Abstract
P-selectin and intercellular adhesion molecule-1 (ICAM-1) participate in inflammatory processes by promoting adhesion of leukocytes to vascular wall endothelium. Their soluble levels have been associated with adverse cardiovascular events. To identify loci affecting soluble levels of P-selectin (sP-selectin) and ICAM-1 (sICAM-1), we performed a genome-wide association study in a sample of 4115 (sP-selectin) and 9813 (sICAM-1) individuals of European ancestry as a part of The Cohorts for Heart and Aging Research in Genome Epidemiology consortium. The most significant SNP association for sP-selectin was within the SELP gene (rs6136, P = 4.05 × 10−61) and for sICAM-1 levels within the ICAM-1 gene (rs3093030, P = 3.53 × 10−23). Both sP-selectin and sICAM-1 were associated with ABO gene variants (rs579459, P = 1.86 × 10−41 and rs649129, P = 1.22 × 10−15, respectively) and in both cases the observed associations could be accounted for by the A1 allele of the ABO blood group. The absence of an association between ABO blood group and platelet-bound P-selectin levels in an independent subsample (N = 1088) from the ARIC study, suggests that the ABO blood group may influence cleavage of the P-selectin protein from the cell surface or clearance from the circulation, rather than its production and cellular presentation. These results provide new insights into adhesion molecule biology.
INTRODUCTION
Cardiovascular diseases (CVD), including coronary heart disease, stroke and the renal and peripheral complications of diabetes are leading causes of morbidity and mortality in the USA and elsewhere (1). Inflammation has a critical role in their pathogenesis (2–4). During vascular inflammation, leukocytes attach to and migrate into the vascular wall. This crucial step is mediated by adhesion molecules, which are expressed on leukocytes and vascular endothelial cells in response to inflammatory stimuli (5,6).
Adhesion molecules are found in two forms, membrane-bound and soluble. The membrane bound forms mediate leukocyte attachment and internalization (7). The soluble form arises as a result of shedding or enzymatic cleavage of the membrane bound form, and in some cases as alternatively spliced molecules lacking the transmembrane domain (8). The precise proteolytic mechanism responsible for cleavage is not fully understood. The soluble form may inhibit additional leukocyte adhesion thus having a modulating effect on leukocyte recruitment (9,10). Although there are limited data available, it is thought that soluble levels of the adhesion molecules reflect differences in cellular levels (11). Soluble levels of multiple adhesion molecules, including P-selectin and intercellular adhesion molecule-1 (sICAM-1), have been associated with coronary heart disease (12,13) and other cardiovascular conditions (14–16).
P-selectin is a member of the selectin family of adhesion molecules and is expressed mainly at the surface of platelets and endothelial cells. It promotes leukocyte rolling and mediates interactions of leukocytes and platelets with the endothelium (17). ICAM-1 is a member of the immunoglobulin gene superfamily of adhesion molecules and is expressed on leukocytes, fibroblasts, epithelial cells and endothelial cells (18). Soluble forms of these two molecules, sP-selectin and sICAM-1 have high heritabilities (45–70 and 34–59%, respectively; 19–23), suggesting a strong genetic influence. Linkage studies have shown evidence of quantitative trait loci for P-selectin levels on chromosome 15 (LOD = 3.8), chromosome 12 (LOD = 2.6) (24) and chromosome 1 (LOD = 1.73) (22). ICAM-1 levels have shown linkage to the ICAM gene cluster on chromosome 19 (25,26). In a large-scale genomic study in 6578 women, multiple SNPs within the ICAM gene and a SNP in the ABO locus were associated with plasma sICAM-1 concentrations (21). No similar studies have been carried out on sP-selectin levels.
The Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) was established as a consortium to identify genetic variants influencing CVD and its risk factors (27). In the framework of CHARGE, we present the results of a genome-wide association study (GWAS) on a full HapMap set of ∼2.5 million SNPs for both sICAM-1 and sP-selectin levels in a sample of 9813 (sICAM-1) and 4115 (sP-selectin) individuals of European ancestry. For significant loci obtained from sP-selectin GWAS, we also examine the association with cellular P-selectin on platelets in an independent sample of 1,088 individuals of European ancestry from Atherosclerosis Risk in Communities (ARIC) study.
RESULTS
The total study sample for the sP-selectin studies consisted of 4115 individuals. Mean age ranged from 56.6 years in ARIC to 69.4 years in Rotterdam Study (RS) and percentage of women from 35.7% in ARIC to 48.8% in RS. Mean sP-selectin levels ranged from 31.6 ng/ml in RS to 43.9 ng/ml in ARIC. Additional characteristics of the three study samples involved in the sP-selectin analysis are provided in Supplementary Material, Table S1a.
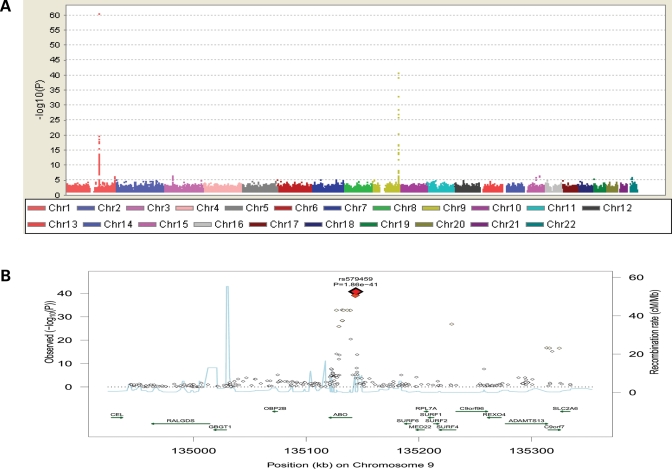
Results of the meta-analysis are shown in Figure 1A. The most significant association was observed in a region on chromosome 1 encompassing the P-selectin precursor gene (SELP; Supplementary Material, Fig. S1a). Apart from SELP, this region also contains the coagulation factor V precursor gene (F5). More than 100 SNPs in this region were above the pre-specified genome-wide significance level (5 × 10−8). The most statistically significant association was detected at rs6136 located in exon 13 of the SELP gene (MAF = 10.3%, P = 4.05 × 10−61) (Table 1). This SNP is a missense variation changing a Threonine to a Proline, and is in complete LD (r2 = 1) with the second most significant SNP, rs9332575 (MAF = 10.3%, P = 4.23 × 10−61), located in intron 5 of the F5 gene. Rs6136 and rs9332575 are in weak LD (r2 < 0.2) with the remaining genome-wide significant SNPs in the region. The other significant SNPs form two almost perfect LD blocks (Supplementary Material, Fig. S1b). Rs760694 (MAF = 46.7%, P = 2.60 × 10−19) is the most significant well-imputed SNP in the left block (3′ end) and rs2235302 (MAF = 48.01%, P = 3.95 × 10−16) is the most significant SNP in the right block (5′ end). After including in the same model all three SNPs representing three distinct LD blocks (rs6136, rs760694 and rs2235302), rs6136 and rs2235302 remained significantly associated with sP-selectin levels (P = 1.26 × 10−41, P = 2.34 × 10−12, respectively). The significance of rs760694 did not reach genome-wide threshold ( P = 6.80 × 10−4).
Figure 1.
(A) Genome wide association plot for sP-selectin; (B) Regional plot of ABO locus for association with sP-selectin. Top SNP and corresponding P-value are indicated.
Table 1.
GWAS results for sP-selectin
| SNP | Chr | Position | Gene | Coded allele | Meta-analysis | ARIC (N = 673) | FHS (N = 3036) | RS (N = 406) | |
|---|---|---|---|---|---|---|---|---|---|
| rs6136 | 1 | 167830575 | SELP | T | P-value | 4.05 × 10−61 | 2.7 × 10−3 | 2.33 × 10−59 | 0.002 |
| βa | 0.226 | 0.358 | 0.242 | 0.117 | |||||
| SE | 0.014 | 0.119 | 0.015 | 0.037 | |||||
| R2 b | 0.8 | 7.9 | 2.2 | ||||||
| rs760694 | 1 | 167835322 | SELP | T | P-value | 2.60 × 10−19 | 0.001 | 3.24 × 10−17 | 0.054 |
| βa | 0.077 | 0.143 | 0.08 | 0.043 | |||||
| SE | 0.009 | 0.044 | 0.01 | 0.022 | |||||
| R2 b | 1.2 | 2.2 | 0.9 | ||||||
| rs2235302 | 1 | rs2235302 | SELP | T | P-value | 3.95 × 10−16 | 0.020 | 8.01 × 10−14 | 0.01 |
| βa | −0.073 | −0.109 | −0.074 | −0.058 | |||||
| SE | 0.009 | 0.047 | 0.01 | 0.024 | |||||
| R2 b | 0.9 | 1.8 | 1.3 | ||||||
| rs579459 | 9 | 135143989 | ABO | T | P-value | 1.86 × 10−41 | 0.00001 | 3.29 × 10−33 | 3.61 × 10−6 |
| βa | 0.140 | 0.223 | 0.139 | 0.125 | |||||
| SE | 0.010 | 0.051 | 0.012 | 0.027 | |||||
| R2 b | 2.7 | 4.6 | 4.7 |
aβ calculated using log-transformed sP-selectin values.
bPercentage of sP-selectin variance explained by the SNP reported.
The second region with genome-wide significant signals for sP-selectin levels was observed on chromosome 9, where rs579459 (MAF = 21.8%, P = 1.86 × 10−41) achieved the most significant P-value. Rs579459 is located ∼4 kb from the 5′ end of ABO blood group gene (Fig. 1B). In total, 30 SNPs reached genome-wide significance in this region, 18 of which were located within the ABO gene. After conditioning on rs579459, none of the variants was significantly associated with sP-selectin.
No other region contained genome-wide significant signals. However, applying the less stringent threshold of P < 4 × 10−7 (expecting one false positive per 2.5 million SNPs), one additional region on chromosome 3 emerged (Supplementary Material, Fig. S1c). In this region the most significant SNP, rs11711824 (MAF = 41.37%, P = 3.72 × 10−7), is located in intron 6 of SCAP—a gene encoding a protein involved in regulation of sterol biosynthesis. However, SNPs with very similar level of significance (P < 10−6) are dispersed across a broad region containing several genes—PTPN23 (protein-tyrosine phosphatase), KIF9 (kinesin family member 9) and TMEM103 (transmembrane protein) (Supplementary Material, Fig. S1c).
The above GWAS results reflect an association with sP-selectin levels that are readily measurable in large cohort studies. However, the more biologically active molecule is thought to be the membrane bound form (7). Therefore, we examined four SNPs from the two regions significantly associated with sP-selectin (rs6136, rs760694, rs2235302, rs579459) in relation to membrane-bound platelet P-selectin (the proportion of cells expressing P-selectin and the average fluorescence of P-selectin on platelets) as measured by flow cytometry in the ARIC carotid MRI substudy (N = 1088, see Materials and Methods). Out of three SELP gene SNPs (rs6136, rs760694 and rs2235302) that were significantly associated with sP-selectin, rs6136 was also associated with platelet P-selectin (P = 0.01 with average fluorescence of P-selectin on platelets and P = 0.02 with the proportion of cells expressing P-selectin). The ABO SNP, rs579459, was not associated with P-selectin on platelets (Table 2).
Table 2.
Association of platelet P-selectin (proportion of cells expressing P-selectin and P-selectin expression assessed by MFI) with three SNPs in SELP and one in ABO associated with sP-selectin
| SNP | Gene | Proportion of cells expressing P-selectin |
P-selectin expression assessed by MFI |
||||
|---|---|---|---|---|---|---|---|
| β | SE | P | β | SE | P | ||
| rs6136 | SELP | 4.55 | 2.01 | 0.02 | 1.31 | 0.53 | 0.01 |
| rs760694 | SELP | 0.13 | 0.73 | 0.86 | −0.01 | 0.22 | 0.95 |
| rs2235302 | SELP | 0.82 | 0.74 | 0.27 | −0.08 | 0.20 | 0.69 |
| rs579459 | ABO | −1.18 | 0.78 | 0.13 | −0.01 | 0.22 | 0.98 |
MFI, median flourescence intensity.
Four of the CHARGE cohorts measured plasma sICAM-1 levels, providing a total sample of 9813 individuals. Mean age ranged from 49.4 years in Framingham Heart Study (FHS) to 72.8 years in Cardiovascular Health Study (CHS), and the percent of women ranged from 38.4% in ARIC to 53.3% in RS. sICAM-1 levels ranged from 222.7 ng/ml in RS to 329.1 ng/ml in CHS. Additional sample characteristics are presented in Supplementary Material, Table S1b.
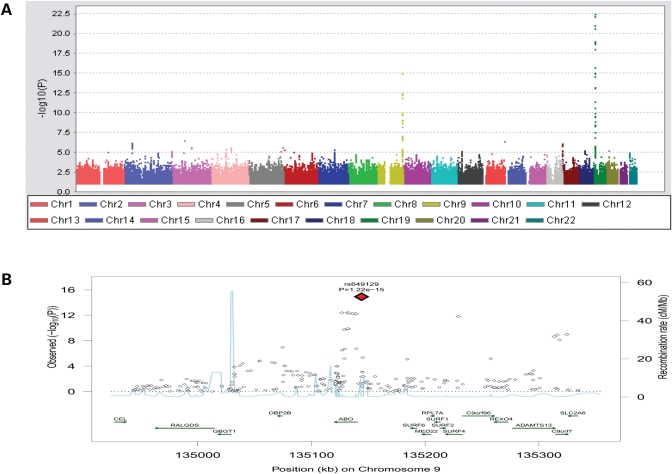
The meta-analysis for the genome-wide association analysis of sICAM-1 levels are shown in Figure 2A. The strongest signal was on chromosome 19 in the region of the ICAM genes cluster (Table 3 and Supplementary Material, Fig. S2). The most significant association was at SNP rs3093030 (MAF = 41.72%, P = 3.53 × 10−23), located ∼100 bp from the 3′ end of ICAM-1 gene. It is located near to previously described non-synonymous coding SNPs, rs5498 (Lys469Glu) and rs1799969 (Gly241Arg). Rs3093030 is in high LD with rs5498 (r2 = 0.98) and in weak LD with rs1799969 (r2 = 0.18). rs5498 and rs1799969 were less significantly associated with sICAM-1 levels than rs3093030 (rs5498, P = 2.5 × 10−21; rs1799969, P = 2.8 × 10−6). In order to determine if rs1799969 is independently associated with sICAM-1 levels from rs3093030, we performed an analysis including both rs1799969 and rs3093030 in the model. Results showed strong independent association of both SNPs (β = 0.126; P = 1.344 × 10−43 for rs1799969 and β = 0.079; P = 1.038 × 10−56 for rs3093030).
Figure 2.
(A) Genome wide association plot for sICAM-1; (B) Regional plot of ABO locus for association with sICAM-1. Top SNP and corresponding P-value are indicated.
Table 3.
GWAS results for sICAM-1
| SNP | Chr | Position | Gene | Coded allele | Meta-analysis | ARIC (N = 829) | FHS (N = 6845) | CHS (N = 1487) | RS (N = 600) | |
|---|---|---|---|---|---|---|---|---|---|---|
| rs3093030 | 19 | 10258403 | ICAM | T | P | 3.53 × 10−23 | 0.011 | 6.24 × 10–19 | 0.00046 | 0.107 |
| βa | 0.0415 | 0.031 | 0.045 | 0.042 | 0.027 | |||||
| SE | 0.0042 | 0.012 | 0.005 | 0.012 | 0.017 | |||||
| R2 b | 0.8 | 1.2 | 1.1 | 0.3 | ||||||
| rs649129 | 9 | 135144125 | ABO | T | P | 1.22 × 10−15 | 0.032 | 2.24 × 10–12 | 0.010 | 0.040 |
| βa | −0.0395 | −0.030 | −0.041 | −0.042 | −0.039 | |||||
| SE | 0.0049 | 0.014 | 0.006 | 0.016 | 0.019 | |||||
| R2 b | 0.5 | 0.8 | 0.6 | 0.5 |
aβ calculated using log-transformed sICAM-1 values.
bPercentage of sICAM-1 variance explained by the SNP reported.
The second region reaching genome-wide significance was in the ABO gene region (Fig. 2B). Rs649129, a SNP in complete LD with rs579459, the most associated one with sP-selectin levels (see above), provided the highest significance level for sICAM-1 levels (P = 1.22 × 10−15). In total, 20 SNPs within this region were above the genome-wide significance level. When viewed together, the association plots for sICAM-1 and sP-selectin in the ABO region overlap, with the only difference being the level of statistical significance (Figs 1B and 2B). The correlation between sICAM-1 and sP-selectin levels was 0.19 in FHS. After adjusting sP-selectin levels for sICAM-1, the ABO SNP rs579459 remained significantly associated with sP-selectin levels (P = 1.62 × 10−28; proportional change in effect size = 7%). After adjusting sICAM-1 for sP-selectin, the same SNP is associated with sICAM with P = 7.98 × 10−4 (with proportional change in effect size of 31%).
We next sought to investigate whether the observed association of the ABO locus with sICAM and sP-selectin could be accounted for by the well-known ABO blood group variants (A1, A2, B and O). The ABO blood group alleles in ARIC and FHS were estimated using halotypes formed out of 3 ABO SNPs—rs8176749, rs8176704 and rs687289 (Table 4). None of these three SNPs are in high LD with the significant GWAS ABO SNP—rs579459 (r2 = 0.02; 0.02 and 0.40, respectively). The estimated ABO blood group allele frequencies are shown in Table 4 and reflect their expected frequencies in populations of European descent. The A1 allele was negatively associated with both traits (meta-analysis of FHS and ARIC results: β =− 0.20, P = 3.88 × 10−46 for sP-selectin; β =− 0.06, P = 2.89 × 10−18 for sICAM-1). The β estimates were higher in models with A1 allele than in those with GWAS ABO hit SNP (rs579459) for both sP-selectin and sICAM. Moreover, the significant association of sP-selectin and sICAM-1 levels with the GWAS SNP rs579459 was no longer significant after inclusion in the model with the ABO blood group alleles. On the other hand, the A1 allele remained significant (Table 4, P-selectin and ICAM). The A2 allele showed the same (negative) direction of effect on sP-selectin and sICAM-1, but the association was only significant for sP-selectin levels. In contrast, none of the ABO blood group alleles were associated with P-selectin on platelets (Table 4, Platelet P-selectin).
Table 4.
Estimated ABO allele frequencies in ARIC and FHS cohorts; the association results of ABO alleles with sP-selectin; sICAM and P-selectin bound on platelets (all analyses are adjusted for age and sex)
| ABO frequencies | |||||||||
| ABO allele | Haplotype | ARIC | FHS | ||||||
| rs8176749 | rs8176704 | rs687289 | |||||||
| O | C | G | G | 0.650 | 0.673 | ||||
| A1 | C | G | A | 0.209 | 0.196 | ||||
| B | T | G | A | 0.073 | 0.077 | ||||
| A2 | C | A | A | 0.068 | 0.054 | ||||
| sP-selectin | |||||||||
| ARIC (N = 653) | FHS (N = 3048) | Meta-analysis | |||||||
| β | SE | P | β | SE | P | β | SE | P | |
| A1 | −0.358 | 0.127 | 0.005 | −0.170 | 0.027 | 2.7E−10 | −0.178 | 0.026 | 1.8E−11 |
| A2 | −0.032 | 0.092 | 0.73 | −0.062 | 0.022 | 0.005 | −0.060 | 0.021 | 0.005 |
| B | 0.042 | 0.090 | 0.64 | −0.015 | 0.019 | 0.421 | −0.013 | 0.019 | 0.499 |
| rs579459 | −0.029 | 0.102 | 0.775 | −0.019 | 0.023 | 0.388 | −0.019 | 0.022 | 0.385 |
| sICAM-1 | |||||||||
| ARIC (N = 829) | FHS (N = 6897) | Meta-analysis | |||||||
| β | SE | P | β | SE | P | β | SE | P | |
| A1 | −0.080 | 0.033 | 0.018 | −0.056 | 0.013 | 1.6E−05 | −0.059 | 0.012 | 9.9E−07 |
| A2 | −0.007 | 0.023 | 0.75 | −0.017 | 0.011 | 0.12 | −0.015 | 0.010 | 0.127 |
| B | 0.042 | 0.025 | 0.10 | 0.0003 | 0.009 | 0.97 | 0.005 | 0.008 | 0.548 |
| rs579459 | −0.029 | 0.027 | 0.29 | −0.0006 | 0.011 | 0.96 | −0.004 | 0.010 | 0.649 |
| Platelet P-selectin | |||||||||
| ARIC (N = 1088) | |||||||||
| Proportion of cells expressing P-selectin | P-selectin expression assessed by MFI | ||||||||
| β | SE | P | β | SE | P | ||||
| A1 | −0.355 | 2.152 | 0.87 | 0.64 | 0.98 | 0.52 | |||
| A2 | 0.831 | 1.509 | 0.58 | −0.32 | 0.41 | 0.43 | |||
| B | 1.337 | 1.445 | 0.36 | 0.61 | 0.86 | 0.45 | |||
| rs579459 | −1.621 | 1.540 | 0.29 | −0.15 | 0.65 | 0.82 | |||
DISCUSSION
We present here the results of meta-analyses of GWAS results, performed in the CHARGE consortium, that identified significant and replicated associations of sICAM-1 and sP-selectin levels with variants in the genes encoding these proteins, ICAM-1 and SELP, respectively. In addition, both markers were associated with variants in the well-known ABO blood group locus, which encodes glycosyltransferase enzymes that transfer sugar residues to the H antigen and determine an individual's blood group (28).
The observed association of sICAM-1 and sP-selectin with ABO variants could be explained by the A1 blood group allele, that determines A blood group. The other A blood group allele, A2, was also associated with decreased levels of sP-selectin, however, with much lower level of significance. Both A1 and A2 alleles encode for the same form of glycosyltransferase that adds the N-acetylgalactosamine to the H antigen, however, with lower activity in case of A2 allele (29). These findings suggest a possible role of glycosylation in ABO's association with sP-selectin and sICAM levels.
There was no observed association between the ABO locus with platelet-bound P-selectin although the flow cytometry analysis was limited to P-selectin on platelets, and does not rule out the possibility that endothelial P-selectin levels may be influenced by the ABO locus. Soluble forms of P-selectin arise in part from shedding or active proteolytic cleavage of membrane molecules (8,15,30). It is known that both soluble and cellular forms of ICAM-1 (31) and probably of P-selectin are glycosylated. The glycosylation is also critical for the binding activity of the P-selectin receptor, P-selectin glycoprotein ligand-1 (32). The observed association of ABO blood group markers with sP-selectin and sICAM-1 and not platelet-bound P-selectin suggests that the ABO gene product related glycosylation influences shedding/cleavage of these markers from the endothelium, probably by glycosylation of P-selectin and ICAM-1. Glycosylation could also affect the clearance rate of sP-selectin and sICAM-1 from blood as has been suggested for von Willebrand factor (vWF), a large circulating glycoprotein involved in hemostasis (33,34), whose levels are also influenced by the ABO locus (34). It is interesting that both P-selectin and vWF are stored in the same granules, Weibel-Palade bodies, prior to being released on endothelial cells (35,36). Moreover, the vWF receptor, glycoprotein (GP)Ibα, has been shown to act as a counter-receptor for P-selectin (37). vWF carries N-linked ABO antigens, and it has been suggested that vWF levels vary according to vWF glycosylation that influence its secretion and/or clearance rate (34). The similarity in P-selectin and vWF physiology underscores the inter-related nature of hemostatic and inflammation responses, both of related to CVD risk and now reported to be influenced by the ABO blood group locus. In a recent GWA study, variation in ABO locus was also associated with serum soluble E-selectin levels (38)—another member of selectin family of adhesion molecules.
Additional insights may come from ABO's association with malaria infection susceptibility and severity (39,40). Recent studies of Plasmodium falciparum infection show that the infected erythrocytes mimic the leukocytes’ attachment to endothelium by binding to ICAM-1 and P-selectin on endothelium (41). Erythrocytes also bind to the A and B antigens (but not O) on endothelial cells as an additional contributor to cytoadherence (42). This functional homology suggests the hypothesis that leukocytes might also interact with ABO antigens on endothelium. The A antigen might promote stronger (longer) binding of leukocytes to P-selectin and ICAM-1 on the vascular wall, thereby protecting the protein from enzymatic cleavage which in turn would lead to decreased levels in circulation. Decreased cleavage of adhesion molecules from endothelial cells associated with A allele would mean more adhesion molecules on the endothelial cells, increased adhesion and inflammation (21). This corresponds well with the findings of studies relating ABO blood group alleles with CVD that have been of interest for many years (43–45) and most of which suggested A allele association with increased risk of CVD (46). However, multiple epidemiologic studies have reported that increased levels of sP-selectin and sICAM-1 are associated with increased risk of CVD (12,15). Consequently, the A allele's association with decreased levels of sICAM-1 and sP-selectin but increased risk of CVD seems as a paradox and underscores the complex nature of CVD, its risk factors and their interrelationships (21).
Apart from ABO, sP-selectin and sICAM-1 levels are largely determined by variants within the genes encoding the two biomarker proteins, SELP and ICAM-1. The strong association of rs6136 with sP-selectin has been previously reported by ourselves (22,47) and others (13,20,47). Although heterogeneity test for any of the top SNPs in our sP-selectin and sICAM-1 meta-analyses was not statistically significant after accounting for multiple comparison, the I2 statistic (percentage of total variation across studies that is due to heterogeneity) for rs6136 was high (82%). This high level of heterogeneity in our meta-analysis for this SNP should be investigated further. rs6136 (Thr715Pro) has a functional effect of changing the conformation of P-selectin near the cleavage site (22). Besides Thr715Pro, the P-selectin gene is characterized by 2 LD blocks that are additionally associated with sP-selectin levels. Another SELP variant, Val599Leu (rs6133), reported to be associated with sP-selectin in previous studies (13,20), was not in high LD with any of the 3 LD blocks and did not reach genome-wide significance in our study. In total, the three SELP regions accounted for 9.3% of the variance of sP-selectin levels in the FHS sample, the largest in our study. However, only Thr715Pro remained significantly associated with platelets P-selectin, a result that has already been reported (48). The lack of association of the other two SNPs could be explained by smaller sample size and insufficient power to detect the association for platelet P-selectin. Alternatively, it is also possible that different variants contribute to cellular and soluble P-selectin levels.
We report here suggestive evidence for a novel locus influencing soluble P-selectin levels on chromosome 3. The region contains SCAP (SREBP cleavage-activating protein), PTPN23 (protein-tyrosine phosphatase), KIF9 (kinesin family member 9) and TMEM103 (transmembrane protein). SCAP cleaves the active domain of SREBP (sterol regulatory element binding protein) which acts as a transcription factor and participates in cellular cholesterol homeostasis (49,50). Given SCAP variants association with sP-selectin, it could be hypothesized that SCAP is also capable of cleaving P-selectin from cell membrane. However, SCAP is only known for its intracellular activities and its extracellular presence is doubtful.
The top SNP associated with ICAM-1 levels, rs3093030, is in the 3′ end of the ICAM- 1 gene and is in high LD with rs5498 (Lys469Glu) that causes an amino-acid change from glutamic acid to lysine and is located in the fifth Ig-like domain of ICAM-1 (51). Lys469Glu has previously been reported to influence sICAM-1 levels (21,51,52). This variant influences the dimerization of the protein which promotes enhanced binding to leukocytes (53). Another variant in ICAM-1 gene (rs1799969 or Gly241Arg) is independently associated with ICAM-1 levels. This variant changes the amino-acid in the third immunoglobulin domain of ICAM-1 protein and has been shown to be of importance in binding to the leukocyte integrin Mac-1 (52,54).
Although this study was well-suited to identify loci with common variation associated with sP-selectin and sICAM-1 levels detectable by GWAS in large samples, it was limited to European ancestry population and its generalizability to other ethnic groups remains to be investigated. As with other GWAS, we were able to narrow down the association of sP-selectin and sICAM-1 to a particular genomic region, but further analyses, beginning with DNA resequencing in large well-phenotyped cohorts, will be necessary to detect functional variants within those genes. Future projects with larger data sets will be required to examine any gene–gene and gene–environment interactions and the identification of additional variants that may have been undetected in this study. Finally, the possible heterogeneity of the effect of rs6136 on sP-selectin levels is a limitation and deserves further attention.
In summary, this study suggests the importance of the ABO blood group in influencing circulating sP-selectin and sICAM-1 levels. The probable mechanism involves glycosylation that interacts with P-selectin/ICAM-1 shedding from the cell membrane. These results contribute to the knowledge of the adhesion molecules physiology that is necessary to understand the link between the inflammation and atherosclerotic process and the development of new anti-inflammatory therapies for cardiovascular complications.
MATERIALS AND METHODS
The CHARGE consortium cohorts (27) included in this study are the ARIC study, CHS, FHS and RS. All participants gave written informed consent for study participation and for use of DNA for genetic research. The Institutional Review Board of each cohort approved the study protocols. All participants in this study are of European ancestry. Detailed descriptions of the participating cohorts are provided in the Supplemental Methods.
Laboratory measurements
sP-selectin and sICAM-1 concentrations were determined by standard ELISA methods. EDTA plasma was used for sP-Selectin in ARIC, FHS and RS, and for sICAM-1 in ARIC, CHS and RS; serum was used for sICAM-1 in FHS. Detailed laboratory methods for each cohort are provided in the Supplemental Methods.
Platelet P-selectin analysis
Flow cytometry was performed on a subsample of the ARIC study, as part of the ARIC Carotid MRI ancillary study that was conducted between 2004 and 2005. Participants were selected from the ARIC cohort based on results of their last carotid artery ultrasound examinations (Visits 3 and 4, 1993–1998) (55). For this study we used the data for the proportion of cells expressing P-selectin and the relative level of P-selectin expression assessed by median fluorescence intensity (MFI). The procedures have been described previously elsewhere in more details (56) and in the Supplemental Methods. We included 1088 individuals with platelet P-selectin measures and genotypes in this study. All individuals were of European ancestry.
ABO blood group allele determination and analysis
The ABO blood group alleles were estimated using the genetic data available in ARIC and FHS cohorts. We assessed the ABO blood group alleles by constructing haplotypes out of three SNPs (rs8176749, rs8176704, rs687289) within the ABO locus (21). Four common haplotypes corresponded to four ABO blood group alleles (A1, A2, B and O). The association analyses of sP-selectin and sICAM with ABO blood group alleles (O is taken as a reference) were performed in ARIC and FHS and the results were combined using inverse-variance weighted meta-analysis.
Genotyping
Genotyping was done in each cohort separately using high-density SNP marker platforms (ARIC—Affymetrix 6.0, CHS—Illumina 370, FHS—Affymetrix 500K, RS—Illumina Infinium HumanHap550) and then imputed to ∼2.5 million HapMap SNPs by using either MACH (57) (ARIC, FHS and RS) or BIMBAM (58) (CHS) software. Detailed information on genotyping and imputation in each cohort are provided in the Supplemental Methods.
Statistical analysis
Association studies of sP-selectin and sICAM-1 concentrations were done separately for each cohort. Concentration values were natural log-transformed before analyses. For the primary analysis, linear regression of concentration on (additive) allele dosage adjusted for age and sex was performed. In CHS, sICAM-1 was measured on a case-cohort subset and the appropriate sampling weights were used in the primary analysis to account for the sampling design. In ARIC, sP-selectin and sICAM-1 were measured in multiple nested case-cohorts samples and for those SNPs reaching genome-wide significance in the meta-analysis, the analyses were repeated with appropriate weights accounting for the sampling design. The results were similar to the unweighted analyses and therefore the results of unweighted analyses are shown throughout. In FHS, which consists of extended families, a linear mixed effects model was employed with a fixed additive effect for the SNP genotype, fixed covariate effects and random family specific additive residual polygenic effects to account for within family correlations. The cohort-specific results were combined using inverse-variance weighted meta-analysis with fixed effect models, as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html). Prior to meta-analysis, imputed SNPs with quality score (ratio of observed to expected variance) less than 0.3 were excluded. To test for heterogeneity between the samples used in the meta-analysis, we performed the Cochran's Q statistic using METAL and calculated I2 statistic that describes the percentage of total variation across studies that is due to heterogeneity (59). Finally, we report as ‘genome-wide significant’ all associations with P < 5 × 10−8. As an alternative criterion, we also applied a threshold of P < 4 × 10−7, that represents an expectation of one false positive result per 2.5 million tests. All studies had a genomic control parameter ≤1.05. For follow-up studies of platelet P-selectin, weighted analysis was performed on the inverse of the sampling fractions in the sampling strata used in the ARIC carotid MRI substudy. Sex and age were added as covariates in the model. The analysis was done using the R software's ‘survey’ package (60).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
ARIC: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. CHS: The research reported in this article was supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant number U01 HL080295 and R01 HL087652 from the National Heart, Lung, and Blood Institute, with additional contribution from the National Institute of Neurological Disorders and Stroke. Genotyping was supported by the Cedars-Sinai General Clinical Research Center grant M01-RR00425 and NIDDK grant DK063491 to the Southern California Diabetes Endocrinology Research Center. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. FHS: This research was conducted in part using data and resources from the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. This work was partially supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). A portion of this research utilized the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The work was also supported in part by grants from the National Institutes of Health HL076784, AG028321 and HL064753 to Dr Benjamin. RS: The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research; the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; The Netherlands Heart Foundation; the Ministry of Education, Culture and Science; the Ministry of Health Welfare and Sports; the European Commission; and the Municipality of Rotterdam. Support for genotyping was provided by the Netherlands Organization for Scientific Research (NWO) (175.010.2005.011, 911.03.012) and Research Institute for Diseases in the Elderly (RIDE). This study was further supported by the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project nr. 050-060-810.
Supplementary Material
REFERENCES
- 1.Rosamond W., Flegal K., Friday G., Furie K., Go A., Greenlund K., Haase N., Ho M., Howard V., Kissela B., et al. Heart disease and stroke statistics-2007 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2007;115:e69–e171. doi: 10.1161/CIRCULATIONAHA.106.179918. [DOI] [PubMed] [Google Scholar]
- 2.Tracy R.P. Emerging relationships of inflammation, cardiovascular disease and chronic diseases of aging. Int. J. Obes. Relat. Metab. Disord. 2003;27(Suppl. 3):S29–S34. doi: 10.1038/sj.ijo.0802497. [DOI] [PubMed] [Google Scholar]
- 3.Hansson G.K. Inflammation, atherosclerosis, and coronary artery disease. N. Engl. J. Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 4.Fornage M., Chiang Y.A., O'Meara E.S., Psaty B.M., Reiner A.P., Siscovick D.S., Tracy R.P., Longstreth W.T., Jr Biomarkers of inflammation and MRI-defined small vessel disease of the brain: the cardiovascular health study. Stroke. 2008;39:1952–1959. doi: 10.1161/STROKEAHA.107.508135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Springer T.A. Adhesion receptors of the immune system. Nature. 1990;346:425–434. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 6.Blankenberg S., Barbaux S., Tiret L. Adhesion molecules and atherosclerosis. Atherosclerosis. 2003;170:191–203. doi: 10.1016/s0021-9150(03)00097-2. [DOI] [PubMed] [Google Scholar]
- 7.Ley K., Laudanna C., Cybulsky M.I., Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 8.Ishiwata N., Takio K., Katayama M., Watanabe K., Titani K., Ikeda Y., Handa M. Alternatively spliced isoform of P-selectin is present in vivo as a soluble molecule. J. Biol. Chem. 1994;269:23708–23715. [PubMed] [Google Scholar]
- 9.Wong C.S., Gamble J.R., Skinner M.P., Lucas C.M., Berndt M.C., Vadas M.A. Adhesion protein GMP140 inhibits superoxide anion release by human neutrophils. Proc. Natl. Acad. Sci. USA. 1991;88:2397–2401. doi: 10.1073/pnas.88.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rieckmann P., Michel U., Albrecht M., Bruck W., Wockel L., Felgenhauer K. Soluble forms of intercellular adhesion molecule-1 (ICAM-1) block lymphocyte attachment to cerebral endothelial cells. J. Neuroimmunol. 1995;60:9–15. doi: 10.1016/0165-5728(95)00047-6. [DOI] [PubMed] [Google Scholar]
- 11.Leeuwenberg J.F., Smeets E.F., Neefjes J.J., Shaffer M.A., Cinek T., Jeunhomme T.M., Ahern T.J., Buurman W.A. E-selectin and intercellular adhesion molecule-1 are released by activated human endothelial cells in vitro. Immunology. 1992;77:543–549. [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang S.J., Ballantyne C.M., Sharrett A.R., Smith L.C., Davis C.E., Gotto A.M., Jr, Boerwinkle E. Circulating adhesion molecules VCAM-1, ICAM-1, and E-selectin in carotid atherosclerosis and incident coronary heart disease cases: the Atherosclerosis Risk In Communities (ARIC) study. Circulation. 1997;96:4219–4225. doi: 10.1161/01.cir.96.12.4219. [DOI] [PubMed] [Google Scholar]
- 13.Barbaux S.C., Blankenberg S., Rupprecht H.J., Francomme C., Bickel C., Hafner G., Nicaud V., Meyer J., Cambien F., Tiret L. Association between P-selectin gene polymorphisms and soluble P-selectin levels and their relation to coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2001;21:1668–1673. doi: 10.1161/hq1001.097022. [DOI] [PubMed] [Google Scholar]
- 14.Rohde L.E., Lee R.T., Rivero J., Jamacochian M., Arroyo L.H., Briggs W., Rifai N., Libby P., Creager M.A., Ridker P.M. Circulating cell adhesion molecules are correlated with ultrasound-based assessment of carotid atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 1998;18:1765–1770. doi: 10.1161/01.atv.18.11.1765. [DOI] [PubMed] [Google Scholar]
- 15.Blann A.D., Nadar S.K., Lip G.Y. The adhesion molecule P-selectin and cardiovascular disease. Eur. Heart. J. 2003;24:2166–2179. doi: 10.1016/j.ehj.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Tzoulaki I., Murray G.D., Lee A.J., Rumley A., Lowe G.D., Fowkes F.G. C-reactive protein, interleukin-6, and soluble adhesion molecules as predictors of progressive peripheral atherosclerosis in the general population: Edinburgh Artery Study. Circulation. 2005;112:976–983. doi: 10.1161/CIRCULATIONAHA.104.513085. [DOI] [PubMed] [Google Scholar]
- 17.Polgar J., Matuskova J., Wagner D.D. The P-selectin, tissue factor, coagulation triad. J. Thromb. Haemost. 2005;3:1590–1596. doi: 10.1111/j.1538-7836.2005.01373.x. [DOI] [PubMed] [Google Scholar]
- 18.Fijnheer R., Frijns C.J., Korteweg J., Rommes H., Peters J.H., Sixma J.J., Nieuwenhuis H.K. The origin of P-selectin as a circulating plasma protein. Thromb. Haemost. 1997;77:1081–1085. [PubMed] [Google Scholar]
- 19.Schnabel R.B., Lunetta K.L., Larson M.G., Dupuis J., Lipinska I., Rong J., Chen M., Zhao Z., Yamamoto J.F., Meigs J.B., et al. The relation of genetic and environmental factors to systemic inflammatory biomarker concentrations. Circ. Cardiovasc. Genet. 2009;2:229–237. doi: 10.1161/CIRCGENETICS.108.804245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner A.P., Carlson C.S., Thyagarajan B., Rieder M.J., Polak J.F., Siscovick D.S., Nickerson D.A., Jacobs D.R., Jr, Gross M.D. Soluble P-selectin, SELP polymorphisms, and atherosclerotic risk in European-American and African-African young adults: the Coronary Artery Risk Development in Young Adults (CARDIA) Study. Arterioscler. Thromb. Vasc. Biol. 2008;28:1549–1555. doi: 10.1161/ATVBAHA.108.169532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pare G., Chasman D.I., Kellogg M., Zee R.Y., Rifai N., Badola S., Miletich J.P., Ridker P.M. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee D.S., Larson M.G., Lunetta K.L., Dupuis J., Rong J., Keaney J.F., Jr, Lipinska I., Baldwin C.T., Vasan R.S., Benjamin E.J. Clinical and genetic correlates of soluble P-selectin in the community. J. Thromb. Haemost. 2008;6:20–31. doi: 10.1111/j.1538-7836.2007.02805.x. [DOI] [PubMed] [Google Scholar]
- 23.Dupuis J., Larson M.G., Vasan R.S., Massaro J.M., Wilson P.W., Lipinska I., Corey D., Vita J.A., Keaney J.F., Jr, Benjamin E.J. Genome scan of systemic biomarkers of vascular inflammation in the Framingham Heart Study: evidence for susceptibility loci on 1q. Atherosclerosis. 2005;182:307–314. doi: 10.1016/j.atherosclerosis.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Hixson J.E., Blangero J. Genomic searches for genes that influence atherosclerosis and its risk factors. Ann. N. Y. Acad. Sci. 2000;902:1–7. doi: 10.1111/j.1749-6632.2000.tb06295.x. [DOI] [PubMed] [Google Scholar]
- 25.Bielinski S.J., Pankow J.S., Foster C.L., Miller M.B., Hopkins P.N., Eckfeldt J.H., Hixson J., Liu Y., Register T., Myers R.H., et al. Circulating soluble ICAM-1 levels shows linkage to ICAM gene cluster region on chromosome 19: the NHLBI Family Heart Study follow-up examination. Atherosclerosis. 2008;199:172–178. doi: 10.1016/j.atherosclerosis.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent J.W., Jr, Mahaney M.C., Comuzzie A.G., Goring H.H., Almasy L., Dyer T.D., Cole S.A., MacCluer J.W., Blangero J. Quantitative trait locus on Chromosome 19 for circulating levels of intercellular adhesion molecule-1 in Mexican Americans. Atherosclerosis. 2007;195:367–373. doi: 10.1016/j.atherosclerosis.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psaty B., O'Donnell C.J., Gudnason V., Lunetta K.L., Folsom A.R., Rotter J.I., Uitterlinden A.G., Harris T.B., Witteman J.C.M., Boerwinkle E. on Behalf of the CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ. Cardiovasc. Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto F., Clausen H., White T., Marken J., Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto F. Molecular biology of ABO genes. Tanpakushitsu Kakusan Koso. 1992;37:1696–1700. [PubMed] [Google Scholar]
- 30.Champagne B., Tremblay P., Cantin A., St Pierre Y. Proteolytic cleavage of ICAM-1 by human neutrophil elastase. J. Immunol. 1998;161:6398–6405. [PubMed] [Google Scholar]
- 31.Otto V.I., Damoc E., Cueni L.N., Schurpf T., Frei R., Ali S., Callewaert N., Moise A., Leary J.A., Folkers G., et al. N-glycan structures and N-glycosylation sites of mouse soluble intercellular adhesion molecule-1 revealed by MALDI-TOF and FTICR mass spectrometry. Glycobiology. 2006;16:1033–1044. doi: 10.1093/glycob/cwl032. [DOI] [PubMed] [Google Scholar]
- 32.Martinez M., Joffraud M., Giraud S., Baisse B., Bernimoulin M.P., Schapira M., Spertini O. Regulation of PSGL-1 interactions with L-selectin, P-selectin, and E-selectin: role of human fucosyltransferase-IV and -VII. J. Biol. Chem. 2005;280:5378–5390. doi: 10.1074/jbc.M410899200. [DOI] [PubMed] [Google Scholar]
- 33.Souto J.C., Almasy L., Soria J.M., Buil A., Stone W., Lathrop M., Blangero J., Fontcuberta J. Genome-wide linkage analysis of von Willebrand factor plasma levels: results from the GAIT project. Thromb. Haemost. 2003;89:468–474. [PubMed] [Google Scholar]
- 34.Vischer U.M. von Willebrand factor, endothelial dysfunction, and cardiovascular disease. J. Thromb. Haemost. 2006;4:1186–1193. doi: 10.1111/j.1538-7836.2006.01949.x. [DOI] [PubMed] [Google Scholar]
- 35.Bonfanti R., Furie B.C., Furie B., Wagner D.D. PADGEM (GMP140) is a component of Weibel-Palade bodies of human endothelial cells. Blood. 1989;73:1109–1112. [PubMed] [Google Scholar]
- 36.Wagner D.D., Olmsted J.B., Marder V.J. Immunolocalization of von Willebrand protein in Weibel-Palade bodies of human endothelial cells. J. Cell. Biol. 1982;95:355–360. doi: 10.1083/jcb.95.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romo G.M., Dong J.F., Schade A.J., Gardiner E.E., Kansas G.S., Li C.Q., McIntire L.V., Berndt M.C., Lopez J.A. The glycoprotein Ib-IX-V complex is a platelet counterreceptor for P-selectin. J. Exp. Med. 1999;190:803–814. doi: 10.1084/jem.190.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paterson A.D., Lopes-Virella M.F., Waggott D., Boright A.P., Hosseini S.M., Carter R.E., Shen E., Mirea L., Bharaj B., Sun L., et al. Genome-Wide association identifies the ABO blood group as a major locus associated with serum levels of soluble E-selectin. Arterioscler. Thromb. Vasc. Biol. 2009;29:1958–1967. doi: 10.1161/ATVBAHA.109.192971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer P.R., Boone P. Short report: severe malaria associated with blood group. Am. J. Trop. Med. Hyg. 1998;58:122–123. doi: 10.4269/ajtmh.1998.58.122. [DOI] [PubMed] [Google Scholar]
- 40.Pathirana S.L., Alles H.K., Bandara S., Phone-Kyaw M., Perera M.K., Wickremasinghe A.R., Mendis K.N., Handunnetti S.M. ABO-blood-group types and protection against severe, Plasmodium falciparum malaria. Ann. Trop. Med. Parasitol. 2005;99:119–124. doi: 10.1179/136485905X19946. [DOI] [PubMed] [Google Scholar]
- 41.Ho M., Hickey M.J., Murray A.G., Andonegui G., Kubes P. Visualization of Plasmodium falciparum–endothelium interactions in human microvasculature: mimicry of leukocyte recruitment. J. Exp. Med. 2000;192:1205–1211. doi: 10.1084/jem.192.8.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cserti C.M., Dzik W.H. The ABO blood group system and Plasmodium falciparum malaria. Blood. 2007;110:2250–2258. doi: 10.1182/blood-2007-03-077602. [DOI] [PubMed] [Google Scholar]
- 43.Garrison R.J., Havlik R.J., Harris R.B., Feinleib M., Kannel W.B., Padgett S.J. ABO blood group and cardiovacular disease: the Framingham study. Atherosclerosis. 1976;25:311–318. doi: 10.1016/0021-9150(76)90036-8. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell J.R. An association between abo blood-group distribution and geographical differences in death-rates. Lancet. 1977;1:295–297. doi: 10.1016/s0140-6736(77)91838-4. [DOI] [PubMed] [Google Scholar]
- 45.Nydegger U.E., Wuillemin W.A., Julmy F., Meyer B.J., Carrel T.P. Association of ABO histo-blood group B allele with myocardial infarction. Eur. J. Immunogenet. 2003;30:201–206. doi: 10.1046/j.1365-2370.2003.00390.x. [DOI] [PubMed] [Google Scholar]
- 46.Wu O., Bayoumi N., Vickers M.A., Clark P. ABO(H) blood groups and vascular disease: a systematic review and meta-analysis. J. Thromb. Haemost. 2008;6:62–69. doi: 10.1111/j.1538-7836.2007.02818.x. [DOI] [PubMed] [Google Scholar]
- 47.Volcik K.A., Ballantyne C.M., Coresh J., Folsom A.R., Wu K.K., Boerwinkle E. P-selectin Thr715Pro polymorphism predicts P-selectin levels but not risk of incident coronary heart disease or ischemic stroke in a cohort of 14595 participants: the Atherosclerosis Risk in Communities Study. Atherosclerosis. 2006;186:74–79. doi: 10.1016/j.atherosclerosis.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Volcik K.A., Catellier D., Folsom A.R., Matijevic N., Wasserman B., Boerwinkle E. SELP and SELPLG genetic variation is associated with cell surface measures of SELP and SELPLG: The Atherosclerosis Risk in Communities (ARIC) carotid MRI study. Clin. Chem. 2009;55:1076–1082. doi: 10.1373/clinchem.2008.119487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima T., Hamakubo T., Kodama T., Inazawa J., Emi M. Genomic structure and chromosomal mapping of the human sterol regulatory element binding protein (SREBP) cleavage-activating protein (SCAP) gene. J. Hum. Genet. 1999;44:402–407. doi: 10.1007/s100380050187. [DOI] [PubMed] [Google Scholar]
- 50.DeBose-Boyd R.A., Brown M.S., Li W.P., Nohturfft A., Goldstein J.L., Espenshade P.J. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 51.Puthothu B., Krueger M., Bernhardt M., Heinzmann A. ICAM1 amino-acid variant K469E is associated with paediatric bronchial asthma and elevated sICAM1 levels. Genes. Immun. 2006;7:322–326. doi: 10.1038/sj.gene.6364302. [DOI] [PubMed] [Google Scholar]
- 52.Ponthieux A., Lambert D., Herbeth B., Droesch S., Pfister M., Visvikis S. Association between Gly241Arg ICAM-1 gene polymorphism and serum sICAM-1 concentration in the Stanislas cohort. Eur. J. Hum. Genet. 2003;11:679–686. doi: 10.1038/sj.ejhg.5201033. [DOI] [PubMed] [Google Scholar]
- 53.Reilly P.L., Woska J.R., Jr, Jeanfavre D.D., McNally E., Rothlein R., Bormann B.J. The native structure of intercellular adhesion molecule-1 (ICAM-1) is a dimer. Correlation with binding to LFA-1. J. Immunol. 1995;155:529–532. [PubMed] [Google Scholar]
- 54.Diamond M.S., Staunton D.E., Marlin S.D., Springer T.A. Binding of the integrin Mac-1 (CD11b/CD18) to the third immunoglobulin-like domain of ICAM-1 (CD54) and its regulation by glycosylation. Cell. 1991;65:961–971. doi: 10.1016/0092-8674(91)90548-d. [DOI] [PubMed] [Google Scholar]
- 55.Folsom A.R., Aleksic N., Sanhueza A., Boerwinkle E. Risk factor correlates of platelet and leukocyte markers assessed by flow cytometry in a population-based sample. Atherosclerosis. 2009;205:272–278. doi: 10.1016/j.atherosclerosis.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catellier D.J., Aleksic N., Folsom A.R., Boerwinkle E. Atherosclerosis Risk in Communities (ARIC) Carotid MRI flow cytometry study of monocyte and platelet markers: intraindividual variability and reliability. Clin. Chem. 2008;54:1363–1371. doi: 10.1373/clinchem.2007.102202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y., Abecasis G.R. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 58.Servin B., Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lumley T. Analysis of complex survey samples. JSS. 2004;9:1–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.