Abstract
Background and Aims
Knowledge on how climate-induced range shifts might affect natural selection is crucial to understand the evolution of species ranges.
Methods
Using historical demographic perspectives gathered from regional-scale phylogeography on the alpine herb Biscutella laevigata, indirect inferences on gene flow and signature of selection based on AFLP genotyping were compared between local populations persisting at the trailing edge and expanding at the leading edge.
Key Results
Spatial autocorrelation revealed that gene flow was two times more restricted at the trailing edge and genome scans indicated divergent selection in this persisting population. In contrast, no pattern of selection emerged in the expanding population at the leading edge.
Conclusions
Historical effects may determine different architecture of genetic variation and selective patterns within local populations, what is arguably important to understand evolutionary processes acting across the species ranges.
Keywords: Amplified fragment length polymorphism, Biscutella laevigata (Brassicaceae), gene flow, genome scan, landscape genetics, range margins, selection, species range shift
INTRODUCTION
Phylogeographic studies over the past decade have uncovered how past climate oscillations have repeatedly impacted on species ranges and influenced the genetic composition of taxa (Hewitt, 1996; Hampe and Petit, 2005; Schönswetter et al., 2005). Across a distribution range, climate changes trigger the regression of populations where conditions become suboptimal and they render new territories suitable for colonization (Ackerly, 2003). Accordingly, two types of margins can be distinguished: the trailing edge, where populations persist despite climate changes, and the leading edge, where populations expand following climate changes. The Alps went through drastic climatic oscillations during the Pleistocene and represent an excellent landscape study system to explore the consequences of climate-induced range-shifts. There is now accumulating evidence demonstrating long-term persistence of several plant taxa, often at the border of the Alps, and postglacial recolonization from the peripheral to the central Alps and towards higher elevation (Schönswetter et al., 2005; Parisod, 2008). Consequently, the Alps harbour populations with a varying, contrasting history over a restricted geographical scale (i.e. similar climatic conditions, landscapes, biogeographical areas), which may greatly help to understand the evolutionary mechanisms underlying patterns of genetic variation under changing climate.
The focus here is on the widespread alpine herb Biscutella laevigata (Brassicaceae). At the regional scale, the influence of past climate changes on the species range was evidenced as a gradient of genetic diversity along recolonization pathways (Parisod and Besnard, 2007). Accordingly, the peripheral Alps represent the historical core of lineages as attested by ample haplotypic variation among localities and presently correspond to trailing-edge conditions. In contrast, recently glaciated areas at high altitude in the central Alps are typically formed of expanding populations fixed for a single haplotype and correspond to the leading edge. Taking advantage of such historical demographic perspective offered by phylogeography, selected marginal populations of B. laevigata within a particular haplotype lineage (E; according to Parisod and Besnard, 2007) were examined at a local scale with amplified fragment length polymorphism (AFLP): one population corresponding to the trailing edge in the peripheral Alps (Parisod and Christin, 2008) and another population corresponding to the leading edge in the central Alps (Parisod and Bonvin, 2008).
Both populations showed similar total genetic diversity and presented mosaic spatial patterns of genetic differentiation, but distinct patterns of within-population genetic structure. The trailing-edge population presented a significantly lower within-plot genetic diversity than the leading edge (Shannon index: 0·093 ± 0·033 vs. 0·270 ± 0·057, respectively; Wilcoxon test, P < 0·001) and a stronger internal genetic structure (between-group eigenanalysis: βST = 0·49 vs. βST = 0·32, respectively). Furthermore, individuals with similar AFLP profiles were consistently associated with ecological factors in the trailing-edge population, suggesting local adaptation with respect to total solar radiation (Parisod and Christin, 2008). In contrast, the leading-edge population manifested signatures of recent expansion since the distribution of both individual abundance and genetic diversity formed a cline along the local recolonization path (Parisod and Bonvin, 2008). Moreover, sampling plots from in between genetically homogeneous patches consistently showed higher genetic diversity, indicating ongoing admixture after recolonization.
The aim of this study is to explore the impact of divergent natural selection in different types of range margins by comparing indirect estimates of gene flow and genome scans for signature of selection between these trailing-edge and leading-edge populations. According to the hypothesis of Jansson and Dynesius (2002) postulating selection for local adaptation in areas with little climate-enforced range shifts, a clear signature of selection in the trailing-edge population and none or only a low one in the leading-edge population is expected.
MATERIALS AND METHODS
Investigated populations
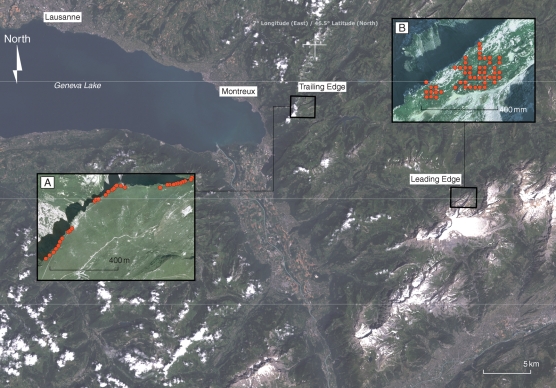
Two populations of Biscutella laevigata L., one lying at the trailing and the other at the leading edge, were chosen for fine-scale surveys because they presented a large number of individuals arranged continuously across comparable altitudinal gradients (Fig. 1). Both populations showed great ecological similarity, sharing climatic conditions and presenting similar environmental heterogeneity as regards to the ecological factors assessed here on a local scale (Table 1; Supplementary data, available online). Individuals are strictly outcrossing, characterized by a sporophytic incompatibility system (Olowokudejo and Heywood, 1984; C. Parisod, pers. obs.). Sampling was achieved across the whole populated area and four individuals were collected in evenly spaced 2 × 2 m plots according to a grid design. The trailing-edge population (JAM in Parisod and Besnard, 2007) was restricted to a linear ridge in the peripheral Alps (Parisod and Christin, 2008) and the leading-edge population (REU in Parisod and Besnard, 2007) was located at the altitudinal limit of the species in a glacier foreland of the central Alps (Parisod and Bonvin, 2008; Table 1). All individuals were genotyped with AFLPs, using the same combinations of fluorescent-labelled primer (M-CAG/E-ACA and M-CTC/E-AGG; Parisod and Bonvin, 2008; Parisod and Christin, 2008). Fragments were visualized after electrophoresis on 6 % Long Ranger denaturing gel using an ABI-PRISM 377 and manually scored (Table 1).
Fig. 1.
Populations of Biscutella laevigata investigated at the trailing versus the leading edge of the species range. The basemap shows the western Swiss Alps (Landsat 7 image, July 1999). (A) The trailing-edge population sampled in the peripheral Alps; (B) the leading-edge population sampled in a glacier foreland in the central Alps. Dots represent the sampling plots.
Table 1.
Description of the populations of Biscutella laevigata investigated at the trailing versus the leading edge of the species range
| Trailing edge | Leading edge | |
|---|---|---|
| Population's details | ||
| Coordinates | 6°58′/46°26′ | 7°12′/46°20′ |
| Historical features | Refugial populations in the peripheral Alps (Parisod and Christin, 2008) | Expanding population in the central Alps (Parisod and Bonvin, 2008) |
| Spatial features | Linear (1000 × 25 m) | Two-dimensional (350 × 200 m) |
| Grid sampling | 31 plots of 4 individuals | 51 plots of 4 individuals |
| Environmental heterogeneity* | ||
| DEM: altitude (m) | (1851–1990); significant SA up to 140 m | (2154–2298); significant SA up to 140 m |
| DDEG: degree-days during growing season (°d) | (1124–1260); significant SA up to 140 m | (839–921); significant SA up to 140 m |
| ETPT: daily average evapotranspiration (mm d−1) | (2·5–6·6); low or no SA | (2·1–5·6); low or no SA |
| PDAY: number of precipitation days (d) | (62·9–63·75); low or no SA | (43–43); low or no SA |
| SRAD: daily average radiation (kJ d−1) | (18057–75418); low or no SA | (17763–63928); low or no SA |
| Slope (°) | (2–69); low or no SA | (7–56); low or no SA |
| Genetic dataset | ||
| Number of polymorphic AFLP bands | 102 | 113 |
| Error rate (number of replicates) | 1·8 % (20) | 2·4 % (10) |
* Eco-climatic parameters discussed here are shown with their range of variation (in parenthesis) and a description of spatial autocorrelation (SA) over the sampling area (for details, see Table S1 in Supplementary data).
Comparative gene flow
Sp statistics are based on the spatial autocorrelation of genetic relatedness coefficients among individuals and represent convenient measures of gene flow within population, even under various sampling scheme such as linear vs. two-dimensional populations (Vekemans and Hardy, 2004). Autocorrelation was assessed as the average relationship coefficient among individuals at increasing distance intervals with balanced number of pairwise comparisons, using SPAGeDi 1·2 (Hardy and Vekemans, 2002). Confidence intervals (95 %) were calculated after 999 permutations of individual locations among all individuals. In order to obtain a robust measure of relative gene flow in the trailing (A) and the leading-edge (B) populations, comparative Sp statistics were computed as the ratio
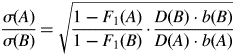 |
where F1 is the relationship coefficient in the first distance class, D the effective density as estimated from field density (i.e. 3 and 2·5 individuals m−2 in A and B, respectively), and b is the slope of the regression between the relationship coefficient and the logarithm of spatial distance. The surveyed populations are autotetraploid (i.e. with four segregating alleles per locus; Parisod et al., 2010) and the relationship coefficient is thus increased by an unknown constant. This comparative approach has the advantage of circumventing such uncertainties. A 95 % confidence interval for the comparative Sp statistics was assessed by calculating the ratio again using opposite interval boundaries of the parameters (F1 and b) for the two populations. In addition, autocorrelation and comparative Sp statistics were computed with all AFLP markers to deduce the realized gene flow and with neutral markers only (i.e. excluding those AFLP markers showing signature of selection as indicated by BayeScan, see below) to infer gene dispersal.
Detection of loci under selection
To detect signature of natural selection and to find eco-climatic parameters putatively responsible for selection within the trailing- and the leading-edge populations of B. laevigata, two complementary approaches were used: (1) a Bayesian outlier locus approach estimating the posterior probability of each locus to be under selection using BayeScan software (Foll and Gaggiotti, 2008); and (2) an approach estimating the association between genetic data and environmental variables through logistic regressions as implemented in Sam (Joost et al., 2007, 2008). BayeScan is relying on estimates of genetic structure among sampling plots with the rationale that loci under positive selection present a significantly higher genetic differentiation than the multitude of neutral loci over the whole genome, while loci under purifying selection show lower differentiation. Accordingly, BayeScan was used on the subdivided populations (31 and 51 plots, according to the sampling scheme), running 150,000 iterations. Simulation studies demonstrated the robustness of BayeScan to a wide range of demographic scenarios violating underlying assumptions (Foll and Gaggiotti, 2008). In contrast, Sam does not rely on genetic models and works at the individual level, comprehensively inspecting correlations between loci and environmental variables by univariate logistic regressions (Joost et al., 2007). The eco-climatic parameters taken from the swisstopo 25-m-resolution digital elevation model and from Zimmermann and Kienast (1999) were used (see Table 1 for details). Although a formal comparison of the methods is beyond the scope of this paper, BayeScan and Sam profitably complement each other since the former authenticates signature of selection on theoretical grounds, while Sam identifies the potentially effective selection pressure from the environment.
RESULTS
The relationship coefficient among nearby individuals (F1), as well as the slope of the regression between the relationship coefficient and the logarithm of spatial distance (b), was different in the trailing- and the leading-edge populations of B. laevigata (Table 2). In addition, F1 and b remained similar after having removed putatively non-neutral loci from the dataset (see below), although the slope of the regression was lower in the trailing-edge population. Comparative Sp statistics computed from the whole AFLP dataset resulted in a ratio of 0·463 (95 % confidence interval: 0·251; 0·908), indicating that realized gene flow was significantly more restricted in the trailing-edge population. Similarly, comparative Sp statistics computed from the neutral AFLP markers only resulted in a ratio of 0·468 (95 % confidence interval: 0·281; 0·994).
Table 2.
Spatial autocorrelation of AFLP genotypes in the trailing-edge and the leading-edge populations of Biscutella laevigata
| Trailing edge – all loci (95 % CI*) | Trailing edge – neutral loci (95 % CI*) | Leading edge – all loci (95 % CI*) | Leading edge – neutral loci (95 % CI*) | |
|---|---|---|---|---|
| F1† | 0·2280 (0·2109; 0·2510) | 0·2094 (0·1925; 0·2307) | 0·1526 (0·0765; 0·2145) | 0·1531 (0·0769; 0·2153) |
| b‡ | −0·0397 (−0·0429; −0·0368) | −0·0337 (−0·0369; −0·0311) | −0·0069 (−0·0111; −0·0033) | −0·0071 (−0·0123; −0·0039) |
* 95 % confidence interval (CI) as computed by 999 permutations.
† Relationship coefficient in the first distance class.
‡ Slope of the regression between the relationship coefficient and the logarithm of spatial distance.
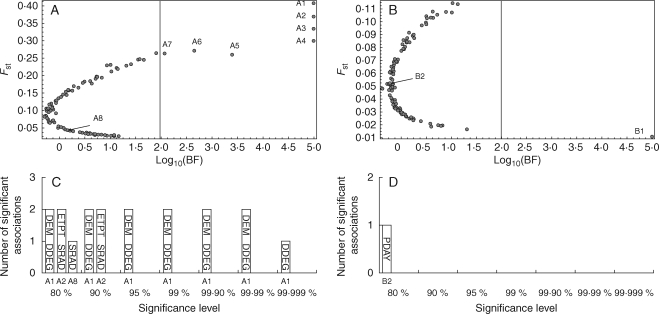
In the trailing-edge population, BayeScan identified seven AFLP markers possibly under selection (Fig. 2A): A1 (posterior probability = 1; log10 of Bayes Factor = 5); A2 (pb = 1; log10 BF = 5); A3 (pb = 1; log10 BF = 5); A4 (pb = 1; log10 BF = 5); A5 (pb = 1; log10 BF = 3·4); A6 (pb = 0·998; log10 BF = 2·66); A7 (pb = 0·992; log10 BF = 2·07). Sam identified three loci putatively under selection (A1, A2 and A8) by total solar radiation, degree-days during growing season, altitude (possibly correlated with other ecological determinants) (Fig. 2C). Two of the strongest outlier loci (A1 and A2) were clearly detected by both BayeScan and Sam. In the leading edge, BayeScan identified only one AFLP marker with strong evidence of purifying selection (Fig. 2B): B1 (pb = 1; log10 BF = 5). Sam identified a different AFLP locus (B2) with a maximum confidence level of 80 % (Fig. 2D).
Fig. 2.
Genome scans for loci under selection in a trailing-edge population (A, C) and a leading-edge population (B, D) of Biscutella laevigata. (A) and (B): BayeScan analysis, with the posterior probability for a locus to be under natural selection on the x-axis (log scale). The vertical line indicates the threshold beyond which there is a decisive evidence for selection. (C) and (D) Sam analysis, with significant associations between particular AFLP loci and eco-climatic parameters indicated on bars (DEM, altitude; DDEG, degree-days during growing season; ETPT, daily average evapotranspiration; PDAY, number of precipitation days during growing season; SRAD, daily average radiation). Sections on the x-axis refer to different significance levels. Loci detected by BayeScan and/or Sam are called A1 to A8 and B1 and B2.
DISCUSSION
Genetic structure, as assessed here from the spatial distribution of AFLP genotypes through comparative Sp statistics, indicated that gene flow was above two times more restricted in the trailing-edge than in the leading-edge population of B. laevigata (ratio: 0·463). Despite differences according to uncontrolled factors (e.g. linear vs. two-dimensional habitats), the populations here surveyed presented great ecological similarity and were mostly distinguished by historical features (Table 1), suggesting an association between spatial genetic structure and the location at either the trailing edge or the leading edge.
Results from genome scans (BayeScan and Sam) pointed to a differential efficiency of selective processes in the two types of marginal populations (Fig. 2). Although such inferences in real populations have to be interpreted with caution (see Nielsen, 2005), the methods adopted here are reputed to provide reliable inferences under complex biological scenarios. The robustness of the BayeScan procedure against non-equilibrium situation was demonstrated, especially when assessing a reasonably large number of groups (Foll and Gaggiotti, 2008). Furthermore, Sam works at the individual level and is independent of any theoretical genetic model (Joost et al., 2007). Comparative frameworks inspecting different populations with exactly the same approach, such as used here, should further alleviate from potential drawbacks since the main limitations are expected to apply equally to the contrasted situations. Finally, we argue that congruent results from totally independent approaches (BayeScan and Sam) bring decisive evidence favouring a reliable detection of the loci putatively under selection. Accordingly, the present results from genome scans indicate that, all else being equal, divergent selection was more efficient at the trailing edge than at the leading edge.
In the trailing-edge population, Sam highlighted loci (A1 and A2) referred as under directional selection by BayeScan to be significantly correlated to degree-days during growing season and total solar radiation. These ecological factors related to habitat thermal conditions were already associated with the genetic structure in this population (Parisod and Christin, 2008). The present results thus further indicate that selection by climatic factors may participate in maintaining pervasive genome-wide divergence at a local scale. Accordingly, since both BayeScan and Sam detected putative adaptive loci (Fig. 2), the stronger genetic structure observed in the trailing-edge population is linked to selection against migrants to a certain extent (Nosil et al., 2008). Since realized gene flow assessed with all AFLP markers was slightly lower than gene dispersal inferred with neutral markers only (Table 2), the loci detected here as under divergent selection might contribute to incipient reproductive isolation and, together with other barriers, sustain genome-wide association to environmental heterogeneity (Nosil et al., 2005; Parisod and Christin, 2008). In marked contrast, few loci putatively under selection were detected in the leading-edge population, and BayeScan and Sam provided incongruent results. No signature of divergent selection emerged in the leading-edge population of B. laevigata, although it was investigated with a stronger sampling than the trailing-edge one. This evidence supports the hypothesis that selective processes were less effective across the leading-edge population. Since expansion going on with high levels of admixture was detected in this population (Parisod and Bonvin, 2008), the lack of apparent selective pattern might be due to either comparatively intense recombination, limiting detection of selective patterns with genome scans, or to relatively high gene dispersal, hampering selection.
In this study, restricted gene flow at the trailing edge was observed with patterns of selection, while relatively high gene flow at the leading edge was associated with a lack of selection signature. Considering that species ranges experience strong historical heterogeneity due to past climate changes (Hampe and Petit, 2005), time passed since the establishment of populations may be an expected driver of spatial patterns of genetic variation. Indeed, gene pools persisting at the trailing edge obviously had long time periods to diversify and reach equilibrium, which suggests a strong influence of selective processes in sorting genotypes. In contrast, leading-edge populations are younger and characterized by non-equilibrium conditions, suggesting that dispersal processes may be prevalent in shaping genetic variation. Accordingly, the dynamic processes acting during expansion into empty habitats could promote either a thinning of genetic diversity, with an accordingly low potential of adaptation (Pujol and Pannell, 2008), and/or an increased gene flow, possibly preventing local adaptation (Parisod and Bonvin, 2008).
Replication at the population level is required to ascertain that historical fluctuation in population sizes and/or differential selection regimes shaped distinctive spatial genetic structure in trailing vs. leading-edge populations. However, equilibrium should not be expected across the entire range of species (Vucetich and Waite, 2003) and the present work stresses the need to rely on dynamic perspectives when investigating processes acting on populations at range margins. We advise using demographic insights from phylogeography to distinguish between trailing- and leading-edge populations in future studies, promising a deeper understanding of evolution and adaptation across the range of various species.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
We thank Rolf Holderegger, Ovidiu Paun and anonymous reviewers for insightful comments. C.P. acknowledges financial support by the Fondation Hainard.
LITERATURE CITED
- Ackerly DD. Community assembly, niche conservatism, and adaptive evolution in changing environments. International Journal of Plant Sciences. 2003;164:S165–S184. [Google Scholar]
- Foll M, Gaggiotti OE. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe A, Petit RJ. Conserving biodiversity under climate change: the rear edge matters. Ecology Letters. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2:618–620. [Google Scholar]
- Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996;58:247–276. [Google Scholar]
- Jansson R, Dynesius M. The fate of clades in a world of recurrent climatic change: Milankovitch oscillations and evolution. Annual Review of Ecology and Systematics. 2002;33:741–777. [Google Scholar]
- Joost S, Bonin A, Bruford MW, et al. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Molecular Ecology. 2007;16:3955–3969. doi: 10.1111/j.1365-294X.2007.03442.x. [DOI] [PubMed] [Google Scholar]
- Joost S, Kalbermatten M, Bonin A. Spatial analysis method (SAM): a software tool combining molecular and environmental data to identify candidate loci for selection. Molecular Ecology Resources. 2008;8:957–960. doi: 10.1111/j.1755-0998.2008.02162.x. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Nosil P, Egan SP, Funk DJ. Heterogeneous genomic differentiation between walking-stick ecotypes: ‘isolation by adaptation’ and multiple roles for divergent selection. Evolution. 2008;62:316–336. doi: 10.1111/j.1558-5646.2007.00299.x. [DOI] [PubMed] [Google Scholar]
- Nosil P, Vines TH, Funk DJ. Perspective: reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- Olowokudejo JD, Heywood VH. Cyto-taxonomy and breeding system of the genus Biscutella (Cruciferae) Plant Systematics and Evolution. 1984;145:291–309. [Google Scholar]
- Parisod C. Postglacial recolonisation of plants in the western Alps of Switzerland. Botanica Helvetica. 2008;118:1–12. [Google Scholar]
- Parisod C, Besnard G. Glacial in situ survival in the Western Alps and polytopic autopolyploidy in Biscutella laevigata L. (Brassicaceae) Molecular Ecology. 2007;16:2755–2767. doi: 10.1111/j.1365-294X.2007.03315.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Bonvin G. Fine-scale genetic structure and marginal processes in an expanding population of Biscutella laevigata L. (Brassicaceae) Heredity. 2008;101:536–542. doi: 10.1038/hdy.2008.95. [DOI] [PubMed] [Google Scholar]
- Parisod C, Christin PA. Genome-wide association to fine-scale ecological heterogeneity within a continuous population of Biscutella laevigata (Brassicaceae) New Phytologist. 2008;178:436–447. doi: 10.1111/j.1469-8137.2007.02361.x. [DOI] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. Evolutionary consequences of autopolyploidy. New Phytologist. 2010 doi: 10.1111/j.1469-8137.2009.03142.x. in press. doi:10.1111/j.1469-8137.2009.03142.x. [DOI] [PubMed] [Google Scholar]
- Pujol B, Pannell JR. Reduced responses to selection after species range expansion. Science. 2008;321:96. doi: 10.1126/science.1157570. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Stehlik I, Holderegger R, Tribsch A. Molecular evidence for glacial refugia of mountain plants in the European Alps. Molecular Ecology. 2005;14:3547–3555. doi: 10.1111/j.1365-294X.2005.02683.x. [DOI] [PubMed] [Google Scholar]
- Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology. 2004;13:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- Vucetich JA, Waite TA. Spatial patterns of demography and genetic processes across the species' range: null hypotheses for landscape conservation genetics. Conservation Genetics. 2003;4:639–645. [Google Scholar]
- Zimmermann NE, Kienast F. Predictive mapping of alpine grasslands in Switzerland: species versus community approach. Journal of Vegetation Science. 1999;10:469–482. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.