Abstract
Plasma soluble leptin receptor (sOB-R) levels were inversely associated with diabetes risk factors, including adiposity and insulin resistance, and highly correlated with the expression levels of leptin receptor, which is ubiquitously expressed in most tissues. We conducted a genome-wide association study of sOB-R in 1504 women of European ancestry from the Nurses' Health Study. The initial scan yielded 26 single nucleotide polymorphisms (SNPs) significantly associated with sOB-R levels (P < 5 × 10−8); all mapping to the leptin receptor gene (LEPR). Analysis of imputed genotypes on autosomal chromosomes revealed an additional 106 SNPs in and adjacent to this gene that reached genome-wide significance level. Of these 132 SNPs (including two non-synonymous SNPs, rs1137100 and rs1137101), rs2767485, rs1751492 and rs4655555 remained associated with sOB-R levels at the 0.05 level (P = 9.1 × 10−9, 0.0105 and 0.0267, respectively) after adjustment for other univariately associated SNPs in a forward selection procedure. Significant associations with these SNPs were replicated in an independent sample of young males (n = 875) residing in Cyprus (P < 1 × 10−4). These data provide novel evidence revealing the role of polymorphisms in LEPR in modulating plasma levels of sOB-R and may further our understanding of the complex relationships among leptin, leptin receptor and diabetes-related traits.
INTRODUCTION
Leptin receptor (OB-R) plays an essential role in the pleiotropic effects of leptin on body weight regulation, reproduction and metabolic pathways (1–3). In db/db mice and fa/fa rats that express truncated OB-R lacking a functional intracellular domain, a phenotype characteristic of early onset of obesity and extreme resistance to insulin develops (4). Likewise, in humans, a homozygous mutation in the human leptin receptor gene (LEPR) located on chromosome 1p31 results in truncated OB-R and, subsequently, early onset of severe obesity, as well as lack of pubertal development and decreased secretion of growth hormone and thyroid-stimulating hormone (5). In humans with normal LEPR gene expression, several isoforms of membrane-bound OB-R with identical extracellular and transmembrane domains but a variable intracellular domain are expressed on the surface of a wide spectrum of cells in almost all tissues through posttranscriptional alternative RNA splicing (6). According to the length of the intracellular domain, these isoforms can be classified into a long form (OB-RL) primarily expressed in the hypothalamus and several short forms (OB-RS) primarily expressed in peripheral tissues (6,7). The OB-RL mediates leptin's effects on the central neural system, such as body weight control and appetite regulation, whereas the function of OB-RS is not entirely clear (6,7). Soluble OB-R (sOB-R) is a special isoform of leptin receptor in the circulation that lacks both transmembrane and intracellular domains (6). sOB-R is formed by ectodomain shedding of OB-R expressed in peripheral tissues (8,9). sOB-R levels are highly correlated with the cell surface expression of OB-R (r = 0.80) (10), suggesting sOB-R levels may serve as a surrogate of the cell surface expression levels of OB-R in peripheral tissues. Studies have documented inverse correlations of sOB-R levels with adiposity and insulin resistance (11–14). Consistently, we recently found a striking inverse association between sOB-R levels and risk of developing type 2 diabetes (15). While evidence regarding the role of sOB-R in leptin metabolism and diabetes etiology is accumulating, thus far, little data existed regarding the genetic determinants of sOB-R levels. Although a few functional variants in LEPR region have been hypothesized to be such determinants, in a Japanese population two non-synonymous single nucleotide polymorphisms (SNPs) in LEPR region were not associated with variations of sOB-R levels (14). We, therefore, performed a genome-wide association (GWA) study of sOB-R levels in 1504 women from the Nurses' Health Study (NHS) to elucidate on the genetic variants predicting sOB-R levels.
RESULTS
The study population consisted of 684 type 2 diabetes cases and 820 diabetes-free controls from NHS that had both sOB-R measurements and genotype data. Distribution of age, body mass index (BMI), and other diabetes lifestyle and dietary risk factors is shown in Supplementary Material, Table S1. All participants had European ancestry as defined using the principal component of genetic variation based on the full NHS type 2 diabetes GWA study (which includes subjects with self-described African American and Asian ancestry) and the HapMap CEU, YRI and CHB/JPT samples. The mean age of this study sample was 56.2 years (SD = 6.9 years) and the mean level of sOB-R was 31.4 ng/ml (SD = 10.5 ng/ml). The majority (77.7%) of our study participants were postmenopausal. We used Affymetrix Genome-Wide Human 6.0 array to measure genotypes. In the initial scan, 704 409 SNPs passing quality control filters were tested for association with log-transformed sOB-R levels using linear regression. Since sOB-R levels are strongly correlated with BMI and diabetes status (12,13,16) and further influenced by fasting status (11), we adjusted for these covariates together with age, menopausal status and postmenopausal hormone use in the regression analysis. In the initial GWA scan, we identified 26 SNPs mapping to the LEPR region on chromosome 1p31 that accounted for the excess P-values < 5 × 10−8 (Table 1). All are intronic SNPs with the exception of rs1137100 (Lys109Arg). SNP rs12062820 was most strongly associated with sOB-R levels (P = 1.56 × 10−14) and has a minor allele frequency (MAF) of 17%. Each copy of the minor allele (C, non-ancestral) was associated with an increase in covariate-adjusted sOB-R levels.
Table 1.
Top genotyped SNPs (P < 5 × 10−8) for plasma sOB-R levelsa in the Nurses' Health Study
| SNPb | n | Position (bp)c | Alleles (+/−) | MAFd | Geometric LS mean (95% CI), ng/mle |
Betaf | SE | P-valuef | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | ||||||||
| rs12062820 | 1504 | 65743083 | T/C | 0.17 | 29.0 (28.5–29.5) | 31.7 (30.9–32.5) | 36.1 (33.2–39.1) | 0.096 | 0.012 | 1.56E−14 |
| rs11208656 | 1504 | 65743375 | A/G | 0.17 | 29.0 (28.5–29.5) | 31.7 (30.9–32.5) | 36.1 (33.2–39.1) | 0.096 | 0.012 | 1.56E−14 |
| rs1782755 | 1504 | 65759392 | C/A | 0.17 | 29.0 (28.5–29.5) | 31.6 (30.9–32.4) | 36.1 (33.4–39.0) | 0.095 | 0.012 | 1.91E−14 |
| rs1751492 | 1504 | 65765213 | T/C | 0.30 | 31.3 (30.7–31.8) | 29.0 (28.4–29.6) | 26.9 (25.8–28.1) | −0.076 | 0.010 | 6.42E−13 |
| rs2154381 | 1504 | 65768289 | A/G | 0.30 | 31.3 (30.7–31.8) | 29.0 (28.4–29.6) | 26.9 (25.7–28.1) | −0.075 | 0.010 | 7.41E−13 |
| rs1186403 | 1503 | 65773121 | T/C | 0.30 | 31.3 (30.7–31.8) | 29.0 (28.4–29.6) | 26.9 (25.7–28.1) | −0.075 | 0.010 | 8.51E−13 |
| rs6697315 | 1504 | 65766144 | T/C | 0.36 | 31.4 (30.8–32.1) | 29.3 (28.7–29.9) | 27.4 (26.4–28.4) | −0.069 | 0.010 | 7.14E−12 |
| rs10889556 | 1504 | 65742140 | A/G | 0.27 | 31.1 (30.6–31.7) | 28.9 (28.3–29.5) | 26.7 (25.3–28.1) | −0.075 | 0.011 | 7.23E−12 |
| rs11208674 | 1504 | 65811905 | T/C | 0.29 | 31.2 (30.6–31.7) | 29.0 (28.4–29.7) | 27.0 (25.8–28.2) | −0.072 | 0.010 | 9.27E−12 |
| rs4655518 | 1503 | 65812446 | T/G | 0.29 | 31.2 (30.6–31.7) | 29.0 (28.4–29.7) | 27.0 (25.8–28.2) | −0.072 | 0.010 | 1.16E−11 |
| rs1475398 | 1495 | 65755845 | G/C | 0.27 | 31.1 (30.5–31.7) | 28.9 (28.3–29.5) | 26.7 (25.4–28.1) | −0.075 | 0.011 | 1.62E−12 |
| rs10493379 | 1504 | 65818515 | G/A | 0.28 | 31.1 (30.5–31.7) | 29.0 (28.4–29.6) | 26.9 (25.7–28.2) | −0.071 | 0.011 | 3.09E−11 |
| rs3790425 | 1504 | 65815700 | A/G | 0.28 | 31.1 (30.5–31.6) | 29.0 (28.4–29.6) | 26.9 (25.7–28.2) | −0.071 | 0.011 | 3.23E−11 |
| rs11208679 | 1502 | 65822326 | G/A | 0.28 | 31.1 (30.5–31.7) | 29.0 (28.4–29.6) | 26.9 (25.7–28.2) | −0.071 | 0.011 | 3.78E−11 |
| rs1171261 | 1504 | 65773990 | C/T | 0.27 | 31.1 (30.5–31.7) | 28.9 (28.3–29.5) | 26.9 (25.6–28.4) | −0.073 | 0.011 | 3.83E−11 |
| rs2104564 | 1495 | 65742018 | T/C | 0.27 | 31.1 (30.6–31.7) | 28.9 (28.3–29.5) | 26.7 (25.4–28.2) | −0.073 | 0.011 | 4.52E−11 |
| rs10158279 | 1504 | 65806284 | G/T | 0.50 | 28.2 (27.4–28.9) | 29.9 (29.4–30.5) | 31.8 (31.0–32.6) | 0.061 | 0.009 | 1.12E−10 |
| rs1137100 | 1504 | 65809029 | A/G | 0.27 | 31.0 (30.5–31.6) | 29.0 (28.4–29.6) | 26.8 (25.4–28.2) | −0.071 | 0.011 | 1.71E−10 |
| rs10789184 | 1504 | 65809550 | G/A | 0.27 | 31.0 (30.5–31.6) | 29.0 (28.4–29.6) | 26.8 (25.4–28.2) | −0.071 | 0.011 | 1.71E−10 |
| rs12405556 | 1504 | 65835705 | G/T | 0.26 | 31.0 (30.5–31.6) | 28.9 (28.3–29.5) | 27.0 (25.7–28.5) | −0.069 | 0.011 | 2.88E−10 |
| rs10889567 | 1504 | 65829638 | T/C | 0.45 | 31.4 (30.7–32.2) | 29.9 (29.4–30.5) | 27.9 (27.0–28.7) | −0.059 | 0.009 | 5.54E−10 |
| rs10789190 | 1504 | 65836168 | G/A | 0.45 | 31.4 (30.7–32.2) | 29.9 (29.4–30.5) | 27.9 (27.0–28.7) | −0.059 | 0.009 | 5.54E−10 |
| rs10128072 | 1504 | 65729684 | A/C | 0.13 | 29.3 (28.8–29.7) | 31.8 (31.0–32.7) | 34.9 (31.6–38.5) | 0.084 | 0.014 | 1.43E−9 |
| rs1475397 | 1456 | 65755746 | C/T | 0.26 | 29.1 (28.6–29.6) | 30.4 (29.8–31.1) | 34.6 (32.9–36.4) | 0.067 | 0.011 | 1.64E−9 |
| rs7602 | 1480 | 65670539 | G/A | 0.20 | 29.0 (28.5–29.5) | 31.4 (30.7–32.1) | 33.1 (31.2–35.1) | 0.071 | 0.012 | 1.93E−9 |
| rs17127673 | 1478 | 65728313 | A/G | 0.13 | 29.3 (28.8–29.7) | 31.8 (31.0–32.7) | 34.7 (31.6–38.2) | 0.084 | 0.014 | 2.42E−9 |
asOB-R levels were log-transformed.
bAll SNPs are in LEPR gene. Except rs1137100, which is a missense SNP (Lys109Arg) in LEPR, all SNPs are in introns of LEPR.
cPosition based on NCBI build 36.3.
dMinor allele frequency among study participants.
eLeast square geometric means according to the number of minor allele copies were adjusted for age at blood draw, diabetes case–control status, fasting status, BMI, menopausal status and postmenopausal hormone use.
fRegression coefficients and P-values for every one copy of minor allele were estimated from linear regression models adjusted for the same set of covariates.
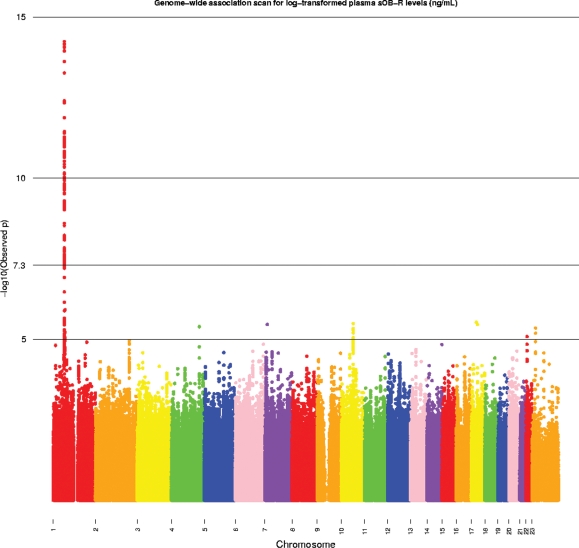
To assess whether any stronger genetic markers of sOB-R levels exist in the LEPR region, we expanded our genome scan by imputing 2 543 887 SNPs spanning chromosomes 1–22. One hundred and six additional SNPs achieved GWA significance (P< 5 × 10−8). Each was imputed with high accuracy (MACH r2 ≥ 0.70) and all mapped to LEPR or regions adjacent to LEPR (Fig. 1 and Supplementary Material, Table S2). The strongest association was found for rs2767485 (P = 5.77 × 10−15); an intronic SNP with a MAF of 17%. A second non-synonymous SNP, rs1137101 (Gln223Arg; P = 1.91 × 10−10), in the gene was also identified. These genotyped and imputed SNPs with GWA significance span six haplotype blocks in LEPR (CEU HapMap Release 22). We searched for LEPR expression levels for SNPs in LEPR region in a database (mRNA by SNP Browser V1.0.1) (17,18). Five SNPs in LEPR region (rs10158579, rs1171278, rs2025805, rs970467 and rs7602) were significantly associated with average LEPR expression levels (all P-values were ≤ 1.0 × 10−4).
Figure 1.
Genome-wide scan of plasma sOB-R levels in the Nurses' Health Study. P-values were adjusted for age at blood draw, fasting and diabetes case–control status, BMI, menopausal status and postmenopausal hormone use. The Y-axis was minus log-transformed observed P-values; the value of 7.3 corresponded to a P-value of 5 × 10−8. P-values for loci on chromosome X (i.e. chromosome 23) were based on genotyped SNPs only.
In secondary analyses that were restricted to non-diabetic women produced similar results, although P-values were somewhat augmented probably due to decreased statistical power (Supplementary Material, Table S3). When we conducted GWA scan in cases and controls separately and pooled the results of these two groups using a fixed-effects linear regression model, we observed a largely similar set of SNPs in LEPR region or regions adjacent to LEPR (Supplementary Material, Table S4). In addition, the directions of the associations for these SNPs were remarkably consistent between cases and controls. The quantile–quantile plot suggests no evidence of systematic bias in the distribution of P-values for the analysis; the genomic inflation factor λ = 1.006 (Supplementary Fig. S1A). Similar results were observed with further adjustment for residual population structure within this European-ancestry sample: λ = 1.010 (Supplementary Fig. S1B).
To identify SNPs independently associated with sOB-R levels, we performed a forward selection procedure of all genome-wide significant SNPs. We forced age at blood draw, BMI, diabetes case-control and fasting status, menopausal status, postmenopausal hormone use, as well as the two non-synonymous SNPs (rs1137100 and rs1137101) into the regression model. This procedure yielded three SNPs that were independently associated with sOB-R levels at the 0.05 significance level: rs2767485 (P = 9.08 × 10−9), rs1751492 (P = 0.0105) and rs4655555 (P = 0.0267) (Table 2). We calculated the percent of variation of sOB-R independently explained by the adjusted covariates and SNPs in the final model. This percent was 0.51% for age at blood draw, 15.60% for BMI, 3.34% for diabetes status, 0.02% for fasting status, 1.25% for menopausal status, 0.55% for postmenopausal hormone use, 0.14% for rs1137100, 0.17% for rs1137101, 2.19% for rs2767485, 0.44% for rs1751492 and 0.33% for rs4655555. Jointly, these five SNPs explained 4.59% of the total variation of sOB-R levels.
Table 2.
Covariates and selected SNPs in LEPR region in relation to plasma sOB-R levels
| Model 1a |
Model 2b |
Model 3c |
Model 4d |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (SE) | P-value | r2 (%)e | Beta (SE) | P-value | r2 (%)e | Beta (SE) | P-value | r2 (%)e | Beta (SE) | P-value | r2 (%)e | |
| Age (year) | 0.004 (0.001) | 0.0044 | 0.54 | 0.004 (0.001) | 0.0048 | 0.53 | 0.004 (0.001) | 0.0046 | 0.54 | 0.003 (0.001) | 0.0057 | 0.51 |
| BMI (kg/m2) | −0.02 (0.001) | 4.79 × 10−55 | 15.08 | −0.02 (0.001) | 5.15 × 10−56 | 15.33 | −0.02 (0.001) | 1.27 × 10−56 | 15.50 | −0.02 (0.001) | 5.46 × 10−57 | 15.60 |
| Diabetes status | 0.11 (0.01) | 2.81 × 10−13 | 3.50 | 0.10 (0.01) | 8.24 × 10−13 | 3.38 | 0.10 (0.01) | 1.09 × 10−12 | 3.34 | 0.10 (0.01) | 1.09 × 10−12 | 3.34 |
| Fasting status | 0.005 (0.01) | 0.7394 | 0.0001 | 0.008 (0.01) | 0.6492 | 0.0002 | 0.008 (0.01) | 0.5740 | 0.02 | 0.008 (0.01) | 0.5548 | 0.02 |
| Menopausal status | 0.03 (0.007) | 1.99 × 10−5 | 1.21 | 0.03 (0.007) | 9.66 × 10−6 | 1.30 | 0.03 (0.007) | 1.21 × 10−5 | 1.28 | 0.03 (0.007) | 1.45 × 10−5 | 1.25 |
| Postmenopausal hormone use | −0.06 (0.02) | 0.0053 | 0.52 | −0.06 (0.02) | 0.0052 | 0.52 | −0.06 (0.02) | 0.0046 | 0.54 | −0.06 (0.02) | 0.0041 | 0.55 |
| rs1137100 | −0.04 (0.01) | 0.0032 | 0.58 | −0.04 (0.01) | 0.0046 | 0.54 | 0.02 (0.03) | 0.3998 | 0.05 | 0.04 (0.03) | 0.1428 | 0.14 |
| rs1137101 | −0.04 (0.01) | 0.0038 | 0.56 | −0.02 (0.01) | 0.1909 | 0.11 | −0.02 (0.01) | 0.0922 | 0.19 | −0.02 (0.01) | 0.1093 | 0.17 |
| rs2767485 | – | – | – | −0.08 (0.01) | 1.72 × 10−9 | 2.40 | −0.07 (0.01) | 6.57 × 10−8 | 2.02 | −0.07 (0.01) | 9.08 × 10−9 | 2.19 |
| rs1751492 | – | – | – | – | – | – | −0.07 (0.02) | 0.0082 | 0.47 | −0.06 (0.02) | 0.0105 | 0.44 |
| rs4655555 | – | – | – | – | – | – | – | – | – | 0.04 (0.02) | 0.0267 | 0.33 |
aIndependent variables included age, BMI, diabetes case–control status (yes/no), fasting status (≥8 or <8 h since last meal), rs1137100 and rs1137101.
bBased on model 1, rs2767485 was further included in the model as an independent variable.
cBased on model 2, rs1751492 was further included.
dBased on model 3, rs4655555 was further included.
er2 was calculated as model sum of squares divided by corrected total sum of squares in the linear regression and represents the proportion of variability of sOB-R independently explained by each variable.
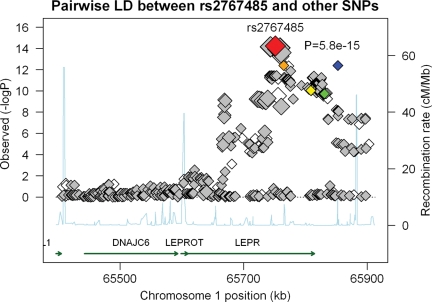
We estimated pairwise LD of each SNP with rs2767485 in the NHS sample (Fig. 2). All SNPs with P < 1 × 10−13 are in strong to perfect LD with rs2767485 (r2 ≥ 0.79), whereas rs1751492, rs4655555 and the two functional non-synonymous SNPs are only in weak LD with rs2767485 (r2 < 0.20). With respect to pairwise LD among rs2767485, rs1751492, rs4655555 and the two non-synonymous SNPs, with exception of the strong LD between rs1751492 and rs1137100 (r2 = 0.84), these SNPs are only in weak to moderate LD with each other (r2 ≤ 0.52). Based on these five SNPs, we were able to construct six haplotypes. None of these haplotypes provided additional predictive value for sOB-R levels beyond these SNPs (data not shown).
Figure 2.
P-values and pairwise LDs with rs2767485 for SNPs in and adjacent to LEPR region between position 65400 kb and 65900 kb on chromosome 1. SNPs included in this panel included all SNPs [108 genotyped ( ) and 384 imputed (
) and 384 imputed ( ) SNPs, including rs2767485 (
) SNPs, including rs2767485 ( ), rs1751492 (
), rs1751492 ( ), rs1137100 (
), rs1137100 ( ), rs1137101 (
), rs1137101 ( ), and rs4655555 (
), and rs4655555 ( )] within the region. Recombination rates in this region were plotted in the background in light blue. Pairwise LDs between rs2767485 and other SNPs were estimated using HapMap LD data. For 12 SNPs that are covered only in Affymetrix 6.0, we calculated the pairwise LDs using our observed data. The size of the diamonds represents the strength of LD; the largest diamonds represent a LD (r2) ≥ 0.80, whereas other sizes (from large to small) represent a LD of ≥0.5, ≥0.2 and <0.2, respectively.
)] within the region. Recombination rates in this region were plotted in the background in light blue. Pairwise LDs between rs2767485 and other SNPs were estimated using HapMap LD data. For 12 SNPs that are covered only in Affymetrix 6.0, we calculated the pairwise LDs using our observed data. The size of the diamonds represents the strength of LD; the largest diamonds represent a LD (r2) ≥ 0.80, whereas other sizes (from large to small) represent a LD of ≥0.5, ≥0.2 and <0.2, respectively.
We further examined associations between these SNPs in relation to other diabetes-related traits, including BMI, waist-to-hip ratio, high-molecular-weight adiponectin and inflammatory biomarkers (Supplementary Material, Table S5). We found a significant inverse association between rs2767485 and fasting insulin levels (P = 0.008) and a significant positive associate between rs4655555 and CRP levels (P = 0.005). P-values for other associations were not significant or with borderline significance.
In an independent sample of 875 adolescent males (mean age = 18.4 years; SD = 0.6 years) studied during an initial health screening in preparation for their 2-year mandatory service in the Cyprus Army, SNPs rs1137100, rs1137101, rs2767485, rs1751492 and rs4655555 were each significantly associated with sOB-R levels after adjustment for age and BMI (Table 3). The direction of these associations was also consistent with those observed in the NHS sample.
Table 3.
Relation of selected SNPs in LEPR region in relation to plasma sOB-R levels in a replication populationa
| SNP | n | Position (bp)b | Alleles (+/−) | MAFc | Geometric LS mean (95% CI), ng/mld |
Betae | SE | P-valuee | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | ||||||||
| rs1137100 | 875 | 65809029 | A/G | 0.11 | 23.0 (22.5–23.4) | 20.7 (20.2–21.3) | 21.6 (19.2–24.3) | −0.089 | 0.019 | 3.83E−6 |
| rs1137101 | 875 | 65831101 | A/G | 0.33 | 23.7 (23.3–24.2) | 21.7 (21.1–22.4) | 21.0 (20.2–21.8) | −0.071 | 0.012 | 2.05E−8 |
| rs2767485 | 875 | 65751505 | T/C | 0.30 | 21.6 (21.0–22.2) | 23.2 (22.8–23.7) | 24.3 (23.3–25.5) | 0.065 | 0.013 | 6.27E−7 |
| rs1751492 | 875 | 65765213 | T/C | 0.14 | 23.1 (22.6–23.6) | 20.9 (20.4–21.4) | 21.3 (19.6–23.1) | −0.086 | 0.017 | 9.37E−7 |
| rs4655555f | 875 | 65852857 | T/A | 0.06 | 22.9 (19.9–26.6) | 20.7 (17.9–23.7) | 21.9 (21.9–22.0) | −0.100 | 0.025 | 6.32E−5 |
asOB-R levels were log-transformed.
bPosition based on NCBI build 36.3.
cMinor allele frequency among study participants in the replication sample.
dLeast square geometric means according to the number of minor allele copies were adjusted for age at blood draw and BMI.
eRegression coefficients and P-values for every one copy of minor allele were estimated from linear regression models adjusted for the same set of covariates.
fLeast square means could not be obtained due to rare homozygous minor alleles. Median (interquartile range) was provided instead.
In addition to the top SNPs with GWA significance, we also identified a number of genotyped and imputed SNPs on various chromosomes that reached suggestive significance level (P < 1 × 10−5) (Supplementary Tables S6 and S7). After excluding SNPs that are in or adjacent to LEPR region (65000 and 67000 kb on chromosome 1), we re-plotted the quantile–quantile plot before (Supplementary Fig. S2A) and after (Supplementary Fig. S2B) adjustment for the residual population structure and we observed genomic inflation factors (λ = 1.006 and 1.009, respectively) that were similar to those based on the complete list of SNPs.
Lastly, since the two non-synonymous SNPs (rs1137100 and rs1137101) may potentially influence the affinity of antibody used in ELISA assay to detect sOB-R levels, we conducted several sensitivity analyses to evaluate whether the strong association between LEPR region and sOB-R levels can be explained by these two SNPs. First, we searched PolyPhen online database (http://genetics.bwh.harvard.edu/pph/) for predicted functional consequences associated with the two non-synonymous SNPs (19–21), and the results indicated that these two non-synonymous SNPs would unlikely lead to significant structural or functional changes. Second, we examined LD between these two functional SNPs and the remaining significant SNPs in LEPR region. Of the remaining 131 significant SNPs, 46 (35.1%) was in strong LD (r2 ≥ 0.8), 23 (17.6%) were in moderate LD (r2, 0.5–0.7), 37 (28.2%) were in weak LD (r2, 0.2–0.4), and 25 (19.1%) were not in LD (r2 < 0.2) with rs1137100. With respect to LD with rs1137101, these figures were 10 (7.6%), 11 (8.4%), 85 (64.9%) and 25 (19.1%), respectively. In addition, we examined the association between SNPs in LEPR region and sOB-R levels within participants who were homozygotes of the ancestral allele of both or either SNPs, respectively. Although P-values were increased due to reduced power, the top SNP, rs2767485, remained highly significant. The P-values were 2.86 × 10−5 for participants who were homozygotes of the ancestral allele of both SNPs, 8.14 × 10−7 for participants who were homozygotes of the ancestral allele of rs1137100, and 1.63 × 10−5 for participants who were homozygotes of the ancestral allele of rs1137101. Several SNPs that were in strong LD with rs2767485 (i.e. rs12079231, rs12062820, rs11208656, rs1782755, rs1627238, rs1171275, rs1180445, rs1171278 and rs1171276) were associated with sOB-R levels at a similar significance level in these analyses.
DISCUSSION
According to the length of intracellular domain, isoforms of OB-R can be classified into short forms, long form and soluble OB-R (7). The long form OB-R (OB-RL) with the full length of intracellular domain is primarily expressed in the hypothalamus, whereas multiple short forms of OB-R (OB-RS) with various length of intracellular domain are primarily expressed in peripheral tissues (7). Although, in comparison to OB-RL, isoforms of OB-RS lack the motif regulating energy homeostasis, increasing evidence suggests that OB-RS may be involved in the modulation of insulin sensitivity and other peripheral effects through the insulin receptor substrate/phosphatidylinositol-3-OH kinase (IRS/PI3K) pathway (22–26). In humans, circulating sOB-R is formed by ectodomain shedding of membrane-anchored OB-R (primarily OB-RS) (8,9), which are expressed on the surface of a wide variety of cells in almost all peripheral tissues (6). The strong correlation between sOB-R levels and the cell surface expression levels of OB-R (10) suggests that sOB-R may reflect the total amount or activity of OB-RS in peripheral tissues. sOB-R levels are inversely associated with diabetes risk or diabetes-related environmental risk factors (12,13,15,16). The genetic determinants of sOB-R, however, are unclear. To our knowledge, only one study has investigated the role that common genetic variation plays in modulating sOB-R levels: in a Japanese population consisting of 127 men and 90 women, the two functional LEPR SNPs (rs1137100 and rs1137101) identified in the current study were not associated with sOB-R levels (14). However, homozygotes of the minor allele of these two SNPs are much rarer in Japanese than western populations; according to the HapMap data, less than 1% of Japanese are homozygous of the minor allele of either SNPs, whereas in populations with European ancestry this proportion is ∼40% for rs1137100 and 20% for rs1137101 (27). Together with a modest sample size, this study had a fairly limited statistical power to detect any significant differences in sOB-R levels by the genotypes.
To our knowledge, the current study is the first GWA investigation on plasma sOB-R levels. We replicated two functional and three model-selected SNPs in an independent sample with characteristics quite different from the NHS population. In addition, several observations strongly support that associations between LEPR variants and sOB-R levels are likely to be causal. First, biological plausibility for our findings is strongly supported by a priori evidence generated from basic biological research. Secondly, all identified genome-wide significant SNPs are localized to LEPR, two of which have been previously examined in candidate studies. In addition, a few SNPs in LEPR region were significantly associated the LEPR expression levels (17,18) and, thus, likely lead to a significant change in the levels of sOB-R. Thirdly, although all identified SNPs are in LEPR region, not all of them are in strong LD, indicating that the current finding is unlikely a pure play of chance. Lastly, when we restricted our GWA scan within controls only, although the diminished power made the top SNPs less significant, we essentially obtained the same set of SNPs. Furthermore, results independently generated from cases and controls were remarkably consistent.
Several caveats in the current study deserve discussion as well. First, although we cannot entirely exclude the possibility that the associations for the two functional SNPs were due to the potentially altered affinity of antibody induced by the non-synonymous SNPs, the structural and functional change of protein associated with these SNPs is estimated to be small. Furthermore, when we restricted the analysis within homozygotes of the ancestral alleles of the non-synonymous SNPs, the top SNPs were still highly significantly associated with sOB-R levels. Nevertheless, further investigation is warranted to shed light on this issue. Second, we only measured sOB-R levels once. Measurement error due to random assay variability and/or within-person variation of sOB-R levels is inevitable. Although the long-term variation of sOB-R is not well documented, data show that the 24 h within-person variation of sOB-R closely resembles that of high-molecular-weight adiponectin (28), for which the long-term within-person variation is small (29). In addition, sOB-R levels were measured in a blinded way that genotype information had no influence on the measurements. Therefore, the measurement error of sOB-R is more likely to be non-differential and, therefore, more likely to cause a bias to the null in our results. Third, to minimize genotyping error, we only utilized SNPs that passed standard quality control criteria which were developed for our GWA study. The major reasons for the exclusion were rare SNPs (MAF < 0.02) and technical error (call rate < 98%). Although we cannot exclude the possibility that some of the excluded SNPs may actually correlate with sOB-R levels, the identified genome-wide significant associations are unlikely the consequence of genotyping error. Fourth, our study population exclusively consisted of women with genetically determined European ancestry and the replication sample was exclusively composed of young men with European ancestry. Whether the results in the current study can be generalized to other ethnicities deserves further investigation. Lastly, our moderate sample size restrained our ability to detect loci in other gene regions that regulate sOB-R levels as well. The loci found in the current study that reached suggestive GWA significance level warrant further investigation in future studies. The strengths of the current GWA study included relatively large sample size, high quality genotype data, careful quality control and minimal population stratification.
In the current study, two non-synonymous SNPs in LEPR were associated with plasma sOB-R levels at the genome-wide significance level. Several candidate gene studies have examined these functional SNPs in relation to body weight and type 2 diabetes-related traits. Mixed associations were documented for these SNPs in relation to insulin sensitivity or type 2 diabetes (30–34). Similarly, the association between these SNPs and body weight or BMI was also inconclusive (35–37). Likewise, in our study, these two functional SNPs were not associated with type 2 diabetes status or diabetes-related traits. Since these two SNPs can only explain a minimal amount of variation of sOB-R levels, this minor change of OB-R activities associated with these variants may not translate into a dramatic effect on insulin sensitivity or other diabetes-related risk factors. However, it is likely that OB-R expression levels are determined by many other common functional SNPs which jointly may explain a substantial amount of sOB-R, and subsequently affect diabetes-related traits. In the current study, in addition to the significant SNPs in LEPR region, we also found a few SNPs in other regions that were associated with sOB-R levels at suggestive genome-wide significance level. These regions certainly warrant more detailed investigation in future studies with larger sample size. As in many other GWA studies, the identified top SNP (rs2767485) with the best P-value in our study is located in the intron region of the LEPR gene. Interestingly, we documented a significant inverse association between this SNP and fasting insulin levels, probably because this SNP explained larger amount of variation of sOB-R levels than other SNPs. Whether the observed associations for this SNP are causal or not warrants further investigation.
In summary, we identified multiple SNPs in the LEPR gene associated with plasma sOB-R levels and confirmed the role of LEPR as a candidate gene for regulating sOB-R levels. Although further investigation is warranted, we provided first evidence for genetic determinants for sOB-R levels that may play an important role in insulin metabolism. These novel findings will likely shed light on the complex relationship among leptin, sOB-R and metabolic traits.
MATERIALS AND METHODS
Study population
The NHS was established in 1976 when 121 700 female registered nurses aged 30–55 years and residing in 11 large US states completed a mailed questionnaire on their medical history and lifestyle characteristics. Every 2 years since baseline, follow-up questionnaires have been sent to update information on exposures and newly diagnosed illnesses. Beginning in 1980, on a 2–4 year cycle, dietary information has been updated using validated semi-quantitative food frequency questionnaires. Blood was collected from 32 826 NHS members between 1989 and 1990. Demographics and health status of participants who provided blood samples were generally similar to those who did not. Participants for the current study were a subset of those included in a nested case–control study of sOB-R in relation to type 2 diabetes in the NHS (38). Briefly, between blood draw and June 2004, we identified 1038 incident type 2 diabetes cases and randomly selected 1136 healthy controls using risk-set sampling. Cases and controls were matched for age at blood draw, date of blood collection, fasting status (≥8 versus <8 h since last meal) and ethnicity. We restricted the current analysis to unrelated, genetically defined women of primarily European ancestry who had high quality genotyping data. After excluding participants with missing phenotype and covariate data, 1504 participants were included in the final analysis. All study participants have signed and returned consent forms. The study protocol was approved by the institutional review board of the Brigham and Women's Hospital and the Human Subjects Committee Review Board of Harvard School of Public Health.
Replication population
The replication population was comprised of 18-year-old males enrolled during a health screening prior to performing their 2-year mandatory service in the Cyprus Army. Participants were randomly selected from the July 2006 and July 2007 enrollment cycles. Exclusion criteria included health reasons for which a person was unable to participate in the military service. In total, 1056 volunteers were recruited and were representative of the 18-year-old male population of Cyprus. Overnight fasting blood samples were collected among these participants. In the current study, we excluded participants with missing genotype and sOB-R data, leaving 875 participants for the analysis. All participants completed a written consent form and the study protocol was approved by the Cyprus National Bioethics Committee and the Human Subjects Committee Review Board of Harvard School of Public Health.
Laboratory procedures
In the original case–control study, each diabetes case was matched with one to two controls for age, blood draw date, ethnicity, and fasting status. Each diabetes case–control set was shipped in the same batch to the laboratory and analyzed in the same run. Within each batch, samples were assayed by the same technicians in random sequence under identical conditions. sOB-R was measured by ELISA technique (R&D Systems, Minneapolis, MN, USA) with a sensitivity of 0.06 ng/ml. This assay utilizes a monoclonal antibody specific to human sOB-R (amino acid position 20 to 839). Laboratory control samples (n = 20) were run along with the case–control samples. Based on the measurements of these control samples, the average intra-assay coefficient of variation (CV%) was 7.3% for the sOB-R assay. In the replication sample, sOB-R levels were measured using the same assay with a similar accuracy (CV% = 6.1%). In both samples, assessment of sOB-R levels was independent of genotyping procedures.
GWAS genotyping, imputation and quality control
For the GWA study samples, DNA was extracted from white blood cells using the QIAmpTM (QIAGEN Inc., Chatsworth, CA, USA) blood protocol. Genotyping was done using the Affymetrix Genome-Wide Human 6.0 array and the Birdseed calling algorithm (39). Genotypic data for a total of 3429 NHS samples passed laboratory technical quality control criteria which included SNP fingerprints for sample tracking and early detection of sample misidentification, missing call rate (MCR), the use of HapMap controls to check genotype quality independent of study samples and the tracking of reagent and instrumental performance.
Relatedness was evaluated using pairwise identity-by descent using 80 k SNPs in a method of moments approach implemented in PLINK software (40). Five pairs of duplicate samples were identified and removed. One pair of full siblings and eight sets (six pairs and two triplets) of possible first cousins were also identified. Gender was confirmed by examining the mean of the intensities of SNP probes on the X and Y chromosomes. One male sample was mis-identified as a female sample and was excluded. Twenty-seven subjects with gross chromosomal anomalies, determined by analyzing relative intensity (‘LogRRatio’) and allelic imbalance (‘BAlleleFreq’, BAF) (41), and 22 samples having a MCR ≥2% were also removed.
More than 96% (879 071) of the 909 622 SNP probes on the array passed the quality control standards of the genotyping center (Broad Institute of MIT and Harvard) for NHS samples. We further excluded SNPs which were monomorphic, had a MCR ≥2%, more than one discordance, a HWE P-value <1 × 10−4 or a MAF <0.02. Duplicate SNPs (assayed with different probes) were also removed. A total of 704 409 SNPs passed quality control and were included for the analysis.
We used MACH (http://www.sph.umich.edu/ csg/abecasis/MACH) to impute 2 543 887 SNPs on chromosome 1 to 22 with NCBI build 36 of Phase II HapMap CEU data (release 22) as the reference panel. Imputation results were expressed as ‘allele dosages’ (fractional values between 0 and 2). Imputed SNPs with MAF < 0.02 and/or with poor imputation quality scores (MACH r2 ≤ 0.30) were filtered from the analysis.
Replication sample genotyping
In the replication sample, genotyping was carried out using the TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA). The primers and probes were ordered from Applied Biosystems. The fluorescence of PCR products was detected by the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA). A random 10% samples were included as duplicates for quality control. The overall genotyping success rate was 98.7% and the concordance rate for duplicate samples was 99.5%.
Determination of population structure
Population structure was investigated by principal component analysis (42). We used a set of 12 021 SNPs selected to have very low levels of LD and to have MAF >0.05 in populations with European ancestry (43). These SNPs were chosen using a three-step algorithm out of a pool of 40 817 SNPs on autosomal chromosomes that (i) are common to Affymetrix 500k, Illumnia HumanHap300 and Illumina HumanHap550 platforms; (ii) have a MAF >5%; (iii) have Hardy–Weinberg proportion exact test P> 10−3 in two independent control samples (43). Study subjects passing quality control (n = 3369) were analyzed together with a set of 209 HapMap II founders (59 CEU, 60 YRI, 45 JPT and 45 CHB) (Supplementary Material, Fig. S2). Subjects with first and second principal components in the box defined by the means of the first and second principal components among self-described whites ±3 SD were classified as having primarily European ancestry. We found a high concordant rate of self-described European ancestry with genetically identified European ancestry: 99.2% of participants with self-described European ancestry were confirmed using these principal components.
Statistical analysis
The associations between each SNP and log-transformed plasma sOB-R levels were evaluated using linear regression adjusting for age, BMI, diabetes and fasting status, menopausal status and postmenopausal hormone use. We assumed an additive model for each SNP. Observed genotypes were analyzed using PLINK software (40), while imputed data (expressed as allele dosage) were analyzed using ProbABEL software (44).
We used a forward-selection regression procedure to identify genome-wide significant SNPs that were associated with sOB-R levels independent of other SNPs. We forced age, BMI, diabetes and fasting status, menopausal status and postmenopausal hormone use, as well as two missense SNPs (rs1137100 and rs1137101) into the linear regression model. The selection criterion was P less than 0.05 for a SNP to enter the final model. We further estimated the proportion of variation of sOB-R that was explained by selected SNPs in the linear regression analyses. This proportion was measured by r2, which is the difference of model sum of squares between models with and without the SNP(s) of interest divided by the corrected total sum of squares of the full model. These analyses were performed in SAS 9.1 (SAS Institute, Inc., Cary, NC, USA).
SUPPLEMENTARY MATERIAL
FUNDING
The NHS type 2 diabetes GWA study (U01HG004399) is a component of a collaborative project that includes 13 other GWA studies (U01HG004738, U01HG004422, U01HG004402, U01HG004729, U01HG004726, U01HG004735, U01HG004415, U01HG004436, U01HG004423, U01HG004728 and RFAHG006033 funded by the National Human Genome Research Institute, as well as U01DE018993 and U01DE018903 funded by the National Institute of Dental & Craniofacial Research) as part of the Gene Environment-Association Studies (GENEVA) under the National Institutes of Health Genes, Environment and Health Initiative (GEI). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Genotyping was performed at the Broad Institute of Massachusetts Institute of Technology and Harvard University, with funding support from the National Institutes of Health GEI (U01HG04424), and Johns Hopkins University Center for Inherited Disease Research, with support from the National Institutes of Health GEI (U01HG004438) and the National Institutes of Health contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C). Additional funding for the current research was provided by the National Cancer Institute (P01CA087969, P01CA055075), the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK058845), the Merck & Co., Inc. and the Cyprus Research Promotion Foundation (EPYE/0205/10). Q.S. is supported by the Pilot and Feasibility program sponsored by the Boston Obesity Nutrition Research Center (DK46200). M.C.C. is a recipient of a Canadian Institutes of Health Research Fellowship. L.Q. is supported by National Institutes of Health grants RO1 HL71981, American Heart Association Scientist Development Award and the Boston Obesity Nutrition Research Center (DK46200). C.S.M. and the performance of sOB-R assays were supported by National Institutes of Health grants DK58785, DK79929, DK81913, and a discretionary grant from the Beth Israel Deaconess Medical Center.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the contributions of Drs Costas Christophi, Michael Petrou and Stefanos Kales to the replication study.
Conflict of Interest statement. C.J.G. is an employee of the Merck & Co., Inc. Other authors had no financial or personal conflict of interest to disclose.
REFERENCES
- 1.Farooqi I.S., O'Rahilly S. Leptin: a pivotal regulator of human energy homeostasis. Am. J. Clin. Nutr. 2009;89:980S–984S. doi: 10.3945/ajcn.2008.26788C. [DOI] [PubMed] [Google Scholar]
- 2.Friedman J.M., Halaas J.L. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 3.Harris R.B. Leptin—much more than a satiety signal. Annu. Rev. Nutr. 2000;20:45–75. doi: 10.1146/annurev.nutr.20.1.45. [DOI] [PubMed] [Google Scholar]
- 4.Chua S.C., Jr, Chung W.K., Wu-Peng X.S., Zhang Y., Liu S.M., Tartaglia L., Leibel R.L. Phenotypes of mouse diabetes and rat fatty due to mutations in the OB (leptin) receptor. Science. 1996;271:994–996. doi: 10.1126/science.271.5251.994. [DOI] [PubMed] [Google Scholar]
- 5.Clement K., Vaisse C., Lahlou N., Cabrol S., Pelloux V., Cassuto D., Gourmelen M., Dina C., Chambaz J., Lacorte J.M., et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 6.Tartaglia L.A. The leptin receptor. J. Biol. Chem. 1997;272:6093–6096. doi: 10.1074/jbc.272.10.6093. [DOI] [PubMed] [Google Scholar]
- 7.Fruhbeck G. Intracellular signalling pathways activated by leptin. Biochem. J. 2006;393:7–20. doi: 10.1042/BJ20051578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge H., Huang L., Pourbahrami T., Li C. Generation of soluble leptin receptor by ectodomain shedding of membrane-spanning receptors in vitro and in vivo. J. Biol. Chem. 2002;277:45898–45903. doi: 10.1074/jbc.M205825200. [DOI] [PubMed] [Google Scholar]
- 9.Chua S.C., Jr, Koutras I.K., Han L., Liu S.M., Kay J., Young S.J., Chung W.K., Leibel R.L. Fine structure of the murine leptin receptor gene: splice site suppression is required to form two alternatively spliced transcripts. Genomics. 1997;45:264–270. doi: 10.1006/geno.1997.4962. [DOI] [PubMed] [Google Scholar]
- 10.Maamra M., Bidlingmaier M., Postel-Vinay M.C., Wu Z., Strasburger C.J., Ross R.J. Generation of human soluble leptin receptor by proteolytic cleavage of membrane-anchored receptors. Endocrinology. 2001;142:4389–4393. doi: 10.1210/endo.142.10.8442. [DOI] [PubMed] [Google Scholar]
- 11.Chan J.L., Bluher S., Yiannakouris N., Suchard M.A., Kratzsch J., Mantzoros C.S. Regulation of circulating soluble leptin receptor levels by gender, adiposity, sex steroids, and leptin: observational and interventional studies in humans. Diabetes. 2002;51:2105–2112. doi: 10.2337/diabetes.51.7.2105. [DOI] [PubMed] [Google Scholar]
- 12.Laimer M., Ebenbichler C.F., Kaser S., Sandhofer A., Weiss H., Nehoda H., Aigner F., Patsch J.R. Weight loss increases soluble leptin receptor levels and the soluble receptor bound fraction of leptin. Obes. Res. 2002;10:597–601. doi: 10.1038/oby.2002.81. [DOI] [PubMed] [Google Scholar]
- 13.Ogier V., Ziegler O., Mejean L., Nicolas J.P., Stricker-Krongrad A. Obesity is associated with decreasing levels of the circulating soluble leptin receptor in humans. Int. J. Obes. Relat. Metab. Disord. 2002;26:496–503. doi: 10.1038/sj.ijo.0801951. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa T., Hirose H., Yamamoto Y., Nishikai K., Miyashita K., Nakamura H., Saito I., Saruta T. Relationships between serum soluble leptin receptor level and serum leptin and adiponectin levels, insulin resistance index, lipid profile, and leptin receptor gene polymorphisms in the Japanese population. Metabolism. 2004;53:879–885. doi: 10.1016/j.metabol.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Sun Q., van Dam R.M., Meigs J.B., Franco O.H., Mantzoros C.S., Hu F.B. Leptin and Soluble Leptin Receptor Levels in Plasma and Risk of Type 2 Diabetes in US Women: A Prospective Study. Diabetes. 2010;59:611–618. doi: 10.2337/db09-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandhofer A., Laimer M., Ebenbichler C.F., Kaser S., Paulweber B., Patsch J.R. Soluble leptin receptor and soluble receptor-bound fraction of leptin in the metabolic syndrome. Obes. Res. 2003;11:760–768. doi: 10.1038/oby.2003.106. [DOI] [PubMed] [Google Scholar]
- 17.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 18.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 19.Ramensky V., Bork P., Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunyaev S., Ramensky V., Koch I., Lathe W., Kondrashov A.S., Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 21.Sunyaev S., Ramensky V., Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 22.Huan J.N., Li J., Han Y., Chen K., Wu N., Zhao A.Z. Adipocyte-selective reduction of the leptin receptors induced by antisense RNA leads to increased adiposity, dyslipidemia, and insulin resistance. J. Biol. Chem. 2003;278:45638–45650. doi: 10.1074/jbc.M304165200. [DOI] [PubMed] [Google Scholar]
- 23.Morton G.J., Gelling R.W., Niswender K.D., Morrison C.D., Rhodes C.J., Schwartz M.W. Leptin regulates insulin sensitivity via phosphatidylinositol-3-OH kinase signaling in mediobasal hypothalamic neurons. Cell Metab. 2005;2:411–420. doi: 10.1016/j.cmet.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhao A.Z., Shinohara M.M., Huang D., Shimizu M., Eldar-Finkelman H., Krebs E.G., Beavo J.A., Bornfeldt K.E. Leptin induces insulin-like signaling that antagonizes cAMP elevation by glucagon in hepatocytes. J. Biol. Chem. 2000;275:11348–11354. doi: 10.1074/jbc.275.15.11348. [DOI] [PubMed] [Google Scholar]
- 25.Anderwald C., Muller G., Koca G., Furnsinn C., Waldhausl W., Roden M. Short-term leptin-dependent inhibition of hepatic gluconeogenesis is mediated by insulin receptor substrate-2. Mol. Endocrinol. 2002;16:1612–1628. doi: 10.1210/mend.16.7.0867. [DOI] [PubMed] [Google Scholar]
- 26.Huang W., Dedousis N., Bhatt B.A., O'Doherty R.M. Impaired activation of phosphatidylinositol 3-kinase by leptin is a novel mechanism of hepatic leptin resistance in diet-induced obesity. J. Biol. Chem. 2004;279:21695–21700. doi: 10.1074/jbc.M401546200. [DOI] [PubMed] [Google Scholar]
- 27.The National Center for Biotechnology Information. dbSNP build 129. Available: http://www.ncbi.nlm.nih.gov/projects/SNP/ (accessed 07 July 2009) [Google Scholar]
- 28.Gavrila A., Peng C.K., Chan J.L., Mietus J.E., Goldberger A.L., Mantzoros C.S. Diurnal and ultradian dynamics of serum adiponectin in healthy men: comparison with leptin, circulating soluble leptin receptor, and cortisol patterns. J. Clin. Endocrinol. Metab. 2003;88:2838–2843. doi: 10.1210/jc.2002-021721. [DOI] [PubMed] [Google Scholar]
- 29.Pischon T., Hotamisligil G.S., Rimm E.B. Adiponectin: stability in plasma over 36 hours and within-person variation over 1 year. Clin. Chem. 2003;49:650–652. doi: 10.1373/49.4.650. [DOI] [PubMed] [Google Scholar]
- 30.Chiu K.C., Chu A., Chuang L.M., Saad M.F. Association of leptin receptor polymorphism with insulin resistance. Eur. J. Endocrinol. 2004;150:725–729. doi: 10.1530/eje.0.1500725. [DOI] [PubMed] [Google Scholar]
- 31.Wauters M., Mertens I., Rankinen T., Chagnon M., Bouchard C., Van Gaal L. Leptin receptor gene polymorphisms are associated with insulin in obese women with impaired glucose tolerance. J. Clin. Endocrinol. Metab. 2001;86:3227–3232. doi: 10.1210/jcem.86.7.7682. [DOI] [PubMed] [Google Scholar]
- 32.Ukkola O., Tremblay A., Despres J.P., Chagnon Y.C., Campfield L.A., Bouchard C. Leptin receptor Gln223Arg variant is associated with a cluster of metabolic abnormalities in response to long-term overfeeding. J. Intern. Med. 2000;248:435–439. doi: 10.1046/j.1365-2796.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- 33.Salopuro T., Pulkkinen L., Lindstrom J., Eriksson J.G., Valle T.T., Hamalainen H., Ilanne-Parikka P., Keinanen-Kiukaanniemi S., Tuomilehto J., Laakso M., et al. Genetic variation in leptin receptor gene is associated with type 2 diabetes and body weight: The Finnish Diabetes Prevention Study. Int. J. Obes. (Lond.) 2005;29:1245–1251. doi: 10.1038/sj.ijo.0803024. [DOI] [PubMed] [Google Scholar]
- 34.Lakka T.A., Rankinen T., Weisnagel S.J., Chagnon Y.C., Lakka H.M., Ukkola O., Boule N., Rice T., Leon A.S., Skinner J.S., et al. Leptin and leptin receptor gene polymorphisms and changes in glucose homeostasis in response to regular exercise in nondiabetic individuals: the HERITAGE family study. Diabetes. 2004;53:1603–1608. doi: 10.2337/diabetes.53.6.1603. [DOI] [PubMed] [Google Scholar]
- 35.Paracchini V., Pedotti P., Taioli E. Genetics of leptin and obesity: a HuGE review. Am. J. Epidemiol. 2005;162:101–114. doi: 10.1093/aje/kwi174. [DOI] [PubMed] [Google Scholar]
- 36.Heo M., Leibel R.L., Boyer B.B., Chung W.K., Koulu M., Karvonen M.K., Pesonen U., Rissanen A., Laakso M., Uusitupa M.I., et al. Pooling analysis of genetic data: the association of leptin receptor (LEPR) polymorphisms with variables related to human adiposity. Genetics. 2001;159:1163–1178. doi: 10.1093/genetics/159.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinton N.D., Lee A.J., Ross R.J., Eastell R., Blakemore A.I. A single nucleotide polymorphism (SNP) in the leptin receptor is associated with BMI, fat mass and leptin levels in postmenopausal Caucasian women. Hum. Genet. 2001;108:233–236. doi: 10.1007/s004390100468. [DOI] [PubMed] [Google Scholar]
- 38.Heidemann C., Sun Q., van Dam R.M., Meigs J.B., Zhang C., Tworoger S.S., Mantzoros C.S., Hu F.B. Total and high-molecular-weight adiponectin and resistin in relation to the risk for type 2 diabetes in women. Ann. Intern. Med. 2008;149:307–316. doi: 10.7326/0003-4819-149-5-200809020-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korn J.M., Kuruvilla F.G., McCarroll S.A., Wysoker A., Nemesh J., Cawley S., Hubbell E., Veitch J., Collins P.J., Darvishi K., et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peiffer D.A., Le J.M., Steemers F.J., Chang W., Jenniges T., Garcia F., Haden K., Li J., Shaw C.A., Belmont J., et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Petersen K.F., Oral E.A., Dufour S., Befroy D., Ariyan C., Yu C., Cline G.W., DePaoli A.M., Taylor S.I., Gorden P., et al. Leptin reverses insulin resistance and hepatic steatosis in patients with severe lipodystrophy. J. Clin. Invest. 2002;109:1345–1350. doi: 10.1172/JCI15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aulchenko Y.S., Ripke S., Isaacs A., van Duijn C.M. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.