantly associated with non-diabetic ESKD among Hispanic Americans. No significant association of these SNPs with diabetic ESKD was found in either Hispanic or African Americans (Supplementary Material, Table S4.6), as also noted in previous studies for African Americans (4,5).
Table 2.
Association of MYH9 SNPs with non-diabetic ESKD in Hispanic Americans
| SNP | Mode | Odds (95% CI) | P | FDR corrected |
|---|---|---|---|---|
| rs7078 | Rec. | 1.58 (0.99–2.53) | 0.11 | 0.1680 |
| rs12107a | Rec. | Undefined | ||
| rs735853b | Rec. | <1 | ||
| rs5756129 | Rec. | 1.53 (1.00–2.35) | 0.10 | 0.1680 |
| rs5756130 | Rec. | 1.91 (1.02–3.56) | 0.09 | 0.1680 |
| rs4821480 | Rec. | 3.70 (1.67–8.20) | 0.01 | 0.0417 |
| rs2032487 | Rec. | 3.55 (1.60–7.89) | 0.01 | 0.0417 |
| rs4821481 | Rec. | 3.66 (1.65–8.11) | 0.01 | 0.0417 |
| rs3752462 | Rec. | 1.11 (0.67–1.84) | 0.74 | 0.9439 |
| rs5756152 | Rec. | 2.23 (0.69–7.25) | 0.26 | 0.3677 |
| rs1557539a | Rec. | Undefined | ||
| rs1005570 | Rec. | 2.76 (1.26–6.05) | 0.03 | 0.1161 |
| rs16996674 | Rec. | 9.56 (1.50–60.91) | 0.04 | 0.1258 |
| rs16996677 | Rec. | 4.29 (1.02–18.07) | 0.10 | 0.1680 |
aSNPs rs12107, rs1557539 did not appear in their risk state from Freedman et al. (6) in our Hispanic non-diabetic ESKD cases and were therefore inestimable in terms of risk.
bSNP rs735853 gave a slight negative estimate in the logistic regression model, implying no added risk and its results are therefore not shown.
Comparing our results to previous analyses in African Americans (4–6), we note that the odds ratios (ORs) we estimate for Hispanic Americans (effect size of the individual SNPs) for non-diabetic ESKD are about 1.5-fold those observed in African American. For example, for the three first E-1 SNPs (rs4821480, rs2032487, rs4821481), Freedman et al. (6), estimated a consistent OR of about 2.4, although our estimates are between 3.56 and 3.69. In contrast, our estimated OR in African Americans for the same SNPs are lower than those reported in Freedman et al. (6). However, these differences are not statistically significant, likely due to the smaller sample size and lower number of non-diabetic ESKD patients in our sample set. Nevertheless, this analysis successfully and importantly replicates a positive association of these previously reported MYH9 polymorphisms with non-diabetic ESKD under a recessive risk inheritance model in a new population sample, with a different African–European ancestry admixture proportion.
A comprehensive association map within the MYH9 gene
Our primary analysis compared all non-diabetic ESKD cases to all controls. Therefore, with the aim of searching for stronger associations with the non-diabetic ESKD phenotype and attempting to narrow regions containing a potential causative polymorphism, we extended the genotyping to a total of 42 SNPs within the MYH9 gene, for the entire sample set of African Americans and Hispanic Americans. Supplementary Material, Table S4.1 summarizes the list of newly genotyped and previously reported SNPs, their physical positions (where exon 1 was designated as the first non-coding exon in the 5′-UTR of MYH9). Supplementary Material, Tables S4.2 and S4.3 present the genotype and the allele frequencies of each SNP in the different groups, and Supplementary Material, Table S4.4 presents the χ2 test for deviation from Hardy–Weinberg proportions of each SNP for each of the population categories. Of the various population categories, based on the demographic history and number of generations of admixture, the African American controls are expected to be in Hardy–Weinberg equilibrium, as has been demonstrated previously (5). Indeed, taking into account multiple test correction, no significant deviation was observed for the African American control group. Omission of the three SNPs in Supplementary Material, Table S4.4 (rs739101, rs1009150, rs1557536) with borderline individual significance for deviation from Hardy–Weinberg equilibrium without correction for multiple testing, would not affect the associations observed (data not shown). Supplementary Material, Table S4.5 presents the ORs, confidence intervals and allele association P-values after adjustment for global and local ancestry for each inheritance mode (recessive, additive and dominant) for the non-diabetic Hispanic American and non-diabetic African American groups and for diabetic Hispanic American, and diabetic African American groups (Supplementary Material, Table S4.6). For each SNP-risk model combination, we also combined the two scores for the African and Hispanic populations using Fisher's χ2 approximation (18), and show this as a combined P-value estimate for the SNP. We then performed a FDR correction for positively correlated tests (17), considering the total number of tests of MYH9 SNPs (42 × 3 = 126). We note that the FDR correction represents a conservative choice due to the positive correlation between the tests. We prefer this conservative approach because it always guarantees a false discovery that is maintained at a level that is no higher than the desired 0.05 level. The SNP-mode combinations for which association testing remained significant following this correction are shown in bold in Supplementary Material, Table S4.5. Using this approach, a total of fifteen SNPs yielded significant associations.
We concentrate our discussion on the 10 most significantly associated SNPs (Table 3), of which five have been previously reported (4–6) as having significant association with ESKD in African Americans (rs4821480, rs2032487, rs4821481, rs16996674, rs16996677), and five are newly reported herein (rs11912763, rs5750248, rs2413396, rs5750250, rs2239784). Table 3 summarizes the association results based on the primary analysis for these 10 SNPs, which yielded any P-values of <0.05 with any combination of risk inheritance model (recessive, dominant or additive) in the non-diabetic groups (African Americans, Hispanic Americans). A secondary analysis comparing ESKD diabetic patients to controls yielded no significant correlations: no P-value for any of the 42 SNPs was <0.01 before multiple comparisons correction and none of these could be considered significant under FDR or any other reasonable multiple comparison correction (Supplementary Material, Table S4.6).
Table 3.
Association of extended MYH9 SNP clusters with non-diabetic ESKD in African and Hispanic Americans
| SNP | Designation | African Americans |
Hispanic Americans |
Meta analysis, P | FDR corrected | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mode | Odds (95% CI) | P | Mode | Odds (95% CI) | P | ||||
| rs11912763 | F-1 | Rec. | 1.96 (0.97,3.99) | 1.180E−01 | Rec. | 4.84 (1.16,20.25) | 6.99E−02 | 4.78E−02 | 1.37E−01 |
| Add. | 1.55 (1.17,2.06) | 1.163E−02 | Add. | 3.71 (2.13,6.44) | 9.61E−05 | 1.64E−05 | 4.37E−04 | ||
| Dom. | 1.67 (1.17,2.39) | 1.721E−02 | Dom. | 4.59 (2.44,8.61) | 6.88E−05 | 1.73E−05 | 4.37E−04 | ||
| rs4821480 | E-1 | Rec. | 2.00 (1.37,2.92) | 2.576E−03 | Rec. | 3.70 (1.67,8.20) | 6.88E−03 | 2.11E−04 | 2.96E−03 |
| Add. | 1.63 (1.23,2.16) | 4.658E−03 | Add. | 1.80 (1.13,2.86) | 3.85E−02 | 1.72E−03 | 1.14E−02 | ||
| Dom. | 1.43 (0.82,2.51) | 2.867E−01 | Dom. | 1.21 (0.69,2.12) | 5.69E−01 | 4.59E−01 | 6.15E−01 | ||
| rs2032487 | E-1 | Rec. | 1.90 (1.30,2.76) | 4.947E−03 | Rec. | 3.55 (1.60,7.89) | 8.94E−03 | 4.88E−04 | 4.73E−03 |
| Add. | 1.57 (1.19,2.09) | 8.453E−03 | Add. | 1.75 (1.10,2.78) | 4.88E−02 | 3.63E−03 | 1.99E−02 | ||
| Dom. | 1.39 (0.79,2.44) | 3.380E−01 | Dom. | 1.19 (0.68,2.08) | 6.09E−01 | 5.31E−01 | 6.83E−01 | ||
| rs4821481 | E-1 | Rec. | 1.91 (1.31,2.79) | 4.475E−03 | Rec. | 3.66 (1.65,8.11) | 7.42E−03 | 3.76E−04 | 3.94E−03 |
| Add. | 1.60 (1.21,2.12) | 5.547E−03 | Add. | 1.74 (1.09,2.77) | 5.23E−02 | 2.65E−03 | 1.52E−02 | ||
| Dom. | 1.51 (0.87,2.60) | 2.177E−01 | Dom. | 1.15 (0.66,2.02) | 6.77E−01 | 4.30E−01 | 5.88E−01 | ||
| rs5750248 | S-1 | Rec. | 2.35 (1.60,3.45) | 2.421E−04 | Rec. | 4.21 (1.83,9.66) | 4.41E−03 | 1.57E−05 | 4.37E−04 |
| Add. | 1.72 (1.33,2.24) | 6.387E−04 | Add. | 1.61 (1.03,2.53) | 8.01E−02 | 5.57E−04 | 5.01E−03 | ||
| Dom. | 1.55 (0.96,2.50) | 1.353E−01 | Dom. | 1.06 (0.62,1.83) | 8.55E−01 | 3.65E−01 | 5.25E−01 | ||
| rs2413396 | S-1 | Rec. | 2.18 (1.50,3.18) | 6.658E−04 | Rec. | 3.19 (1.40,7.29) | 2.07E−02 | 1.68E−04 | 2.64E−03 |
| Add. | 1.59 (1.23,2.06) | 2.697E−03 | Add. | 1.60 (1.03,2.47) | 7.78E−02 | 1.99E−03 | 1.24E−02 | ||
| Dom. | 1.38 (0.86,2.21) | 2.668E−01 | Dom. | 1.22 (0.73,2.05) | 5.27E−01 | 4.16E−01 | 5.76E−01 | ||
| rs5750250 | S-1 | Rec. | 2.53 (1.72,3.72) | 7.890E−05 | Rec. | 3.87 (1.67,8.97) | 8.12E−03 | 9.77E−06 | 4.37E−04 |
| Add. | 1.79 (1.38,2.33) | 2.266E−04 | Add. | 1.50 (0.96,2.35) | 1.39E−01 | 3.58E−04 | 3.94E−03 | ||
| Dom. | 1.59 (1.00,2.54) | 1.031E−01 | Dom. | 1.00 (0.58,1.73) | 9.88E−01 | 3.35E−01 | 5.14E−01 | ||
| rs2239784 | – | Rec. | 1.95 (1.34,2.82) | 3.217E−03 | Rec. | 2.49 (1.26,4.93) | 2.80E−02 | 9.28E−04 | 6.88E−03 |
| Add. | 1.70 (1.32,2.20) | 5.829E−04 | Add. | 1.46 (1.00,2.13) | 1.00E−01 | 6.27E−04 | 5.27E−03 | ||
| Dom. | 2.04 (1.29,3.23) | 1.028E−02 | Dom. | 1.20 (0.72,2.01) | 5.50E−01 | 3.49E−02 | 1.07E−01 | ||
| rs16996674 | F-1 | Rec. | 2.02 (0.99,4.09) | 1.030E−01 | Rec. | 9.56 (1.50,60.91) | 4.49E−02 | 2.95E−02 | 9.78E−02 |
| Add. | 1.53 (1.15,2.03) | 1.416E−02 | Add. | 4.01 (2.29,7.03) | 4.52E−05 | 9.76E−06 | 4.37E−04 | ||
| Dom. | 1.62 (1.13,2.30) | 2.570E−02 | Dom. | 4.31 (2.33,7.97) | 8.95E−05 | 3.22E−05 | 6.76E−04 | ||
| rs16996677 | F-1 | Rec. | 1.82 (0.99,3.35) | 1.043E−01 | Rec. | 4.29 (1.02,18.07) | 9.62E−02 | 5.62E−02 | 1.49E−01 |
| Add. | 1.39 (1.06,1.82) | 4.319E−02 | Add. | 3.30 (1.95,5.58) | 1.86E−04 | 1.02E−04 | 1.84E−03 | ||
| Dom. | 1.43 (1.00,2.02) | 9.641E−02 | Dom. | 3.73 (2.08,6.71) | 2.18E−04 | 2.48E−04 | 3.12E−03 | ||
Boldface indicates statistically significant P values according to the criteria described in the text.
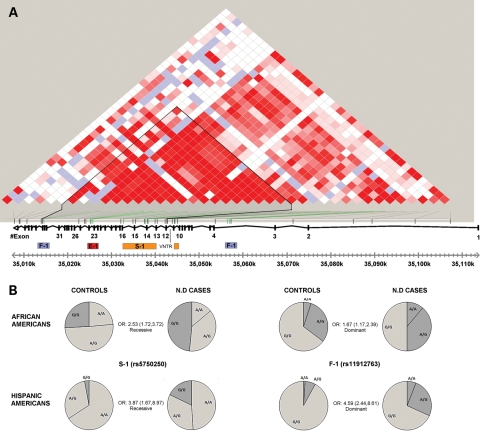
Interestingly, these 10 SNPs can be divided into three clusters and one additional SNP (Fig. 2 and Table 3). As previously reported for the E-1 haplotype (5), each of the clusters in the current study is comprised of a set of SNPs that are in very high LD and in which each SNP within a cluster carries the identical information as the others regarding disease risk, and therefore are designated as haplotypes. This grouping facilitates inferences narrowing regions of association. The first set contains three of the four previously reported E-1 haplotype SNPs located in intron 23 (rs4821480, rs2032487, rs4821481), all of which are in very high LD (Supplementary Material, Fig. S1). In the current analysis, rs3752462 (intron 13) does not have the same level of LD or carry the same association as the three E-1 SNPs noted above, and therefore the designation ‘E-1 haplotype’ in the current study refers to rs4821480, rs2032487, rs4821481. A second cluster comprised three SNPs, rs5750248, rs2413396, rs5750250, also in very high LD (r2 = 0.77, D′ = 0.93 for the latter two in Yoruba phased haplotypes from HapMap), and are designated herein as the S-1 haplotype. This haplotype is also in relatively high LD with the first set (r2 = 0.58, D′ = 0.89 between rs2413396 and rs4821481 in HapMap (Yoruba), and see Supplementary Material, Fig. S1). These two haplotypes show a similar pattern of association with non-diabetic ESKD, in both recessive and additive modes of inheritance.
Figure 2.
MYH9 LD plot and ESKD associated SNPs. (A) Color scheme (D′/LOD) of the 42 MYH9 SNPs that were genotyped in this study based on the African American control samples (n = 140) was generated using the program HaploView (35). Bright red squares present SNPs with linkage LOD ≥ 2 and D′ = 1. The plot overlaps the actual locations of the SNPs tested herein and the physical map of the MYH9 gene. The SNPs demonstrating the highest peaks for association and their clustering into the haplotypes S-1 (rs5750248, rs2413396, rs5750250), E-1 (rs4821480, rs2032487, rs4821481) and F-1 (rs11912763, rs16996674, rs16996677) and the solo tenth SNP (rs2239784) are marked. A block of reduced recombination, bordered at its 5′ end with a 53 bp variable number tandem repeat in intron 11, determined using the program PHASE 2.1 (36), corresponds to the black triangle outline in the LD plot. (B) Pie charts of genotypes frequencies in non-diabetic ESKD cases versus controls for two SNPs representing the novel haplotypes S-1 (rs5750250) and F-1 (rs11912763) are shown for African and Hispanic Americans. The significant OR value for each SNP of the specified tested model is also indicated.
The third set, which we term the F-1 haplotype, comprises three additional SNPs: rs16996674, rs16996677, rs11912763. The first two of these are in close physical proximity to each other and within intron 3 (631 bp apart) and are located more 5′ within the gene compared with any of the SNPs previously reported to be in association with the disease phenotype (Fig. 2). In contrast, SNP rs11912763 at nucleotide position 35 014 668, located in intron 33, is the most 3′ associated SNP found so far. Of note, rs11912763 has an allele distribution similar to that of rs16996674, and these two SNPs are also in high LD in the HapMap Yoruba samples (r2 = 0.54, D′ = 0.81). This risk haplotype is considerably less frequent compared with the E-1 and S-1 haplotypes. The P-values for risk association for the F-1 SNPs are significant for both additive and dominant inheritance modes, and therefore not consistent with the recessive inheritance mode, for which the other disease risk SNPs are significantly associated. Moreover, the strength of the association and the OR related to this set, suggests a higher degree of LD with the causative mutation. In addition to these three haplotypes, one additional SNP within intron 10, rs2239784, has a statistically significant association with non-diabetic ESKD status. It is located at nucleotide position 35 044 581, on the 5′ side of the E-1 and S-1 SNPs, but still 3′ to the rs16996674 and rs16996677 F-1 associated SNPs. It is not in high LD with any of the other SNPs (r2 < 0.3, D′ < 0.65 with the five other SNPs in HapMap reported Yoruba phased haplotypes). It should be noted that the set of SNPs previously designated as L-1 in Freedman et al. (6) (rs7078, rs12107, rs735853 and rs5756129) are in LD with the E-1, S-1 and F-1 SNPs, but were not found to be significantly associated with the disease phenotype in the current study. We interpret this as indicating a weaker association of the SNPs comprising the L-1 haplotype, which therefore do not show up as significant in our smaller sample sets.
Although the frequency of the African ancestry risk variants is lower among Hispanic Americans compared with African Americans, it is of interest (Table 3 and Supplementary Material, Table S4.5), that there is a higher OR for the risk genotypes in the Hispanic compared with African American cohorts. For example, the estimated odds attributable to having one copy of SNP rs11912763 is 1.67 (67% higher risk) in African Americans, compared with 4.59 in Hispanic Americans. These differences persist across multiple associated SNPs, as evidenced by non-overlapping confidence intervals and significant P-values between Hispanic and African Americans for all three F-1 SNPs in Table 3. A formal t-test gives non-corrected two-sided P-values of between 0.011 and 0.025 for the six tests for F-1 SNPs in additive and dominant modes, which lose significance when a conservative multiple comparisons correction is applied.
Identification of additional sets of risk SNPs
To more clearly understand the nature of association of identified risk SNPs and the disease phenotype we performed two additional analyses. First, for each highly associated SNP, we examined whether it is sufficient to fully explain the correlation between global and local African ancestry and the prevalence of ESKD (4). Second, for each highly associated SNP, we examined which additional highly associated SNPs contain ‘independent’ information about ESKD status, as expressed by their statistically significant additive effect when included in the logistic regression. Each one of the highly associated SNPs in our analysis rendered both local and global ancestry insignificant in determining ESKD status, when considered together in a logistic regression model. Thus, we can conclude that at least within our limited dataset we are capturing the major share of the information about ESKD susceptibility in the MYH9 gene by including any of the highly correlated SNPs in the model. This is consistent with the findings of Kao et al. (4), who found that each of the three E-1 SNPs was sufficient to explain fully the effect of local ancestry on ESKD risk.
To examine the excess information in pairs of SNPs, we chose one representative SNP from each of the sets E-1 (rs4821481), S-1 (rs5750250), F-1 (rs11912763), and for each such pair, modeled association using both SNPs according to the risk inheritance mode which yielded the most significant association (recessive for E-1 and S-1, dominant for F-1). As expected, the E-1/S-1 SNPs mostly compensate for each others’ effects, and so are less significant in such a model when considered together. In a meta-analysis combining both African American and Hispanic American results of the model comprising both E-1 and S-1, the P-value for E-1 was 0.53 and for S-1 0.022. Thus, S-1 may contain a little information beyond E-1, but this conclusion is very fragile in the face of possible multiple comparison corrections. On the other hand, when combining one SNP from E-1 or S-1 with the F-1 SNP, both are clearly significant in the resulting model, indicating that the E-1/S-1 group and the F-1 group carry distinctly different information about the risk for non-diabetic ESKD. In a similar meta-analysis of the model comprising both S-1 and F-1, the corresponding P-values were 0.00081 and 0.00095, respectively.
DISCUSSION
ESKD is a major and growing health concern worldwide, especially in the developing regions of the world, resulting in reduced duration and quality of life, and necessitating ongoing renal replacement therapy to sustain life. Population discrepancies in the incidence and prevalence rates of ESKD within the USA and other constituencies have motivated the successful search for and the recent identification of genetic susceptibility factors, which provide important insights in developing disease preventive measures. Specifically, African ancestry sequence variants in the MYH9 gene have been shown to confer major disease risk susceptibility for a number of non-diabetic etiologies of ESKD (4–6). More recently, association between MYH9 and albuminuria was detected in hypertensive African Americans (13), and association between other polymorphisms in the same gene with serum creatinine concentrations has been reported in Europeans (12). Additionally, numerous rare exonic variants have already been reported to cause ESKD in the context of a set of syndromes (Giant Platelet Syndromes) with autosomal dominant inheritance (19). These observations suggest a fundamental role of variants in the MYH9 gene in the pathogenesis of non-diabetic ESKD. The association of a set of common polymorphic variants, whose frequency differs among ancestral populations with more common forms of non-diabetic ESKD, points to the existence of one or more causative variants with which these associated SNPs are in LD. This raises the question of whether multiple common causative disease susceptibility variants within the same gene are responsible for the same ESKD phenotype, and whether they interact with each other. Conversely, it is important to establish whether the same causative variant confers risk due to shared ancestry among different populations.
The Hispanic American population demonstrates an intermediary pattern of ESKD susceptibility when compared with African and Europeans Americans (1), but the pathogenetic basis for this observation remains unknown. The previously reported partial African ancestry among Hispanic Americans has led us to hypothesize that the same ESKD risk polymorphisms reported among African Americans might also explain the elevated susceptibility to kidney disease observed among Hispanic Americans. Our results show that ∼30% of the global genetic ancestry observed among the Hispanic Americans in this study can be attributed to an African component confirming their partially shared ancestry with African Americans. Interestingly, local African ancestry measures for chromosome 22, where the MYH9 gene is located, were highly associated with ESKD in Hispanic Americans, whereas only marginally associated in African Americans. This observation suggests that analysis of the Hispanic American population according to local chromosome 22 ancestry estimators might better identify the subpopulations included under the self-identified designation of Hispanic American, at higher risk for ESKD. A further proof of the important role that the African ancestry component contributes to the higher susceptibility to ESKD observed among Hispanic Americans, is evident from the positive confirmation of the association between the reported E-1 risk haplotype and non-diabetic ESKD among Hispanic Americans after corrections for both global and local ancestries. Cumulatively, our results show that, similarly to the case of African Americans, the higher proportions of non-diabetic ESKD observed among Hispanic Americans in the current study are MYH9-associated. Therefore, the overall higher incidence rate of ESKD observed among Hispanic Americans when compared with European Americans is likely to be attributable at least in part, to the same African population common causative variant with which the E-1 risk polymorphisms are in strong LD. Likewise, the overall lower incidence of ESKD cases observed among Hispanic Americans when compared with African Americans is explained by the overall lower African ancestry component, and hence lower frequencies of the African MYH9 risk alleles among Hispanic Americans. Moreover, our findings and analyses are entirely consistent with the existence of an African ancestry causative variant with which the highly associated SNPs are in LD, and which is responsible for the excess disease phenotype risk. This inference is greatly strengthened by the finding that the very same African ancestry SNPs are the most highly associated with the corresponding relevant ESKD phenotypes in two different populations with varying genomic backgrounds and degrees of African ancestry admixture. It is likely that this African ancestry causative variant shared between the populations studied, is necessary but not sufficient for pathogenesis of the relevant ESKD phenotypes of interest and that epistatic interactions with other genomic regions and/or gene-by-environment interactions are additionally necessary for penetrance of the disease phenotype.
In this regard, the observation of higher ORs for risk genotypes in Hispanic Americans compared with African Americans warrants consideration. Since these associations are obtained using logistic regression—this is not an expected effect of lower levels of African ancestry and lower disease prevalence among the Hispanic Americans. One possible explanation that motivates further study is that the African genomic background may contain as yet unidentified or untested ESKD risk variants at other loci but with lesser effect, and which are less frequent in the European genomic background. Thus, in the Hispanic American ESKD cases the lower frequency of observed African-risk variants of MYH9 may be acting on a mostly ‘protective’ European genomic background, in contrast to African Americans where the African-risk variants of MYH9 are acting on the background of a higher genomic risk for ESKD. Therefore, the relative effect of risk MYH9 variant in Hispanic Americans would be more prominent than in African Americans. Differences in ESKD risk allele frequencies at multiple loci within a population could arise through a variety of mechanisms, including selective effects. This would motivate the search for additional proposed ESKD risk loci (20–25).
The confirmation of the association between the previously reported E-1 haplotype and non-diabetic ESKD among Hispanic Americans lends further support to the role of the MYH9 in the phenotype of non-diabetic ESKD, but does not facilitate progress towards identifying a risk causing mutation. Additional genotyping within the MYH9 gene to a total of 42 SNPs within the MYH9 gene, does however both strengthen the association of the gene with the disease phenotype, and also provides potential insights pertaining to the region containing the causative mutation and its mode of inheritance. In this regard, our results yielded a total of 15 significantly associated SNPs, of which the 10 most significant clustered into three sets (Fig. 2). These results clearly show that the S-1 SNPs (rs5750250, rs2413396, rs5750248) and F-1 SNPs (rs16996674, rs16996677, rs11912763), have a stronger association with non-diabetic ESKD when compared with the SNPs reported to comprise the E-1 haplotype. Of interest, these SNPs are found on both the 5′ and 3′ sides of the E-1 haplotype SNPs, spanning a region of 42 kb. The S-1 SNPs which are the most highly associated, are clustered in a smaller region of only 5.5 kb between intron 13 to intron 15 and like the E-1 SNPs have the strongest association in the recessive inheritance mode. The F-1 SNPs show a significant association with disease risk under a dominant or additive inheritance mode, even though they are located furthest 5′ and 3′ in relation to the SNPs comprising the E-1 and S-1 risk haplotypes, which demonstrate a significant disease risk association in the recessive or additive inheritance mode. This finding may suggest that even one copy of the causative mutation may already confer some of the increased risk for this disease phenotype. Stronger association of the E-1 and S-1 SNPs in the recessive mode may reflect the fact that in the presence of two risk alleles, the likelihood of recombination between the causative mutation variant and the E-1 or S-1 risk variant occurring on both parental chromosomes is greatly reduced. Thus, it will be difficult to draw inferences about the physical location of the causative variant based only on the strength of association in a recessive inheritance mode, without a more complete phylogenetic resolution or functional studies of candidate causative mutations at the molecular and mechanistic levels.
Some potential limitations of the study relate primarily to classification of subjects in the various disease status categories. Thus, it is as yet unclear whether the MYH9 associations relate primarily to FSGS, in which case inclusion of hypertensive ESKD without a biopsy diagnosis of FSGS, would weaken any inferences of association with FSGS per se. Conversely, inclusion of subjects with undiagnosed FSGS among those with a medical record diagnosis of diabetic nephropathy, or inclusion of early chronic kidney disease patients among the controls would certainly not mitigate, and also only weaken any inferences of positive associations. Clearly, the greater accuracy regarding the state and underlying etiology of chronic kidney disease would only render the estimates of strengths of association more secure and accurate. Unfortunately, clinical practice, in which kidney biopsy is often not indicated or carried out prior to initiation of renal replacement therapy, leaves the etiology as presumed, solely on the basis of clinical background.
The potential clinical relevance of the results, presented previously and herein, with respect to clinical and public health warrants some consideration. Thus questions regarding optimization in kidney transplant donation can be considered, and thresholds for instituting antiretroviral therapy in HIV-infected individuals might be influenced by consideration of MYH9 genotypic risk for HIVAN. The current considerations for a physician counseling a patient with normal kidney function and a genetic risk profile for chronic kidney disease at the MYH9 gene are complex, since the actual disease-risk causative variant is as yet unknown, and as only a minority of individuals bearing the MYH9 risk haplotypes will eventually develop ESKD likely reflecting the contribution of epistatic genomic as well as environmental factors (26,27). Of note, the current results comparing MYH9 risk allele effects in African and Hispanic Americans highlight the contribution of differing population genomic backgrounds in MYH9 associated ESKD risk. Nevertheless, it seems reasonable for a physician to include the status of the MYH9 risk haplotype as one of the many considerations used in ESKD risk assessment.
In summary, our results demonstrate that the same MYH9 risk haplotypes are associated with non-diabetic ESKD in both African and Hispanic Americans as a result of their partially shared African ancestry. The finding of this association within the Hispanic American population, in which the overall global African ancestry is significantly lower when compared with African Americans, predicts a functional impact of the MYH9 local African ancestry to ESKD susceptibility, and adds very strong support for the existence and hence motivates the search for a causative disease phenotype risk variant. We have further identified additional SNPs in the MYH9 gene, grouped in clusters designated as S-1 and F-1 that show the strongest associations reported to date with ESKD in both African and Hispanic Americans. Moreover, the finding of significant independent associations in both dominant and additive inheritance modes for the F-1 SNPs, and recessive and additive inheritance modes for the S-1 SNPs, means that the search for the causative mutation should take into account the possibility that a single copy may confer a functional effect. Taken together, these findings strongly motivate the search for a functional African ancestry causative MYH9 variant, which will be facilitated by a deeper understanding of the phylogenetic history of the locus.
MATERIALS AND METHODS
Subjects
The study included a total of 1425 individuals collected at dialysis clinics (ESKD) and ambulatory clinics and nursing centers (controls) affiliated with the Cabrini Medical Center, New York City, USA, and approved by its Institutional Review Board for both ESKD patients and controls. All study participants provided written informed consent. It should be noted that none of the samples reported in the current study overlap those reported in the companion study by Nelson et al. (28).
Participants self reported their ancestry to be African or Hispanic American. A total of 894 African Americans were recruited, of whom 754 were ESKD patients and 140 were controls. Likewise, a total of 531 Hispanic Americans were enrolled, of whom 223 and 308 subjects were ESKD and control subjects, respectively (Table 1). During recruitment, the following information was collected for each of the potential participants by direct questioning and by reviewing the participant's medical records: height, weight, age, gender, place of birth, parental place of birth, self-reported ethnicity, kidney function status (ESKD or plasma creatinine concentration), kidney biopsy diagnosis when available, ESKD duration, and known high blood pressure, diabetes or HIV diagnosis preceding initiation of renal replacement therapy.
As noted from the subject and parental place of birth information, the self-identified Hispanic American subjects in the current study, were mostly of Puerto Rican, Dominican and other Caribbean parental ancestry, with few subjects of Mexican-American affiliation, consistent with the recruitment location in New York City. This was also evident in the ancestry marker panel analysis, indicating a relative paucity of Native American ancestry markers. Thus, use of the term Hispanic American in the context of the current study refers to Hispanic Americans mostly of Caribbean origin, who inherit predominant European and African ancestry. This is in contrast to the use of the term Hispanic American in other studies, which often include many Mexican Americans who inherit predominant European and Native American ancestry. Since the current study refers to European and African ancestry, the Hispanic American sub-groups in our sample set are of particular interest for the current analysis.
Hypertension associated kidney disease refers to patients who have a medical history of hypertension preceding initiation of renal replacement therapy, but who do not have diabetes, HIV infection or other known specific etiologies of kidney disease, and in whom renal biopsy information was usually not available. In a small subset of these cases a renal biopsy diagnosis of ‘hypertensive nephrosclerosis’ was available. Diabetes (type 1 and 2) was defined as self-reported and/or prevalent treatment with insulin and/or oral hypoglycemic agents or diabetes mellitus documented in the medical history. The diagnosis of HIVAN was made in the presence of known HIV infection and was usually not confirmed by a renal biopsy. The diagnosis of idiopathic FSGS was made only in the presence of compatible renal biopsy information. Non-diabetic ESKD cases in our sample set correspond to the union of H-ESKD patients as described in the companion paper (28) with FSGS, HIVAN and ESKD patients, exclusive of the following etiologies: congenital, obstructive, cancer-related kidney diseases or known monogenic forms of kidney disease (e.g. autosomal dominant polycystic kidney disease, Alport syndrome).
Unrelated control subjects were recruited from the general population without known kidney disease at age 55 or greater. Medical history of hypertension or diabetes was also recorded for each of the control individuals. Creatinine concentrations were available in all of the controls, and individuals with creatinine concentration exceeding 1.7 were excluded. We recognize that these controls may have included some individuals with undiagnosed early stages of chronic kidney disease. Inadvertent classification of such individuals as unaffected would not mitigate and only strengthen the inference of any possible positive association of the affected status with genomic SNPs or haplotypes in the overall analysis.
To estimate ancestral population allele counts for global ancestry we included a total of 284 samples available to us from the Coriell collection and as detailed in Supplementary Material, Table S1.
Genotyping
Genomic DNA was obtained from peripheral blood samples using standard phenol-chloroform extraction protocols (29). The KasPar methodology (30) was used to genotype all 119 SNPs reported herein. For global ancestry estimation we used information from HapMap (31) and novel information from genotyping of 40 genomic AIMs in samples from different world populations (Supplementary Material, Tables S1 and S2). Following that, we genotyped the complete set of samples (n = 1425) in order to appropriately evaluate population ancestry for each individual and the potential of population substructure. For local ancestry estimation, a total of 38 additional SNPs restricted to chromosome 22 were selected for genotyping using the recently reported expected mutual information ancestry measure (32) (Supplementary Material, Table S3). To achieve fine mapping within the MYH9 gene, a total of 42 SNPs were genotyped, as shown in Supplementary Material, Table S4.1. These consisted of 14 SNPs originally reported in Kopp et al. (5) plus an additional 28 SNPs identified in the companion manuscript (28), as potentially informative based upon skewed HapMap population frequencies and providing broad coverage across the MYH9 region, especially the region spanned by the previously identified E-1 haplotype (5). Duplicate samples (7.3%) were distributed across plates to assess for consistency between genotype calls. The error rate observed among the duplicated samples was <0.05%. No contaminating DNA signals were observed in water-only controls. Samples with <90% complete call rate were excluded from analysis.
Ancestry estimation
We generated both global and local African ancestry proportion estimates for each of the samples in our data. To estimate global ancestry, we utilized the 40 AIMs shown in Supplementary Material, Table S2 and a maximum likelihood approach assuming a multinomial distribution for the ancestry of each marker, as described in Tang et al. (33). The population proportion information data used for estimating ancestral population proportions was taken from HapMap (31) (for European and African ancestry) and from our novel dataset (Supplementary Material, Table S1) (European, African and Native American ancestry). To estimate local ancestry, we used the program ANCESTRYMAP, using a total of 38 SNPs restricted to chromosome 22 (Supplementary Material, Table S3) and extracted local ancestry estimates for SNP rs2157257, which is located within the MYH9 gene itself. Since recent admixture is expected to generate long haplotype blocks (34), it is not necessary to estimate local ancestry separately for each SNP being investigated, and a single estimate of local ancestry at the MYH9 locus for each sample was used.
Modeling association for MYH9 polymorphisms
As in previous studies (5,6), we used logistic regression to estimate the increase in risk attributable to the allele variant at each of the genetic markers tested in each of our two populations of African Americans and Hispanic Americans, although controlling for other potential stratification effects, in particular local and global African ancestry and self identification as African American or Hispanic American. We did not control for age since one of our criteria for collecting controls was an older age, in particular the age of all controls was >55 years and as a result controls were significantly older than the ESKD patient cohort (Table 1).
For each SNP we considered three possible modes of genetic effect on risk:
Dominant: one risk allele is sufficient to incur all excess risk.
Additive: each additional risk allele makes a measurable contribution to excess risk. More formally, in the framework of logistic regression this model assumes additivity of the log-odds of disease, i.e. a multiplicative effect on the OR.
Recessive: two risk alleles are required to incur any excess risk.
What differentiates the three modes is the manner in which the SNP is used in the logistic regression. For the dominant mode we introduced a binary variable, equal to 1 if at least one risk allele is present and 0 otherwise. For the additive mode, the number of risk alleles (0, 1, 2) is used as a covariate in the logistic regression. For the recessive mode, we again define a binary variable, but this time it is 1 only if two risk alleles are present and 0 otherwise.
To validate the association between MYH9 polymorphisms and the ESKD phenotype we divided our entire dataset to three phenotypic groups: diabetic ESKD patients, non-diabetic ESKD patients and controls (Table 1). Included in the non-diabetic ESKD patients were hypertension associated ESKD, FSGS and HIVAN patients, as defined above. Controls, as defined above, were either diabetic or non-diabetic, but without ESKD at age greater than 55. One small group of patients (13 individuals) whose medical record was positive for both HIVAN and diabetes were included in the group of non-diabetic ESKD HIVAN patients, following confirmation that their inclusion under diabetic ESKD or exclusion from the analysis did not change our results. In total, under the assumption that the main risk mode of MYH9 is for non-diabetic ESKD (4–6), our primary analysis compared all non-diabetic ESKD cases to all controls. We recognize that some patients with a medical record diagnosis of diabetic ESKD, and without a confirmatory biopsy, may actually have a primary non-diabetic glomerulopathy, including some with undiagnosed FSGS (7). Exclusion of these patients from the ‘affected’ cohort would only weaken our ability to uncover a positive association of SNPs and haplotypes in the MYH9 gene with the ‘affected’ disease phenotype, and therefore not mitigate inferences of a positive association.
Analysis of MYH9 SNPs
Previously published analyses of risk SNPs in MYH9 (4–6) have identified a collection of four SNPs (rs4821480, rs2032487, rs4821481, rs3752462) in strong LD and which together were termed the E-1 haplotype, which was the most highly correlated with non-diabetic ESKD occurrence, under a recessive transmission risk model. In our study, we did not detect a significant association with ESKD for the SNP rs3752462, therefore the E-1 haplotype mentioned herein refers only to the three adjacent SNPs located in intron 23 (rs4821480, rs2032487, rs4821481). An additional 10 MYH9 SNPs (rs7078, rs12107, rs735853, rs5756129, rs5756130, rs5756152, rs1557539, rs1005570, rs16996674, rs16996677) were identified as having weaker associations. We chose all 14 of these SNPs for a confirmatory study in our Hispanic American sample set. On the basis of the inferred mode of hereditary risk transmission for positive association in these previous studies, we chose to model them as following a recessive risk model. We then extended the analysis to the full set of MYH9 SNPs (Supplementary Material, Table S4.1) under the inheritance models noted above.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Canadian and American Technion Societies (Eshagian Estate Fund, Veronique Elek Fund, Dr Sidney Kremer Kidney Disease Research Fund); the Israel Science Foundation (890015), and Legacy Heritage Fund; to K.S. The European Research Commission (MIRG-CT-2007-208019); and the Israel Science Foundation (1227/09); to S.R. The National Cancer Institute, National Institutes of Health (HHSN261200800001E); the Intramural Research Program of National Cancer Institute, Center for Cancer Research (the content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government); and the National Institute for Diabetes, Digestive and Kidney Diseases (Project ZO-1 DK043308), to J.B.K.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank the participating subjects and the physicians affiliated with the Cabrini Medical Center and its affiliates for their participation in sample and data acquisition, and Dr. Alan Templeton for his valuable input.
Conflict of Interest statement. None declared.
REFERENCES
- 1.USRDS. Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; 2008. [Google Scholar]
- 2.Kopp J.B., Winkler C. HIV-associated nephropathy in African Americans. Kidney Int. Suppl. 2003;63:S43–S49. doi: 10.1046/j.1523-1755.63.s83.10.x. [DOI] [PubMed] [Google Scholar]
- 3.Kitiyakara C., Kopp J.B., Eggers P. Trends in the epidemiology of focal segmental glomerulosclerosis. Semin. Nephrol. 2003;23:172–182. doi: 10.1053/snep.2003.50025. [DOI] [PubMed] [Google Scholar]
- 4.Kao W.H., Klag M.J., Meoni L.A., Reich D., Berthier-Schaad Y., Li M., Coresh J., Patterson N., Tandon A., Powe N.R., et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kopp J.B., Smith M.W., Nelson G.W., Johnson R.C., Freedman B.I., Bowden D.W., Oleksyk T., McKenzie L.M., Kajiyama H., Ahuja T.S., et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freedman B.I., Hicks P.J., Bostrom M.A., Cunningham M.E., Liu Y., Divers J., Kopp J.B., Winkler C.A., Nelson G.W., Langefeld C.D., et al. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int. 2009;75:736–745. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freedman B.I., Hicks P.J., Bostrom M.A., Comeau M.E., Divers J., Bleyer A.J., Kopp J.B., Winkler C.A., Nelson G.W., Langefeld C.D., et al. Non-muscle myosin heavy chain 9 gene MYH9 associations in African Americans with clinically diagnosed type 2 diabetes mellitus-associated ESRD. Nephrol. Dial. Transplant. 2009;24:3366–3371. doi: 10.1093/ndt/gfp316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anochie I.C., Eke F.U., Okpere A.N. Human immunodeficiency virus-associated nephropathy (HIVAN) in Nigerian children. Pediatr. Nephrol. 2008;23:117–122. doi: 10.1007/s00467-007-0621-0. [DOI] [PubMed] [Google Scholar]
- 9.Han T.M., Naicker S., Ramdial P.K., Assounga A.G. A cross-sectional study of HIV-seropositive patients with varying degrees of proteinuria in South Africa. Kidney Int. 2006;69:2243–2250. doi: 10.1038/sj.ki.5000339. [DOI] [PubMed] [Google Scholar]
- 10.Nochy D., Glotz D., Dosquet P., Pruna A., Guettier C., Weiss L., Hinglais N., Idatte J.M., Mery J.P., Kazatchkine M., et al. Renal disease associated with HIV infection: a multicentric study of 60 patients from Paris hospitals. Nephrol. Dial. Transplant. 1993;8:11–19. doi: 10.1093/oxfordjournals.ndt.a092263. [DOI] [PubMed] [Google Scholar]
- 11.Behar D.M., Shlush L.I., Maor C., Lorber M., Skorecki K. Absence of HIV-associated nephropathy in Ethiopians. Am. J. Kidney Dis. 2006;47:88–94. doi: 10.1053/j.ajkd.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Pattaro C., Aulchenko Y.S., Isaacs A., Vitart V., Hayward C., Franklin C.S., Polasek O., Kolcic I., Biloglav Z., Campbell S., et al. Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int. 2009;76:297–306. doi: 10.1038/ki.2009.135. [DOI] [PubMed] [Google Scholar]
- 13.Freedman B.I., Kopp J.B., Winkler C.A., Nelson G.W., Rao D.C., Eckfeldt J.H., Leppert M.F., Hicks P.J., Divers J., Langefeld C.D., et al. Polymorphisms in the nonmuscle myosin heavy chain 9 gene (MYH9) are associated with albuminuria in hypertensive African Americans: the HyperGEN study. Am. J. Nephrol. 2009;29:626–632. doi: 10.1159/000194791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedoya G., Montoya P., Garcia J., Soto I., Bourgeois S., Carvajal L., Labuda D., Alvarez V., Ospina J., Hedrick P.W., et al. Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc. Natl. Acad. Sci. USA. 2006;103:7234–7239. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price A.L., Patterson N., Yu F., Cox D.R., Waliszewska A., McDonald G.J., Tandon A., Schirmer C., Neubauer J., Bedoya G., et al. A genomewide admixture map for Latino populations. Am. J. Hum. Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parra E.J., Kittles R.A., Argyropoulos G., Pfaff C.L., Hiester K., Bonilla C., Sylvester N., Parrish-Gause D., Garvey W.T., Jin L., et al. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am. J. Phys. Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 18.Guerra R., Etzel C.J., Goldstein D.R., Sain S.R. Meta-analysis by combining P-values:simulated linkage studies. Genet. Epidemiol. 1999;17(Suppl. 1):S605–S609. doi: 10.1002/gepi.1370170798. [DOI] [PubMed] [Google Scholar]
- 19.Althaus K., Greinacher A. MYH9-related platelet disorders. Semin. Thromb. Hemost. 2009;35:189–203. doi: 10.1055/s-0029-1220327. [DOI] [PubMed] [Google Scholar]
- 20.Boute N., Gribouval O., Roselli S., Benessy F., Lee H., Fuchshuber A., Dahan K., Gubler M.C., Niaudet P., Antignac C. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 21.Gbadegesin R., Hinkes B.G., Hoskins B.E., Vlangos C.N., Heeringa S.F., Liu J., Loirat C., Ozaltin F., Hashmi S., Ulmer F., et al. Mutations in PLCE1 are a major cause of isolated diffuse mesangial sclerosis (IDMS) Nephrol. Dial. Transplant. 2008;23:1291–1297. doi: 10.1093/ndt/gfm759. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan J.M., Kim S.H., North K.N., Rennke H., Correia L.A., Tong H.Q., Mathis B.J., Rodriguez-Perez J.C., Allen P.G., Beggs A.H., et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 23.Kestila M., Lenkkeri U., Mannikko M., Lamerdin J., McCready P., Putaala H., Ruotsalainen V., Morita T., Nissinen M., Herva R., et al. Positionally cloned gene for a novel glomerular protein–nephrin—is mutated in congenital nephrotic syndrome. Mol. Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 24.Kim J.M., Wu H., Green G., Winkler C.A., Kopp J.B., Miner J.H., Unanue E.R., Shaw A.S. CD2-associated protein haploinsufficiency is linked to glomerular disease susceptibility. Science. 2003;300:1298–1300. doi: 10.1126/science.1081068. [DOI] [PubMed] [Google Scholar]
- 25.Winn M.P., Conlon P.J., Lynn K.L., Farrington M.K., Creazzo T., Hawkins A.F., Daskalakis N., Kwan S.Y., Ebersviller S., Burchette J.L., et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308:1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 26.Ward M.M. Socioeconomic status and the incidence of ESRD. Am. J. Kidney Dis. 2008;51:563–572. doi: 10.1053/j.ajkd.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 27.Norris K., Nissenson A.R. Race, gender, and socioeconomic disparities in CKD in the United States. J. Am. Soc. Nephrol. 2008;19:1261–1270. doi: 10.1681/ASN.2008030276. [DOI] [PubMed] [Google Scholar]
- 28.Nelson G.W., Freedman B.I., Bowden D.W., Langerfield C., An P., Johnson R.C., Kopp J.B., Winkler C. Fine mapping of MYH9 localizes the strongest kidney disease associations to the region of introns 13 to 15. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq039. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch E., Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Petkov P.M., Ding Y., Cassell M.A., Zhang W., Wagner G., Sargent E.E., Asquith S., Crew V., Johnson K.A., Robinson P., et al. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thorisson G.A., Smith A.V., Krishnan L., Stein L.D. The International HapMap Project Web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bercovici S., Geiger D., Shlush L., Skorecki K., Templeton A. Panel construction for mapping in admixed populations via expected mutual information. Genome Res. 2008;18:661–667. doi: 10.1101/gr.073148.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 34.Darvasi A., Shifman S. The beauty of admixture. Nat. Genet. 2005;37:118–119. doi: 10.1038/ng0205-118. [DOI] [PubMed] [Google Scholar]
- 35.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 36.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.