Abstract
Background
The genus Spartina exhibits extensive hybridization and includes classic examples of recent speciation by allopolyploidy. In the UK there are two hexaploid species, S. maritima and S. alterniflora, as well as the homoploid hybrid S. × townsendii (2n = 60) and a derived allododecaploid S. anglica (2n = 120, 122, 124); the latter two are considered to have originated in Hythe, southern England at the end of the 19th century.
Methods
Genomic in situ hybridization (GISH) and flow cytometry were used to characterize the genomic composition and distribution of these species and their ploidy levels at Eling Marchwood and Hythe, both near Southampton, southern England.
Key Results
GISH identified approx. 60 chromosomes each of S. maritima and S. alterniflora origin in S. anglica and 62 chromosomes from S. alterniflora and 30 chromosomes from S. maritima in a nonaploid individual from Eling Marchwood, UK. GISH and flow cytometry also revealed that most (94 %) individuals examined at Hythe were hexaploid (the remaining two individuals (6 %) were dodedcaploid; n = 34), whereas hexaploid (approx. 36 % of plants), nonaploid (approx. 27 %) and dodecaploid (approx. 36 %) individuals were found at Eling Marchwood (n = 22).
Conclusions
Nonaploid individuals indicate the potential for introgression between hexaploid and dodecaploid species, complicating the picture of polyploid-induced speciation within the genus. Despite the aggressive ecological habit of S. anglica, it has not out-competed S. × townsendii at Hythe (homoploid hybrids at a frequency of 94 %, n = 34), despite >100 years of coexistence. The success of GISH opens up the potential for future studies of polyploid-induced genome restructuring in this genus.
Keywords: Polyploidy, speciation, Spartina, nonaploid, nonaploid bridge, interspecific hybridization, GISH
INTRODUCTION
Most or all angiosperm species are believed to have experienced at least one round of polyploidy in their ancestry (Otto and Whitton, 2000; Bowers et al., 2003; Mallet, 2007; Soltis et al., 2009), indicating its importance in the evolution of the flowering plants. Certainly polyploidy has been a driving force behind speciation in the genus Spartina (Poaceae) (Ainouche et al., 2004a). All Spartina species reported to date are polyploid with a base chromosome number x = 10 (Marchant, 1968). This genus evolved through two main lineages that diverged approx. 2·5–3 MYA (Christin et al., 2008), including tetraploid and hexaploid species (Baumel et al., 2002b) that are most likely of hybrid origin (Fortune et al., 2008). Four taxa are of interest here: the hexaploids S. alterniflora Loiseleur (2n = 60, 62) and S. maritima Fernald (2n = 60), their homoploid hybrid S. × townsendii H. Groves & J. Groves (2n = 60, 62) and the derived allododecaploid S. anglica C.E. Hubbard (2n = 12x = 120, 122, 124).
The allopolyploid Spartina anglica formed during the 19th century in Hythe, Southampton, after the introduction of S. alterniflora, a perennial grass native to the Atlantic coast of North America. Interspecific hybridization of S. alterniflora with the native British S. maritima (which inhabits the Atlantic coast of Africa and Europe) produced the infertile hybrid S. × townsendii, first identified in 1880 (Groves and Groves, 1880) – there are no reports of seed set as far as the authors are aware. Following this, genome duplication in the hybrid resulted in the formation of the fertile allopolyploid S. anglica (Marchant, 1963), a vigorous salt marsh species that has rapidly expanded in European salt marshes and has now colonized several continents (Marchants, 1967, 1968). Isozyme studies (Raybould et al., 1991), followed by a range of molecular methods (Ayres and Strong, 2001; Baumel et al., 2001, 2002a, 2003; Ainouche et al., 2004a, b), have confirmed the parentage of S. anglica and S. × townsendii, and analysis of the plastid sequences revealed the maternal parent to be S. alterniflora in all European populations investigated to date (Ferris et al., 1997; Baumel et al., 2001).
The emergence of S. anglica in southern Britain is reported to have paralleled the decline of native S. maritima along the entire southern coast of England, and now only a few populations exist on Hayling Island near Southampton and possibly on the Isle of Wight (Raybould et al., 2000). Contrasting with the rapid spread of the new allopolyploid S. anglica, the introduced parental species S. alterniflora remains confined to a few sites near Southampton (Charman, 1990). Since its emergence, S. anglica has gone on to colonize many parts of the world, frequently with severe ecological consequences (Hacker et al., 2001; An et al., 2007; Cottet et al., 2007). Land reclamation is common at sites colonized by S. anglica and, for this reason, many national wildlife authorities have imported this pioneer species as a means of combating land loss due to rising sea levels and erosion of mud flats. These factors have combined to give this young species a diverse ecological range throughout areas of Europe, Australia and China.
Spartina anglica populations exhibit little inter-individual genetic variation (Raybould et al., 1991; Ayres and Strong, 2001; Baumel et al., 2001, 2002b). Such a lack of diversity is thought to have resulted from a genetic bottleneck at the time of formation (Ainouche et al., 2004a). This genetic uniformity indicates a single origin or multiple origins involving similar genotypes (Ayres and Strong, 2001; Baumel et al., 2001, 2003; Yannic et al., 2004). In fact, where hybridization occurs, there is little genetic variation amongst the parental species. Southern England is the northern limit of the range of S. maritima, and populations here do not produce seeds, instead they propagate vegetatively (Yannic et al., 2004). There is also little genetic variation among individuals of S. alterniflora, which were introduced from America, due to founder effects (Baumel et al., 2003). The original population of S. × townsendii, as well as individuals of the derived neopolyploid S. anglica, exhibits additivity of multilocus parental molecular markers (Ayres and Strong, 2001; Baumel et al., 2001, 2002b). In spite of belonging to the same (hexaploid) clade, S. maritima and S. alterniflora exhibit considerable divergence (Baumel et al., 2002a) as through diploidization processes they have retained different paralogous gene copies that arose from ancient polyploidy (Fortune et al., 2008).
While no extensive structural changes have been observed in the homeologous genomes of S. × townsendii and S. anglica (Baumel et al., 2002b; Ainouche et al., 2004a, 2009; Fortune et al., 2008), the hybridization step which gave rise to S. × townsendii has resulted in epigenetic alterations that have also been transmitted to the allopolyploid, S. anglica (Salmon et al., 2005). These changes mostly affect the regions flanking transposable elements (Parisod et al., 2009). Considerable changes to the transcriptome have also accompanied the formation S. × townsendii and S. anglica (Chelaifa et al., 2010). These epigenetic changes most likely explain the phenotypic plasticity observed in natural populations of the allopolyploid (Thompson, 1990).
The extensive and rapid spreading (via both seed production and vegetative means) of the fertile S. anglica has not been seen in the sterile, yet vigorous S. × townsendii (Ainouche et al., 2004a), leading Ainouche and co-workers to propose that the more extensive ecological adaptability of S. anglica compared with its hexaploid progenitors is due to chromosome doubling, rather than interspecific hybridization. Previous studies have documented the distribution of the varying ploidy-levels and species within this genus (Marchant, 1967, 1968; Charman, 1990); however, there have been no recent studies using cytogenetic approaches to interpret the frequency of ploidy levels in mixed populations of Spartina. By extensively sampling Spartina populations in the area where indigenous S. maritima and introduced S. alterniflora originally hybridized, the aim here is to evaluate the relative abundance of the different ploidy levels (homoploid and allopolyploid) encountered in this region and to analyse, for the first time, these populations using molecular cytogenetic approaches.
MATERIALS AND METHODS
Plant material
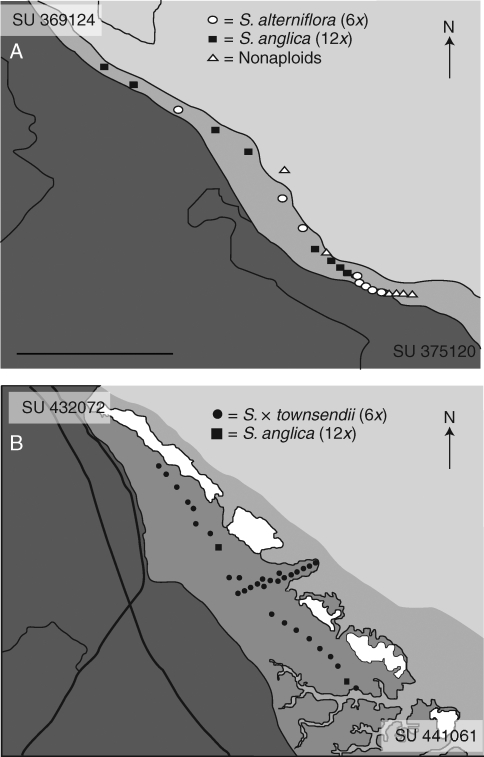
Spartina individuals were taken from two locations near Southampton on the south coast of England. Twenty-two plants along a north-west to south-east transect were collected at Eling Marchwood on the 13 August 2007 (Fig. 1A). Thirty-four plants were also collected at Hythe (around 6 shoreline miles from the site at Eling Marchwood), on 6 August 2007 along a transect following a north-east to south-west direction (Fig. 1B). Samples collected on both days were returned to the laboratory for re-potting and analysis.
Fig. 1.
Transect showing ploidy levels of plants collected at (A) Eling Marchwood and (B) Hythe. Dark shaded areas represent forest or field, medium grey areas represent tidal zone and pale grey represents water. White sections represent cockle shell deposits. Scale bar = 500 m. Ordnance survey grid references are shown at the top-left and bottom-right corners of (A) and (B).
Genomic in situ hybridization (GISH)
Cytological preparations were made from seed embryos or root tips from pot-grown plants. Metaphases were accumulated by pre-treatment of material in saturated Gammexane® (hexachlorocyclohexane; Sigma) in water for 4 h, then fixed for 24 h in 3 : 1 absolute ethanol : glacial acetic acid and stored in 100 % ethanol at –20 °C. Plant material was squashed onto a glass slide after enzyme digestion as described in Sumner and Leitch (1999). GISH was performed using total genomic DNA probes from S. alterniflora (digoxigenin-11-dUTP labelled) and S. maritima (biotin-14-dUTP labelled) labelled by nick translation (Roche Nick Translation Kit) according to the manufacturers instructions. GISH was conducted as described in Lim et al. (1998) after denaturation in 70 % formamide (v/v) in 2× SSC (0·3 m sodium chloride, 0·03 m sodium citrate) at 70 °C for 2 min. The hybridization mix comprised of 50 % (v/v) formamide, 10 % (w/v) dextran sulfate, 0·1 % (w/v) sodium dodecyl sulfate in 2× SSC and 50 µg mL−1 of each genomic probe. After overnight hybridization at 37 °C, slides were washed in 20 % (v/v) formamide in 0·1× SSC at 42 °C at an estimated hybridization stringency of 80–85 %. Sites of probe labelling were detected with 20 µg mL−1 fluorescein-conjugated anti-digoxigenin IgG (Roche Biochemicals Ltd) and 5 µg mL−1 Cy3-conjugated streptavidin (Amersham Pharmacia Biotech), and chromosomes were counterstained with 2 µg mL−1 DAPI (4′,6-diamidino-2-phenylindole) in 4× SSC, mounted in Vectashield (Vector Laboratories) medium. Material was photographed with an Orca ER camera mounted to a Leica DMRA2 epifluorescent microscope. Images were processed with Improvision Openlab software (Improvision, Coventry, UK) and adjusted for colour balance, contrast and brightness uniformly.
Analysis of ploidy levels
Flow cytometry was used to screen for ploidy level using methods described in Hanson et al. (2005) and either a Partec PAII or a Partec CyFlow flow cytometer. Pisum sativum ‘Minerva Maple’ was used as a calibration standard for assessing ploidy levels.
RESULTS
GISH
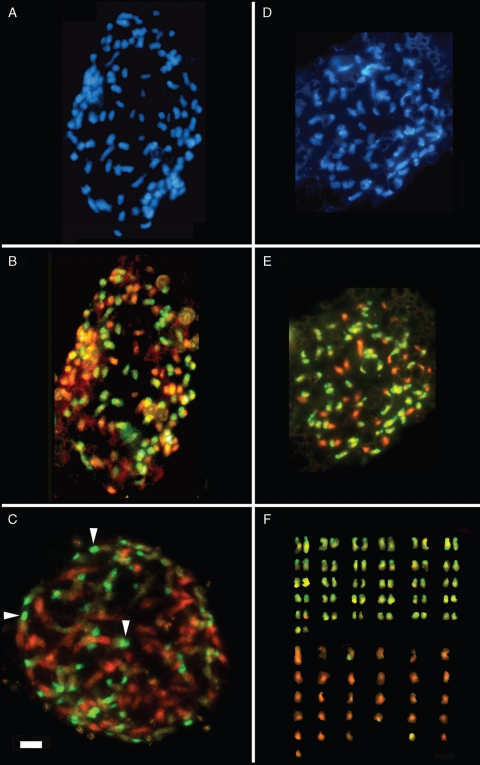
Two Spartina individuals, both originally identified as S. anglica, and thus assumed to be dodecaploids were selected for analysis by GISH. Figure 2A and B shows a GISH-labelled root tip metaphase from a plant collected at Eling Marchwood. The approx. 120 chromosomes resolve into two groups, one comprising approx. 60 chromosomes that predominantly stained with the S. alterniflora probe (green) while the remaining approx. 60 chromosomes were labelled with the S. maritima probe (orange). The individual had a dodecaploid karyotype that could be designated for simplicity AAMM, assuming A and M represent triplicated ancestral genomes from the hexaploid S. alterniflora and S. maritima, respectively. Frequently the short arm and/or centromere regions of the chromosomes of S. alterniflora origin were more heavily labelled, indicative of heterochromatic sequences (see arrowheads on prophase cell in Fig. 2C). In addition the GISH data reveal that recombination between parental sets of chromosomes is likely to be minimal (beyond the resolution of this experiment) or absent, as all chromosomes stained one colour along their whole length.
Fig. 2.
Root tip metaphases (A, B, D–F) and prophase nucleus (C) after DAPI staining (blue fluorescence) (A, D) and GISH (B, C, E, F) using digoxigenin-labelled probes of S. alterniflora genomic DNA (green, detected using FITC-conjugated anti-digoxigenin IgG) and biotin-labelled probes of S. maritima genomic DNA (orange, detected using Cy3-conjugated streptavidin). Shown in (A–C) are cells from an AAMM dodecaploid individual with approx. 120 chromosomes, approx. 60 of which are of S. maritima origin and approx. 60 of S. alterniflora origin. Note, the intense signal in the short arm of chromosomes (arrowheads) of S. maritima origin in the prophase (C). This is indicative of heterochromatic sequences (also seen in E and F). The same metaphase of an AAM nonaploid individual with 92 chromosomes is shown in (D–F). The AAM karyotype (F) has chromosomes organized by size and parental genome. Due to their similar morphologies and staining patterns, chromosomes may not be in natural pairs. Scale bar = 5 µm.
Figure 2D–F shows a GISH-labelled metaphase from another plant collected at Eling Marchwood. There were only 92 chromosomes, 30–32 fewer than expected. Of these, approx. 60 chromosomes preferentially hybridized with the S. alterniflora probe (green) while approx. 30 were labelled with the S. maritima probe (orange) (Fig. 2F). The individual was therefore considered to be a nonaploid (9x), with a genome designation of AAM.
Distribution of Spartina ploidy levels
The flow cytometric analysis of 56 individuals from Hythe and Marchwood revealed hexaploid, nonaploid and dodecaploid individuals. Overall, combined data from both sites showed that nonaploids were the rarest, occurring at a frequency of approx. 10·7 % (= 6 individuals), whereas dodecaploids and hexaploids occurred at higher frequencies of 17·8 % (= 10 individuals) and 71 % (= 40 individuals), respectively. However, the occurrence and distribution of the different ploidy levels and species varied markedly between the two sites.
At Hythe the vast majority of plants analysed (i.e. 32 of the 34 plants = 94 %) were hexaploid and all were identified morphologically as the homoploid hybrid S. × townsendii (Fig. 1B). The occurrence of the other hexaploid species S. maritima or S. alterniflora could be ruled out on morphological grounds. The remaining two individuals were dodecaploids corresponding to S. anglica.
In contrast, at Eling Marchwood three ploidy levels were observed in the 22 plants analysed (Fig. 1A). Eight individuals (36·4 %) identified as S. alterniflora were shown to be hexaploid while six plants (27·2 %) showing considerable morphological diversity ranging between S. anglica and S. alterniflora phenotypes were found to be nonaploid similar to the findings of Marchant (1967, 1968). The remaining eight plants (36·4 %), which were morphologically identified as S. anglica, were dodecaploid. Individuals of different ploidy levels were intermixed and in close association along the transect, although there was a tendency for nonaploids to be more abundant towards the south-east end of the transect (Fig. 1A).
Because there was some variability in the position of the peaks in the flow histograms between individuals at the same ploidy level, the aim was to determine the likelihood that the samples were taken from a population with a continuous distribution of genome sizes. A computer program was developed in Mathematica 5·0 (Wolfram Illinois) to simulate sampling from a continuous population of 1000 individuals with the same mean and standard deviation (mean = 10·709, s.d. = 3·247, n = 56) of the collective genome size estimates of all 56 plants analysed. After 1000 randomizations (each time taking 56 samples) only 22 of the resulting distributions had discontinuities that were similar to the real data. The real data is therefore likely to be discontinuous and structured around integer ploidy levels (P = 0·022).
DISCUSSION
Spartina is one of the classic models of recent polyploidy-induced speciation. Spartina anglica is thought to have been formed like many natural polyploids via genome doubling of an interspecific hybrid, typically the route used to make synthetic polyploid crops. The current consensus is that polyploid formation frequently arises in nature via unreduced gamete formation. The mean frequency of unreduced gametes, based on studies across many eukaryote taxa, including angiosperms, is approx. 5 % (Ramsey and Schemske, 1998), although their frequency is affected by genetic and environmental factors. Recently, the first gene, AtPS1, directly associated with diploid gamete formation was characterized in Arabidopsis thaliana. Mutations in this gene were shown to cause misalignment of the spindle in meiosis II, resulting in astonishingly high levels of diploid gametes (up to 65 %) (d'Erfurth et al., 2008).
In the current study, nonaploids were identified amongst the Eling Marchwood population, occurring at a frequency of 27·2 %. Using GISH the genomic composition of one nonaploid was shown to be AAM, comprised of two triplicated genomes (6x = 60) derived from S. alterniflora (AA) and one (3x = 30) from S. maritima (M; Fig. 2D–F). Marchant (1967, 1968) reported the occurrence of 2n = approx. 90 and 2n = 76 plants at Hythe, although no such ploidy levels were encountered in the present survey at this site.
The nonaploid plants discovered at Eling Marchwood could have arisen in several possible ways. (a) A likely route of formation could involve a cross between regular (reduced) hexaploid gamete (6x = AM) of the dodecaploid S. anglica (AAMM) and a regular gamete (3x = A) of S. alterniflora (AA). This hypothesis is reinforced by the current distribution of plants, where the nonaploid plants are found with S. alterniflora and S. anglica. This was also hypothesized by Marchant (1968) to explain the occurrence of the 2n = 90 plants found at Hythe. However, the nonaploid plants may have formed when distribution patterns were different and alternative hypotheses cannot be ruled out. (b) Given the relatively high frequency of unreduced gametes in many angiosperm species, nonaploids may have arisen via an unreduced gamete from S. alterniflora hybridizing with a normal gamete from S. maritima. This could have occurred if these species grew sympatrically, perhaps at the time when S. × townsendii itself formed (Marchant, 1967, 1968). (c) Another possibility involves an unreduced gamete from S. × townsendii crossing with S. alterniflora, which could have occurred at Hythe, followed by dispersal of the nonaploid hybrids to Eling Marchwood.
Currently, it is not possible to distinguish between these hypotheses because the nonaploids could have formed at any time from the introduction of S. alterniflora to the present day. Indeed, Marchant (1967, 1968) has documented a period of around 20 years when S. alterniflora and S. maritima co-existed at Hythe (the last record of S. maritima at this locality being in 1900) before the formation of the allopolyploid S. anglica, leaving ample time for polyploidy event(s), as described above, to give rise to the nonaploids. It is possible that the nonaploids observed at Eling Marchwood are derived from those previously described by Marchant (1967, 1968). Certainly the distribution of plants can be influenced by plant mobility through the movement of plant fragments or seeds, which in Spartina can float (Huiskes et al., 1995). Alternatively, the nonaploids recorded here may have been formed independently, as frequently observed in recurrently formed hybrids and allopolyploids (Soltis and Soltis, 2009).
Further studies are needed to determine whether other nonaploid cytotypes exist, such as AMM, produced by, for example, a cross between S. anglica and S. maritima. Morphologically intermediate plants are encountered in southern Brittany (France) where intermixed populations of S. anglica and S. maritima plants do occur, but the molecular analyses performed to date have failed to detect hybrids (G. Allard and M.-T. Misset, Rennes University, unpubl. res.). The possibility of backcrossing between S. anglica and its parental species revealed here represents a potentially significant source of genetic diversity in the otherwise genetically depauperate populations of S. anglica.
Backcrosses involving polyploids and their diploid progenitors are commonly reported in natural populations. In Chamerion angustifolium, triploids in mixed ploidy populations (2x, 3x, 4x) provide a means of tetraploid formation, via uniting unreduced triploid gametes (3n) with normal gametes (n). This route to polyploid formation may enable tetraploids to overcome any frequency-dependent mating disadvantages (Husband, 2004). Using computer modelling Husband and Schemske (2000) showed that the presence of so called triploid-bridges could facilitate the maintenance and fixation of tetraploid individuals at the expense of both triploids and diploids. Ayres et al. (2008) speculated that triploid bridges may have played a role in the evolution and recurrent formation of S. anglica. Thus, the Spartina nonaploids identified here might potentially act in such a way by providing a bridge to the formation of new ploidy levels, although no such higher ploidy levels, such as 18x individuals, have been recorded to date either in the literature or in this survey.
Spartina individuals at all three ploidy levels occurred in close proximity at Eling Marchwood, raising the possibility of introgressive hybridization between them. Hybrids involving S. alterniflora (introduced) and S. foliosa (native) have arisen in California (Ayres et al., 1999) where they compete with the native plants. In addition, interspecific hybridization is reported in this area between the hexaploid S. foliosa and the introduced alloheptaploid S. densiflora (Ayres et al., 2008). Furthermore S. densiflora originated in South America following hybridization between a hexaploid species (likely to be S. alterniflora) and a tetraploid species related to S. arundinacea, also from the southern hemisphere (Fortune et al., 2008). Backcrossing of the F1 S. densiflora × S. foliosa hybrids (2n = 66) has resulted in individuals with 2n = 94–96, exhibiting either ‘foliosa’ or ‘alterniflora’ plastid sequence types (Ayres et al., 2008). These studies, as well as data presented here, indicate the extensive reticulate evolution in the genus.
Transects at Eling and Hythe showed dodecaploids at a frequency of approx. 36 % and 6 %, respectively (Fig. 1A and B). This is in contrast to the estimated abundance of S. anglica by Charman (1990), who claimed that the expansion of S. anglica had been concomitant with the reduction of the F1 homoploid hybrid and parental species across the entire southern coast of Britain. It is clear from the data presented here that this broad generalization is likely to be incorrect. At Hythe, only two out of 34 individuals were S. anglica plants and, instead, the site was dominated by S. × townsendii, which comprised the remaining 32 individuals (94 %) of the sample. Furthermore, while S. anglica was more common at Eling Marchwood (36·4 %); it was not the dominant species as S. alterniflora also occurred with similar frequency (i.e. 36·4 %), with the remaining individuals being nonaploids (27·2 %). Perhaps S. anglica has experienced a decline in abundance in the years since Charman's study, as S. anglica ‘die-back’ resulting from age-related decline of vigour has been reported in some other sites in Britain (Gray, 2004). Such a process is not without precedence in newly formed allopolyploids as Senecio eboracensis, an allotetraploid that formed within the last approx. 40 years, is now virtually extinct even though it initially expanded its numbers greatly following its formation (Abbott and Lowe, 2004).
In Spartina all studies to date have failed to identify significant genetic changes following either hybridization or genome doubling (Ainouche et al., 2004a). However, evidence of significant epigenetic reprogramming was recorded in S. × townsendii (Salmon et al., 2005), that mostly affects the regions flanking transposable elements (Parisod et al., 2009). Spartina anglica has inherited almost all these changes. Moreover, gene expression changes are exhibited following both hybridization and polyploidy (Chelaifa et al., 2010). Perhaps epigenetic and regulatory changes triggered by interspecific hybridization, rather than those occurring subsequent to polyploidy, explain why S. × townsendii fares well at its site of origin in the face of competition from S. anglica. In addition it is likely that S. × townsendii has failed to spread so rapidly because of its infertility.
In conclusion, the results reveal that, in spite of fertility and invasive behaviour, the allopolyploid S. anglica has not outcompeted the sterile homoploid hybrid S. × townsendii at the hybridization site. This study also demonstrates the potential of GISH to characterize the genome composition of different ploidy levels in Spartina, including nonaploids. This in turn can be used to assess the possible recurrent formation of polyploids with varying ploidy levels within this group. Furthermore the present studies demonstrate that the history of hybridization and speciation in this complex is more complicated than previously thought with the potential for even more complexity than is already apparent.
ACKNOWLDGEMENTS
We would like to thank Mr R. Joseph, Mr Farhan Kazi and Ms L. Hanson for assistance. We thank Mr Mike Smith (Northney Marina, Hampshire), Mr Martin Rand (BSBI recorder, Hampshire), Mr Andrew Groom (New Forest District Council, Hampshire), Mr Chris Pirie (Natural England Office, Hampshire), Mr Richard Fort (ExxonMobile, Hampshire), Ms Alison Fowler (Chichester Harbour AONB manager, Hampshire) and Ms Sue Simmonite (Associated British Ports, Southampton) for granting permission to collect Spartina plants. We also would like to thank Dr M. Chester and Ms E. W. McCarthy for insightful comments on the manuscript as well as two anonymous referees whose comments have improved the manuscript. M.A. was supported by the French National Research Agency (ANR) through the ‘Polyploidy and Biodiversity’ Programme.
LITERATURE CITED
- Abbott R, Lowe A. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biological Journal of the Linnean Society. 2004;82:467–474. [Google Scholar]
- Ainouche ML, Baumel A, Salmon A. Spartina anglica C. E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biological Journal of the Linnean Society. 2004a;82:475–484. [Google Scholar]
- Ainouche ML, Baumel A, Salmon A, Yannic G. Hybridization, polyploidy and speciation in Spartina (Poaceae) New Phytologist. 2004b;161:165–172. [Google Scholar]
- Ainouche ML, Fortune PM, Salmon A, et al. Hybridization, polyploidy and invasion: lessons from Spartina (Poaceae) Biological Invasions. 2009;11:1159–1173. [Google Scholar]
- An SQ, Gu BH, Zhou CF, et al. Spartina invasion in China: implications for invasive species management and future research. Weed Research. 2007;47:183–191. [Google Scholar]
- Ayres DR, Strong DR. Origin and genetic diversity of Spartina anglica (Poaceae) using nuclear DNA markers. American Journal of Botany. 2001;88:1863–1867. [PubMed] [Google Scholar]
- Ayres DR, Garcia-Rossi D, Davis HG, Strong DR. Extent and degree of hybridization between exotic (Spartina alterniflora) and native (S. foliosa) cordgrass in California, USA determined by randomly amplified polymorphic DNA (RAPDs) Molecular Ecology. 1999;8:1179–1186. doi: 10.1046/j.1365-294x.1999.00679.x. [DOI] [PubMed] [Google Scholar]
- Ayres DR, Grotkopp E, Zaremba K, et al. Hybridization between invasive Spartina densiflora (Poaceae) and native S. foliosa in San Francisco Bay, California, USA. American Journal of Botany. 2008;95:713–719. doi: 10.3732/ajb.2007358. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche ML, Bayer RJ, Ainouche AK, Misset MT. Molecular phylogeny of hybridizing species from the genus Spartina Schreb. (Poaceae) Molecular Phylogenetics and Evolution. 2002a;22:303–314. doi: 10.1006/mpev.2001.1064. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche M, Kalendar R, Schulman AH. Retrotransposons and genomic stability in populations of the young allopolyploid species Spartina anglica CE Hubbard (Poaceae) Molecular Biology and Evolution. 2002b;19:1218–1227. doi: 10.1093/oxfordjournals.molbev.a004182. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche ML, Levasseur JE. Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France) Molecular Ecology. 2001;10:1689–1701. doi: 10.1046/j.1365-294x.2001.01299.x. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche ML, Misset MT, Gourret JP, Bayer RJ. Genetic evidence for hybridization between the native Spartina maritima and the introduced Spartina alterniflora (Poaceae) in South-West France: Spartina × neyrautii re-examined. Plant Systematics and Evolution. 2003;237:87–97. [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Charman K. The current status of Spartina anglica in Britain. In: Gray AJ, Benham PEM, editors. London: HMSO; 1990. Spartina anglica: a research review. Institute of Terrestrial Ecology Research Publication No. 2. [Google Scholar]
- Chelaifa H, Monnier A, Ainouche M. Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina × townsendii and Spartina anglica (Poaceae) New Phytologist. 2010;186 doi: 10.1111/j.1469-8137.2010.03179.x. in press. [DOI] [PubMed] [Google Scholar]
- Christin PA, Besnard G, Samartiani E, et al. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Current Biology. 2008;18:37–43. doi: 10.1016/j.cub.2007.11.058. [DOI] [PubMed] [Google Scholar]
- Cottet M, de Montaudouin X, Blanchet H, Lebleu P. Spartina anglica eradication experiment and in situ monitoring assess structuring strength of habitat complexity on marine macrofauna at high tidal level. Estuarine Coastal and Shelf Science. 2007;71:629–640. [Google Scholar]
- d'Erfurth I, Jolivet S, Froger N, et al. Mutations in AtPS1 (Arabidopsis thaliana Parallel Spindle 1) lead to the production of diploid pollen grains. PLoS Genetics. 2008;4 doi: 10.1371/journal.pgen.1000274. 11 e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris C, King RA, Gray AJ. Molecular evidence for the maternal parentage in the hybrid origin of Spartina anglica. Molecular Ecology. 1997;6:185–187. [Google Scholar]
- Fortune PM, Schierenbeck K, Ayres D, et al. The enigmatic invasive Spartina densiflora: A history of hybridizations in a polyploidy context. Molecular Ecology. 2008;17:4304–4316. doi: 10.1111/j.1365-294X.2008.03916.x. [DOI] [PubMed] [Google Scholar]
- Gray AJ. Will Spartina anglica invade northwards with changing climate? Proceedings of the Third International Conference on Invasive. 2004 Spartina. San Francisco, CA, 8–10 November 2004. [Google Scholar]
- Groves H, Groves J. Sparina townsendii nobis. Reports of the Botanical Society and Exchange Club of the British Isles. 1880:1. [Google Scholar]
- Hacker S, Heimer D, Hellquist C, et al. A marine plant (Spartina anglica) invades widely varying habitats: potential mechanism of invasion control. Biological Invasions. 2001;3:211–217. [Google Scholar]
- Hanson L, Boyd A, Johnson MAT, Bennett MD. First nuclear DNA C-values for 18 eudicot families. Annals of Botany. 2005;96:1315–1320. doi: 10.1093/aob/mci283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiskes AHL, Koustaal P, Herman PMJ, Beeftink WG, Markusse MM, De Munck W. Seed dispersal of haplotypes in tidal salt marshes. Journal of Ecology. 1995;83:559–567. [Google Scholar]
- Husband BC. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biological Journal of the Linnean Society. 2004;82:537–546. [Google Scholar]
- Husband BC, Schemske DW. Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. Journal of Ecology. 2000;88:689–701. [Google Scholar]
- Lim KY, Leitch IJ, Leitch AR. Genomic characterisation and the detection of raspberry chromatin in polyploid Rubus. Theoretical and Applied Genetics. 1998;97:1027–1033. [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Marchant C. Corrected chromosome numbers for Spartina × townsendii and its parent species. Nature. 1963;199:929. [Google Scholar]
- Marchant C. Evolution in Spartina (Gramineae). I. The history and morphology of the genus in Britain. Journal of the Linnean Society (Botany) 1967;60:1–24. [Google Scholar]
- Marchant C. Evolution in Spartina (Gramineae). II. Chromosomes, basic relationships and the problem of Spartina × townsendii agg. Journal of the Linnean Society (Botany) 1968;60:381–409. [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien M-A, Ainouche M. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytologist. 2009;184:1003–1015. doi: 10.1111/j.1469-8137.2009.03029.x. [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics. 1998;29:467–501. [Google Scholar]
- Raybould AF, Gray AJ, Hornby DD. Evolution and currrent status of the salt marsh grass, Spartina anglica. In: Collins M, Ansell K, editors. Solent science: a review. Amsterdam: Elsevier Science; 2000. in the Solent. In. [Google Scholar]
- Raybould AF, Gray AJ, Lawrence MJ, Marshall DF. The evolution of Spartina anglica Hubbard, C.E (Gramineae): genetic variation and status of the parental species in Britain. Biological Journal of the Linnean Society. 1991;44:369–380. [Google Scholar]
- Salmon A, Ainouche ML, Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Molecular Ecology. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annual Review of Plant Biology. 2009;60:562–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Sumner A, Leitch AR. Microscopy of chromosomes. In: Lacey A, editor. Light microscopy in biology. 2nd edn. Oxford: Oxford University Press; 1999. [Google Scholar]
- Thompson J. Morphological variation among natural populations of Spartina anglica. In: Gray AJ, Benham PEM, editors. London: HMSO; 1990. Spartina anglica: a research review. Institute of Terrestrial Ecology Research Publication No. 2. [Google Scholar]
- Yannic G, Baumel A, Ainouche M. Uniformity of the nuclear and chloroplast genomes of Spartina maritima (Poaceae), a salt-marsh species in decline along the Western European Coast. Heredity. 2004;93:182–188. doi: 10.1038/sj.hdy.6800491. [DOI] [PubMed] [Google Scholar]