Abstract
Purified histone H2A.Z from chicken erythrocytes and a sodium butyrate-treated chicken erythroleukemic cell line was used as a model system to identify the acetylation sites (K4, K7, K11, K13, and K15) and quantify their distribution in this vertebrate histone variant. To understand the role played by acetylation in the modulation of the H2A.Z nucleosome core particle (NCP) stability and conformation, an extensive analysis was conducted on NCPs reconstituted from acetylated forms of histones, including H2A.Z and recombinant H2A.Z (K/Q) acetylation mimic mutants. Although the overall global acetylation of core histones destabilizes the NCP, we found that H2A.Z stabilizes the NCP regardless of its state of acetylation. Interestingly and quite unexpectedly, we found that the change in NCP conformation induced by global histone acetylation is dependent on H2A/H2A.Z acetylation. This suggests that acetylated H2A variants act synergistically with the acetylated forms of the core histone complement to alter the particle conformation. Furthermore, the simultaneous occurrence of H2A.Z and H2A in heteromorphic NCPs that most likely occurs in vivo slightly destabilizes the NCP, but only in the presence of acetylation.
In the nucleus of the living cell, DNA interacts with a set of highly basic proteins called histones to form a nucleoprotein complex, which is called chromatin (1). Histones are grouped into two major types: core histones (H2A, H2B, H3, and H4) and linker histones (histone H1). The former organize the protein core around which approximately 146 bp of DNA is wrapped to produce the basic chromatin subunit, the nucleosome core particle (NCP).1 Histones of the H1 family bind to the linker DNA regions connecting adjacent nucleosomes. Both core and linker histones consist of nonallelic variants. Some of these are expressed exclusively during S phase, and the mRNAs transcribed from their genes are not polyadenylated (replication-dependent variants). They are also often termed canonical variants. Other variants are expressed throughout the cell cycle; their mRNAs are polyadenylated, and they replace the replication-dependent variants to specify function or supplement their loss during cell cycle turnover (replacement variants).
Of all core histones, H2A has the largest number of variants. There have been at least 13 canonical H2A variants identified in mammalian Jurkat cells (H2A.A, H2A.C, H2A.E, H2A.G, H2A.L, H2A.M, H2A.O, H2A.Q, Q7L7L0, Q8IUE6, Q96KK5, Q96QV6, and Q9BTM1) (2). The replacement variants include H2A.Z, H2A.X, the vertebrate-specific macroH2A, and H2A. Bbd (see reviews in refs 3 and 4).
In recent years, histone H2A.Z has been the subject of an intensive structural and functional characterization (3). Despite this effort, some of the data have been controversial, and the structural information is still unable to provide a clear answer about how H2A.Z performs its many functions (4).
The functional information gathered so far points to an important physiological role for H2A.Z in the establishment and maintenance of chromatin boundaries (5), including those boundaries that demarcate genes (6) and those that define promoter elements (7). Interestingly, the insulator binding protein CTCF is flanked by arrays of well-positioned nucleosomes that are enriched in H2A.Z as well as several histone PTMs (8). In addition, an enrichment of H2A.Z is found at the 5′ ends of genes both in yeast (9) and in vertebrates (10) where the acetylated form is also present at certain enhancers (11). This contrasts with the involvement of H2A.Z in the postmeiotic assembly of mammalian X and Y chromosomes into facultative heterochromatin that is enriched in H2A.Z as well as at pericentric heterochromatin during early development. It is possible that H2A.Z PTMs may account, at least in part, for these diverse functions. H2A.Z present at the inactive X chromosome of female cells has been shown to be monoubiquitinated, and H2A.Z acetylation appears to be critical for the prevention of heterochromatin spreading into euchromatin domains (12; see reviews in refs 3 and 4). However, the details of the molecular mechanisms involved remain unclear.
The first attempt to characterize acetylation of H2A.Z revealed that the acetylation pattern of this variant was quite different from that of the major canonical variant H2A.1 (13); however, the sites of acetylation could not be determined. A study using the ciliate Tetrahymena thermophila in which H2A.Z (hν1) was found to be preferentially associated with active genes of the macronucleus identified six acetylation sites (K4, K7, K10, K13, K16, and K21) in the N-terminal region (14). In yeast, Htz1 is acetylated at K3, K8, K10, and K14, with K14 being the most abundant acetylated site. In humans, H2A.Z was also shown to be acetylated; three sites [K4, K7, and K11 (15, 16)] have been identified so far, but their relative abundance was not determined.
From a structural perspective, the studies in Tetrahymena revealed that acetylation of H2A.Z works to modulate an essential charge patch (17) that can be replaced by the N-terminal tail of H2A (14). The structural effects that this might have on the nucleosome and the chromatin fiber are unknown. We have previously shown that acetylation of histone H2A.Z by itself and/or in conjunction with acetylation of other core histones destabilizes the association of H2A.Z with chromatin (18). Interestingly, the combination of an unacetylatable H2A.Z and a mutant version of histone H4 that could not be acetylated by NuA4 is lethal in yeast (12), suggesting a concerted action of histone acetylation.
In this study, we characterize the relative abundance and distribution of acetylation in vertebrate H2A.Z before and upon inhibition of HDACs using sodium butyrate. The role this acetylation plays in the conformation and stability of the nucleosome core particle (NCP) is also characterized.
MATERIALS AND METHODS
Native Histones and DNA
Native histones used for NCP reconstitutions were obtained from HeLa cells grown in the absence or presence of 5 mM sodium butyrate and subsequently fractionated by HPLC and reconstituted as described elsewhere (19).
Native H2A.Z in its nonacetylated and acetylated forms was purified from chicken erythrocytes and from chicken MSB cells (chicken erythroleukemic cells transformed by Marek’s virus) (20) grown in the presence of 5 mM sodium butyrate, respectively. Histones were prepared from the isolated nuclei of these two tissues upon HCl extraction as described elsewhere (21). Approximately 30–40 mg of the HCl protein extracts was loaded onto a 1.5 cm × 120 cm BioGel P-60 (Bio-Rad) column and eluted in 50 mM NaCl and 20 mM HCl at a rate of 5 mL per three fractions per hour. In this chromatographic fractionation, H1 and H5 elute first followed by an H2A/H2A.Z peak and subsequent H2B/H3 and H4 peaks. An H2A.Z-enriched H2A fraction corresponding to the late eluting part of the H2A/H2A.Z peak was dialyzed and lyophilized, and H2A.Z was purified using reverse phase HPLC fractionation.
DNA was obtained from (155 ± 5 bp) DNA nucleosome core particles prepared from micrococcal nuclease-digested chicken erythrocyte chromatin (22). DNA purification was conducted by repeated phenol/chloroform extractions and recovered by ethanol precipitation.
Recombinant H2A.Z
The coding region of HsH2AFZ (NM_002106) or mimic mutant forms were cloned into the pET11 expression vector (Novagen). For protein expression, the vector was introduced into BL21 (DE3) Escherichia coli (Novagen), and the bacteria were grown in 1 L of LB medium. Cells were grown to an A600 of 0.8, and isopropyl β-d-thiogalactoside was added to a final concentration of 1 mM. Cells were harvested after 3 h by centrifugation at 5000g for 10 min in 4 °C. Cell pellets were resuspended in buffer [6 M GuHCl, 1 mM EDTA, 1 mM DTT, and 50 mM Tris-HCl (pH 7.5)] and then dounced for 40 strokes in a homogenizer. The homogenized sample was dialyzed against 0.1 M NaCl, 50 mM Tris-HCl (pH 7.5), and 1 mM EDTA for 2 h at 4 °C. After dialysis, HCl was added to a final concentration of 0.5 N, and the cells were centrifuged at 110000g for 15 min at 4 °C. Six volumes of acetone was added to the supernatant, and proteins were precipitated overnight at −20 °C. The next day, the sample was centrifuged at 12000g for 10 min at 4 °C followed by HPLC fractionation.
Chromatin Fractionation with Western Blot Analysis and PCR
Hela S3 cells were grown in suspension to a density of 5 × 105 cells/mL in the presence or absence of 5 mM sodium butyrate. S1, SE, and P chromatin fractions were isolated as described previously (18). Briefly, nuclei at a DNA concentration of 2 mg/mL in 50 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, 10 mM PIPES (pH 6.8) buffer were digested with micrococcal nuclease (Worthingon) at 30 units/mg of DNA for 5 min at 37 °C. The reaction was stopped by addition of EDTA to a final concentration of 10 mM on ice, and the sample was centrifuged at 10000g for 10 min at 4 °C to yield an S1 supernatant and a pellet. The pellet was resuspended and lysed in 0.25 mM EDTA (pH 7.5) and stirred for 1 h at 4 °C. Upon centrifugation as before, a supernatant SE and a final pellet P were thus obtained. Under the experimental conditions used here, S1, SE, and P correspond to approximately 5–10, 25–30, and 60–65%, respectively, of the total nuclear DNA. Western blotting was performed on the protein components of these fractions using SDS–PAGE and standard procedures. Histone H3K9me3 antibody was from Abcam and was used at a 1:5000 dilution; the H4 antibody was produced in house and was used at a 1:10000 dilution, and the acetylated H2A.Z antibody (10) was a kind gift from C. Crane-Robinson and was used at a 1:1000 dilution.
For the PCR analysis, a 147 bp fragment of the human protamine 1 gene was amplified from equal amounts of DNA from the S1, SE, and P fractions using the following two primers: CTT TGC CCT CAC AAT GAC C (forward) and GGC TTG GCT GAA TGC TCA (reverse). The fragment was selected from the chromatin organization of the rat protamine 1 gene (23) but using the DNA sequence of the highly homologous human counterpart. It encompasses a full NCP at the promoter region. Amplification was conducted with 32 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s and final extension at 72 °C for 7 min. The enzyme was platinum Taq DNA polymerase (Invitrogen).
Polyacrylamide Gel Electrophoresis
Proteins were analyzed via 15% SDS–PAGE. Gels were then stained with 0.2% (w/v) Coomassie blue in 25% (v/v) 2-propanol and 10% (v/v) acetic acid and destained in 10% (v/v) 2-propanol and 10% (v/v) acetic acid.
DNA and nucleosome core particles were analyzed by 4.5% native PAGE (29:1 acrylamide:bisacrylamide) in 0.5× TBE (45 mM Tris, 45 mM boric acid, and 1 mM EDTA).
Nucleosome Core Particle Reconstitution
A histone protein titration was carried out using SDS–PAGE to ensure that all histones in the final mixture were present in equimolar amounts. The histone mixture thus obtained was dialyzed overnight against 2 M NaCl, 10 mM Tris-HCl (pH 7.5), 10 mM β-mercaptoethanol, and 0.1 mM EDTA at 4 °C and was mixed with 155 bp random sequence chicken erythrocyte DNA in the same buffer, at a histone:DNA ratio of 1.13:1.0 (w/w). Nucleosome core particle reconstitution was conducted using salt gradient dialysis (19). The integrity of the core particles was analyzed by 4% native PAGE and sedimentation velocity in the analytical ultracentrifuge (see below). Table 1 summarizes the different reconstituted NCPs used in this work.
Table 1.
Summary and Nomenclature of the Reconstituted NCPs Used in This Work According to Their Histone Compositiona
| name | H2A variant | histone complement (H2B, H3, H4) |
|---|---|---|
| 2A-N | H2Ab | native (N)b |
| r2A-N | rH2Ac | native (N) |
| r2AZ-N | rH2A.Zd | native (N) |
| r2A4Q-N | rH2AQ5-Q9-Q13-Q15c | native (N) |
| r2AZ3Q-N | rH2A.ZQ4-Q7-Q11d | native (N) |
| 2A/r2AZ-N | H2Ab/rH2A.Zd | native (N) |
| 2A-N* | H2A | acetylated (N*)e |
| 2A*-N* | Ac*H2Ae | acetylated (N*) |
| 2AZ*-N* | Ac*H2A.Zf | acetylated (N*) |
| r2AZ-N* | rH2A.Z | acetylated (N*) |
| r2A4Q-N* | rH2AQ5-Q9-Q13-Q15 | acetylated (N*) |
| r2AZ3Q-N* | rH2A.ZQ4-Q7-Q11 | acetylated (N*) |
| r2AQ36-N* | rH2AQ36 | acetylated (N*) |
| 2A*/ r2AZ3Q-N* | Ac*H2Ae/ rH2A.ZQ4-Q7-Q11 | acetylated (N*) |
The asterisks denote acetylation.
Native (N) histones obtained from HeLa cells.
Recombinant human H2A.
Recombinant human H2A.Z.
Native acetylated (N*) histones obtained from butyrate-treated HeLa cells.
Native (H2A.Z) obtained from butyrate-treated chicken MSB cells.
Analytical Ultracentrifuge
Reconstituted NCPs were dialyzed against buffers of varying ionic strengths and were subjected to analytical ultracentrifuge analysis as described elsewhere (22). Briefly, sedimentation velocity runs were performed in a Beckman XL-I analytical ultracentrifuge (Beckman-Coulter Instruments, Fullerton, CA) in an An-55 Al aluminum rotor using cells with double-sector aluminum-filled Epon centerpieces. A value of 0.650 cm/g was used for the partial specific volume of NCP (22). Absorbance scans were routinely obtained at 260 nm, and the boundaries were analyzed as described elsewhere (19) with the help of UltraScan 8.0 sedimentation data analysis software (Borries Demeler, Missoula, MT).
Mass Spectrometric Analysis
Upon purification of H2AZ as described above, the H2AZ-containing fractions from chicken erythrocytes and from sodium butyrate-treated MSB cells were lyophilized and reconstituted in 100 mM ammonium bicarbonate (pH 8). An aliquot (10%) of each fraction was treated with propionic anhydride to derivatize ε-amino groups of lysine residues. Chemical derivatization with propionic anhydride converts the amino groups to their corresponding propionyl amides, and these methods have been detailed previously (24). In brief, equal volumes of propionylation reagent (15 μL) and H2AZ (15 μL) were reacted, and derivatization was performed twice to ensure full conversion. The sample was vacuum-dried after each derivatization. The two derivatized H2AZ samples were then digested with 4 ng of trypsin (Promega) for 8 h at 37 °C. Derivatization blocks lysine residues from cleavage, and thus, trypsin cleaves C-terminal to arginine residues only. Following digestion, the samples were again reacted with propionic anhydride to derivatize the amino termini of the trypsin-generated H2A.Z peptides. Both samples were dried a final time in a speed-vac concentrator and were subsequently reconstituted in 0.1% acetic acid.
The resulting H2A.Z peptide mixtures were separately analyzed via tandem mass spectrometry. Each sample was pressure-loaded onto a capillary precolumn (360 μm outside diameter × 75 μm inside diameter fused silica) packed with 4 cm of C18 reverse phase resin (5–20 μm diameter). After being washed for 5 min with 0.1% acetic acid, the precolumn was connected to an analytical column (360 μm outside diameter × 50 μm inside diameter fused silica) packed with 8 cm of C18 (5 μm diameter) resin and equipped with an electrospray emitter tip as previously described (25). H2A.Z peptides were eluted using nanoflow HPLC with an 1100 series HPLC pump (Agilent Technologies), and a flow rate of 60 nL/min was achieved by splitting the flow from the HPLC. The gradient was from 0 to 60% B over 50 min and from 60 to 100% B over 10 min (solvent A, 0.1 M acetic acid; solvent B, 70% acetonitrile and 0.1 M acetic acid). Gradienteluted peptides were ionized using an electrospray ionization source, modified for nanospray, and were analyzed using a hybrid quadrupole linear ion trap Fourier transform (LTQ-FT) mass spectrometer (Thermo Scientific). The LTQ-FT instrument was operated with a data-dependent method, which consisted of acquisition of a full scan mass spectrum using the FT as the analyzer followed by 10 MS/MS scans of the 10 most abundant ions in the initial full scan. Full scan mass spectra were acquired for m/z 300–2000 using a resolution of 100000. Upon collision-activated dissociation (CAD) of precursor ions, MS/MS spectra were acquired using the ion trap as the analzyer. Dynamic exclusion was enabled by which precursor ions were selected for dissociation two times within 20 s before they were added to the exclusion list for 30 s. All CAD MS/MS spectra were interpreted manually for the identification of acetylation sites on the H2A.Z N-terminal peptide, AGGKAGKDSGKAKAKAVSR.
To obtain the relative abundances of all the H2AZ N-terminal species (unacetylated, singly acetylated, doubly acetylated, and triply acetylated), accurate mass measurements taken in the FT were utilized to generate selected ion chromatograms (SICs). A window of ±0.005 Da around the theoretical monoisotopic m/z values of the [M + 2H]2+ and [M + 3H]3+ ions was used to generate the SICs for the four N-terminal H2AZ species. The area under the peak in each of the SICs was used to determine the percent relative abundance of each species with respect to the total area of all N-terminal species.
RESULTS
Chicken H2A.Z Is Equally Acetylated at K4 and K7, and the Extent of Acetylation Gradually Decreases at K11, K13, and K15 of Its N-Terminal Region
A mass spectrometric method was employed to determine the relative extent of acetylation of the lysine residues of the N-terminal tail of H2A.Z isolated from chicken erythrocytes (Table 2A) or sodium buty-rate-treated chicken erythroleukemic cells (Table 2B). Purified H2A.Z was derivatized with propionic anhydride to modify ε-amino groups on lysine residues and thus limit proteolytic cleavage to arginine residues upon digestion with trypsin (24). H2A.Z peptides were then analyzed via LC-coupled tandem mass spectrometry, which facilitated sequence analysis of the N-terminal H2A.Z peptide and allowed for identification of multiple acetylation sites on the N-terminal lysine residues. In addition, the mass spectrometric technique that was used in this study included acquisition of accurate mass measurements of all the acetylated and unacetylated forms of the N-terminal peptide. The accurate masses obtained were subsequently utilized to generate selected ion chromatograms (as described in Materials and Methods) for determination of the relative abundance of all the identified forms.
Table 2.
Sites of Chicken H2A.Z Acetylation and Their Relative Abundances
| modification | percent of total sample |
modified lysine residues |
|---|---|---|
| (A) N-Terminal Peptide of H2A.Z from Native Chicken Erythrocytes (AGGKAGKDSGKAKAKAVSR) | ||
| unmodified | 90 | |
| monoacetylated | 9 | K4 = K7 > K11 > K13 ≫ K15 |
| diacetylated | < 1 | K4 + K7, K7 + K11, K4 + K11 |
| triacetylated | < 0.1 | K4 + K7 + K11 |
| (B) N-Terminal Peptide of H2A.Z from Butyrate-Treated MSB Cells (AGGKAGKDSGKAKAKAVSR) | ||
| unmodified | 40 | |
| monoacetylated | 20 | K4 = K7 > K11 > K13 ≫ K15 |
| diacetylated | 20 | K4 + K7, K7 + K11, K4 + K11 |
| triacetylated | 20 | K4 + K7 + K11 |
In the native chicken erythrocytes, 90% of the H2A.Z N-terminus is unmodified, whereas the unmodified N-terminal peptide has a relative abundance of less than half of this amount in the butyrate-treated cells. While all five lysine residues were found to be acetylated, the incidence of acetylation on K4 occurs with approximately the same frequency as that on K7, and these are both more abundant than acetylation on K11, K13, or K15 (Table 2). This pattern of abundance of acetylation on the N-terminal lysine residues can be seen in both the untreated erythrocytes and the sodium butyrate-treated erythroleukemic cells. In addition to detection of singly acetylated peptides, H2A. Z N-terminal peptides that were concurrently modified with multiple acetyl groups were also observed. The doubly acetylated forms of the N-terminal peptide that were identified included the following: K4Ac + K7Ac, K7Ac + K11Ac, and K4Ac + K11Ac (Table 2). These forms were identified in both the treated and untreated cells. Furthermore, triply acetylated N-terminal peptides were also identified, with the predominant species containing acetyl groups on K4, K7, and K11; this form accounts for more than 95% of all triply acetylated species detected (Figure 1). In the untreated chicken erythrocytes, the triply acetylated species are present at less than 0.1%, whereas these species account for 20% of the N-terminal peptide forms in the sodium butyrate-treated MSB cells.
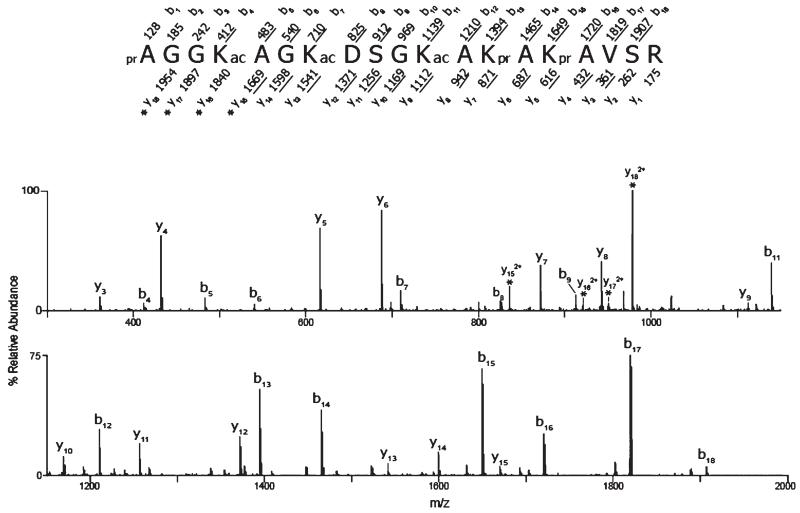
Figure 1.
CAD MS/MS spectrum of triply acetylated H2A.Z peptide 1–19. The ion selected for dissociation was the [M + 2H]2+ ion (m/z 1041.1). The amino acid sequence is shown above the spectrum, and the masses above and below the sequence correspond to the theoretical b- and y-type product ions, respectively. The masses provided are the monoisotopic, nominal masses of the product ions. The observed, singly protonated b- and y-type ions are underlined and are assigned to their corresponding m/z peaks in the spectrum. The observed, doubly protonated ions are denoted with asterisks. The acetylated lysines (K4, K7, and K11) are indicated with ac. The unacetylated amino groups were derivatized with propionic anhydride and are denoted with pr.
The Chromatin Distribution of Acetylated H2A.Z Changes upon Sodium Butyrate Treatment
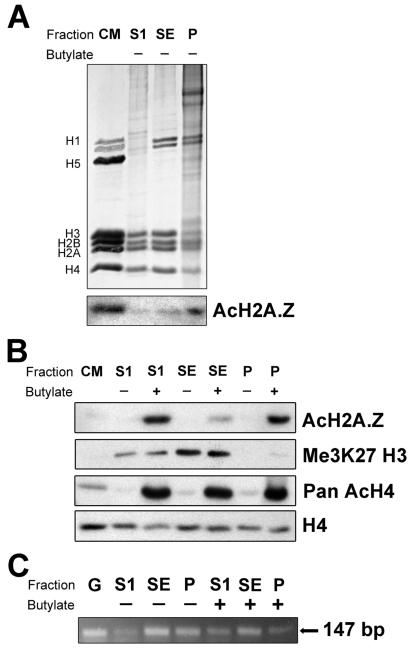
Micrococcal nuclease digestion of chromatin under the conditions described in Materials and Methods leads to a partitioning in three different fractions; S1, SE, and P. Fraction S1 is highly enriched in actively transcribing genes (26, 27) and basically corresponds to the euchromatin fraction. Fraction SE is less readily accessible to nuclease, and as indicated by the Western blot with an anti-trimethyl K27 H3 antibody which is a known marker of facultative heterochromatin (28), it consists mainly of chromatin fragments from transcriptionally inactive regions. The pellet P represents insoluble chromatin material which is resistant to nuclease digestion and is not associated with heterochromatin markers.
Analysis of chromatin fractions generated in this way (Figure 2A) by Western blot using an antibody against triacetylated H2A.Z at K4, K7, and K11 (10) indicates that the levels of this modified histone are dramatically increased when HeLa cells are treated with sodium butyrate (Figure 2B). Furthermore, the butyrate treatment changes the relative fraction distribution of acetylated H2A.Z (compare the anti-Ac H2A.Z Western blot in Figure 2A and the anti-Ac H2A.Z Western blot in Figure 2B). In the butyrate-treated cells, a significant portion of the acetylated H2A.Z fractionates with the most active chromatin of the S1 fraction while a smaller portion is present in the trimethyl-H3 K27-enriched SE fraction (Figure 2B). In contrast, the presence of acetylated H2A.Z increases gradually from the S1 fraction to the P fraction (Figure 2A, Western) in native non-butyrate-treated cells. As expected, the global level of acetylation of H4 also increases dramatically upon butyrate treatment (29). However, the chromatin fraction redistribution is not as pronounced in the case of acetylated H4 as compared to acetylated H2A.Z (see Figure 2B). Interestingly, the acetylated forms of H2A.Z and H4 are also prominent in the P fraction which exhibits an almost complete absence of the H3 K27 heterochromatin 0.01w mark, indicating that this fraction most likely contains active regions of chromatin that are insoluble. It is likely that it consists of oligonucleosomes that are directly bound to components of the nuclear matrix, the nuclear pore, or large protein complexes (30).
Figure 2.
Chromatin partitioning of acetylated H2A.Z. (A) SDS–PAGE of HeLa cell chromatin S1, SE, and P fractions (see Materials and Methods) and Western blot analysis using an antibody against acetylated H2A.Z. (B) Western blot analysis of chromatin fractions obtained from HeLa cell nuclei grown in the presence (+) or absence (−) of 5 mM sodium butyrate (see Materials and Methods) using antibodies against acetylated H2A.Z (AcH2A.Z), methylated H3 (K27me3 H3), acetylated H4 (Pan AcH4), and H4. CM: chicken erythrocyte histones used as a marker. (C) PCR analysis of the S1, SE, and P fractions using primers for a 147 bp fragment of the human protamine 1 gene. G: HeLa genomic DNA.
PCR analysis of DNA isolated from the chromatin fractions indicates that the human protamine 2 gene, which is inactive in Hela cells, as expected is present mainly in the SE chromatin fraction and in the insoluble fraction (Figure 2C). The human protamine genes are present in a cluster that is associated with the nuclear matrix (31). Notice, however, that some gene redistribution takes place (Figure 2B) upon treatment with butyrate, in agreement with the genomic redistribution already described after treatment with HDAC inhibitors (32, 33).
H2A.Z Compacts the NCP and Enhances Its Stability Regardless of Its Acetylated State or That of the Core Histone Complement
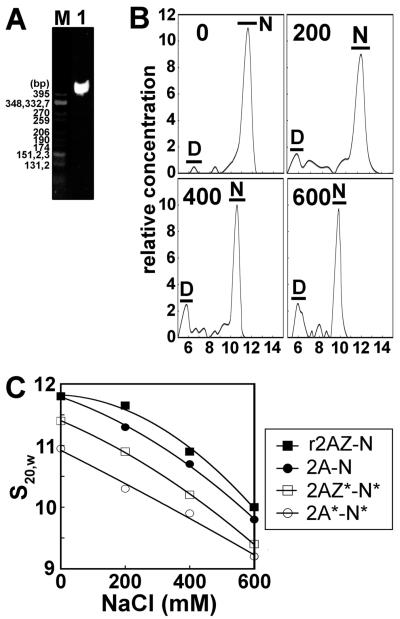
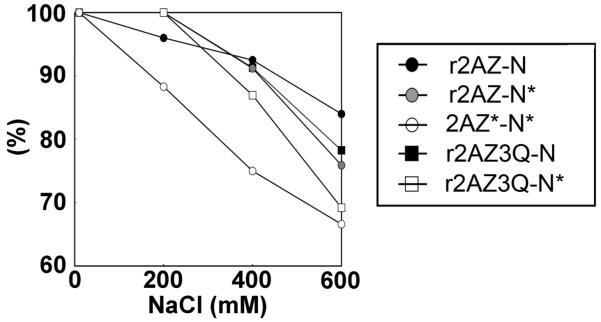
We prepared several forms of reconstituted mononucleosomes by salt gradient dialysis. All nucleosomes were analyzed on 4% native acrylamide gels of which a representative example is shown in Figure 3A. The mononucleosomes were then characterized by sedimentation velocity in the analytical ultracentrifuge where the sedimentation coefficient value can be seen to decrease as the concentration of NaCl increases (Figure 3B) as previously seen (22). To examine the effect of acetylation on the conformation of the nucleosomes, we plotted the s20,w values versus the NaCl concentration of several NCP constructs (Figure 3C). As previously published by our group (18), the H2A.Z-containing particle exhibits slightly higher sedimentation coefficients, especially at low salt levels, which is indicative of a slightly more compact conformation compared to the H2A-containing NCP.
Figure 3.
Sedimentation velocity analysis of acetylated nucleosomes. (A) Native PAGE of nucleosome core particles reconstituted with 155 bp random sequence chicken erythrocyte DNA and histone octamers consisting of native purified H2A and native non-acetylated histone complement. The marker is CfoI-digested pBR322 plasmid DNA. (B) Profile of analytical ultracentrifuge analysis of nucleosomes containing H2A and a native non-acetylated histone complement. The graphs show plots of the relative sample concentration vs the sedimentation coefficient at four different NaCl concentrations (0, 200, 400, and 600 mM). Data were obtained using the histogram envelope analysis from UltraScan (see Materials and Methods). D: free 155 bp DNA. N: nucleosome core particle. (C) Ionic strength dependence of the sedimentation coefficient (s20,W) of reconstituted NCPs that were dialyzed against 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA buffer containing different NaCl concentrations and analyzed by sedimentation velocity at 44000 rpm and 20 °C.
Also in agreement with previously published data (34, 35), it can be observed in Figure 3C that when all components of the histone octamer are acetylated the NCP displays a conformation significantly more relaxed than that of the canonical NCP regardless of whether the particles are reconstituted with H2A or H2A.Z. However, despite this, the trend toward a more compact conformation of the H2A.Z nucleosome persists when H2A.Z and the histone complement are acetylated (Figure 3C).
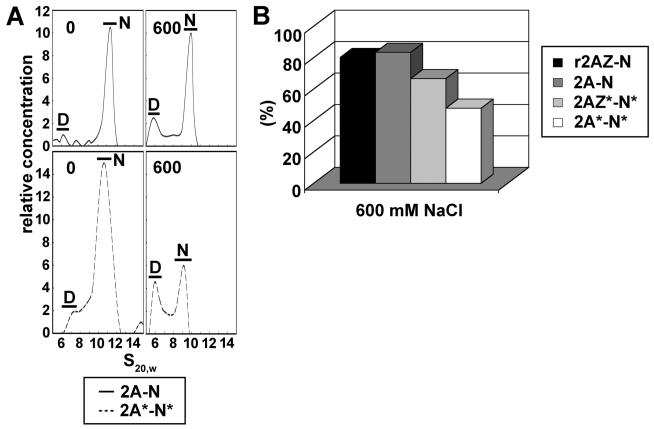
Analytical ultracentrifuge analysis was also used to measure the salt-dependent stability of the various nucleosome constructs by determining the relative amounts of nuclesomes compared to free DNA. Figure 4A shows the profile of H2A with non-acetylated histone complement nucleosomes (H2A/non Ac complement) and native acetylated H2A with acetylated histone complement nucleosomes (AcH2A/Ac complement) at 0 and 600 mM NaCl. In both cases, the relative amount of nucleosomes is decreased at high salt; however, this decrease is much more apparent in the case of the acetylated particles. Acetylation of the histones decreased the stability of the nucleosome irrespective of the presence of H2A or H2A.Z (Figure 4B), and consistent with what was seen in the conformation analysis, the acetylated H2A.Z with acetylated histone complement (AcH2A.Z/Ac complement) nucleosomes were more stable than their acetylated H2A counterparts.
Figure 4.
Salt-dependent stability of acetylated and nonacetylated H2A- and H2A.Z-containing NCPs. (A) Profile of analytical ultracentrifuge analysis of nucleosomes containing H2A with a non-acetylated histone complement or acetylated H2A with an acetylated histone complement at 0 and 600 mM NaCl. D: free 155 bp DNA. N: nucleosome core particle. (B) Nucleosome stability analysis of H2A or H2A.Z with non-acetylated histone complement, acetylated H2A, or acetylated H2A.Z with acetylated complement. Results show the percentage of undissociated nucleosome core particles containing a full histone octamer [calculated from the ratio (%) of area under N in panel A at a given salt level to the area under N in the absence of salt].
Acetylation Mimics Using Glutamine Mutants at Positions K4, K7, and K11 Suggest That Acetylation of the N-Terminal Region of H2A.Z Is Responsible for NCP Destabilization
To determine whether acetylation at lysines 4, 7, and 11 of H2A.Z specifically is involved in the observed differences in the conformation and stability of the nucleosome, we constructed the triple H2A.Z K4Q/K7Q/K11Q mutant (H2A.Z 3Q) and reconstituted nucleosomes with acetylated or non-acetylated histone complement (Figure 5). The H2A.Z 3Q mutant nucleosomes show the same salt dependence of the sedimentation coefficient (s20,w) as native non acetylated H2A.Z nucleosomes (data not shown) which indicates that these glutamine mutations by themselves do not alter the conformation of the nucleosome. As shown in Figure 4B, the AcH2A.Z/Ac complement nucleosome was less stable than the H2A.Z/non Ac complement nucleosome (Figure 5). Figure 5 also indicates that the H2A.Z/Ac complement as well as the H2A.Z 3Q/non Ac complement nucleosomes are approximately as stable as the H2A.Z/non Ac complement nucleosomes, except at 600 mM NaCl. However, the H2A.Z 3Q mutant decreased the stability of the particle in the presence of an acetylated histone complement (H2A.Z 3Q/Ac complement). The H2A.Z 3Q/Ac complement particles were still more stable than nucleosomes reconstituted with purified native acetylated H2A.Z with an acetylated histone complement (H2A.Z/Ac complement), especially at NaCl concentrations of 200 and 400 mM (Figure 5). The decreased stability of the native particles could be due to the presence of other post-translational modifications present on native H2A.Z. Alternatively, it could be the result of the incomplete ability of glutamine mutants to exactly mimic all the effects of acetylation. Regardless, it appears that the nucleosome destabilizing effect of H2A.Z acetylation takes place synergistically with the acetylation of the rest of the core histones.
Figure 5.
Analysis of the salt-dependent stability of H2A.Z-containing NCP constructs. The fraction (%) of undissociated NCPs, determined as described in the legend of Figure 4, is plotted as a function of NaCl concentration.
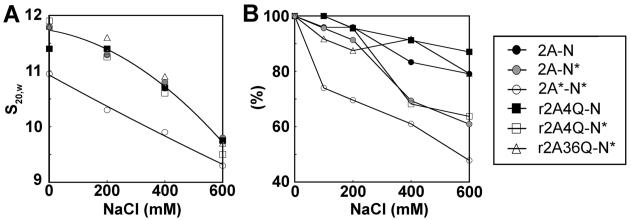
The Change in the Conformation of the Acetylated NCP Is Dependent on an Unidentified Structural Alteration of H2A
We determined that the AcH2A/Ac complement nucleosome core particles adopted the most open conformation (Figure 3C) and were the least stable of all the particles analyzed (Figure 4B). We also determined that nucleosomes reconstituted with non-acetylated H2A and acetylated histone complement (H2A/Ac complement) did not exhibit the open conformation of the fully acetylated AcH2A/Ac complement nucleosomes. Thus, we wanted to investigate whether, in addition to the loss in stability, acetylation of specific lysines within the H2A N-terminal tail was also responsible for the more open conformation of the fully acetylated nucleosomes. We found that neither native AcH2A/non Ac complement NCPs (data not shown) nor H2A/Ac complement NCPs (Figure 6A) were able to induce the more open conformation that is characteristic of the fully acetylated NCP. Furthermore, the H2A K5Q/K9Q/K13Q/K15Q mutant (H2A 4Q) or H2A K36Q mutant nucleosomes reconstituted with acetylated histone complement were equally unable to mediate the conformational change (Figure 6A). A similar observation was made with the H2A.Z 3Q mutant (data not shown). Taken together, these results indicate that native acetylated H2A (but not the H2A K/Q mutant mimics) is essential for producing a fully acetylated nucleosome that has an open conformation (34, 35).
Figure 6.
Structural analysis of H2A NCPs. (A) Ionic strength dependence of the sedimentation coefficient (s20,W) and (B) nucleosome stability analysis at different salt concentrations. The fraction (%) of undissociated NCPs at each salt concentration is indicated.
Nucleosome stability analysis of the H2A-containing constructs indicates the H2A/non Ac complement and the H2A 4Q/non Ac complement have overall high stabilities (Figure 6B). The H2A/Ac complement and H2A 4Q/Ac complement nucleosomes showed an almost equal decrease in stability that was still not as dramatic as the decreased stability of the AcH2A/Ac complement particles (Figure 6B). Therefore, these data suggest that while the H2A 4Q mutant can partially mimic the acetylation-dependent loss of nucleosome stability, it cannot mimic the acetylation-dependent conformational change of the particle.
The Heteromorphic Coexistence of H2A and H2A.Z in a Nucleosome Does Not Have a Major Effect on Its Stability or Its Conformation
Early predictions made from the crystal structure of the H2A.Z-containing nucleosome held that it was unlikely that an H2A.Z-H2A heteromorphic particle could exist (36). However, there is evidence to suggest that the occurrence of homotypic H2A.Z NCPs in vivo is rare (37), and it was recently shown that this hybrid nucleosome can be formed in vitro (38). Therefore, we sought to analyze the conformation and stability of this hybrid with special emphasis on the role played by acetylation.
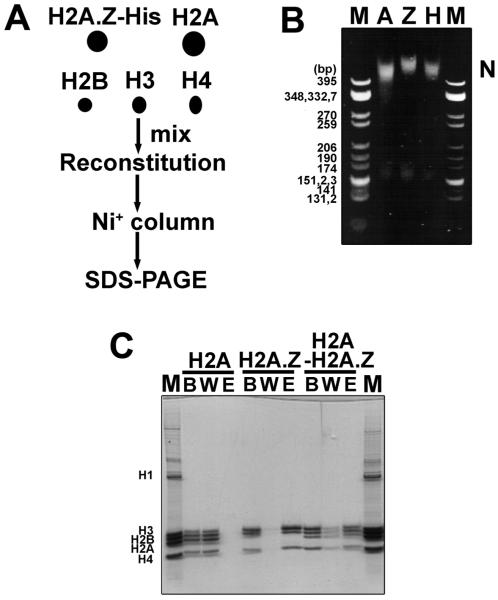
In a first approach, we constructed a His-tagged version of H2A.Z and reconstituted mononucleosomes with untagged H2A, H2B, H3, and H4 and purified these on sucrose gradients. The NCPs were loaded onto a Ni+ column and eluted with imidazole following the protocol described in ref 37 (Figure 7A). Native PAGE indicates that while the His-tagged H2A.Z nucleosome is shifted higher than the untagged H2A nucleosome, this is most likely an effect of the His tag (Figure 7B). The heteromorphic NCP exhibits an intermediate mobility (Figure 7B). The reconstituted NCPs were examined by SDS–PAGE before the wash (B), during the wash (W), and after elution (E) from the Ni+ column (Figure 7C). Since H2A was not His-tagged, no H2A nucleosomes bound to the Ni+ column as indicated by the absence of protein in lane E (Figure 7C). His-tagged H2A.Z NCPs were able to bind to the Ni+ column and be eluted by imidazole as seen in lane E (Figure 7C). The His-tagged H2A.Z comigrates with histone H3. When we performed reconstitutions with the entire mixed population of histones and loaded these purified NCPs onto the Ni+ column, we saw that as expected, the NCPs containing only H2A eluted in the wash fraction while the H2A.Z-containing NCPs bound to the column. Importantly, in the eluted fraction (E) of these mixed nucleosomes, we see a distinct band corresponding to H2A, and we can therefore conclude that these do indeed contain both H2A.Z and H2A with approximately 50% of the particles being heteromorphic (Figure 7C).
Figure 7.
H2A.Z can make a heteromorphic nucleosome with H2A. (A) Scheme for making the heteromorphic nucleosomes. (B) Native PAGE of the reconstituted sucrose gradient-purified nucleosomes loaded onto the Ni+ column. M: CfoI-digested pBR322 plasmid DNA as a marker. A: H2A nucleosome. Z: H2A. Z nucleosome. H: H2A–H2A.Z heteromorphic nucleosome. (C) SDS–PAGE (15%) analysis of nucleosomes after they had passed through the Ni+ column. M: chicken erythrocyte histone marker. B: nucleosomes before loading onto the Ni+ column. W: wash fractions. E: eluted fractions. Mix: H2A–H2A.Z heteromorphic nucleosome.
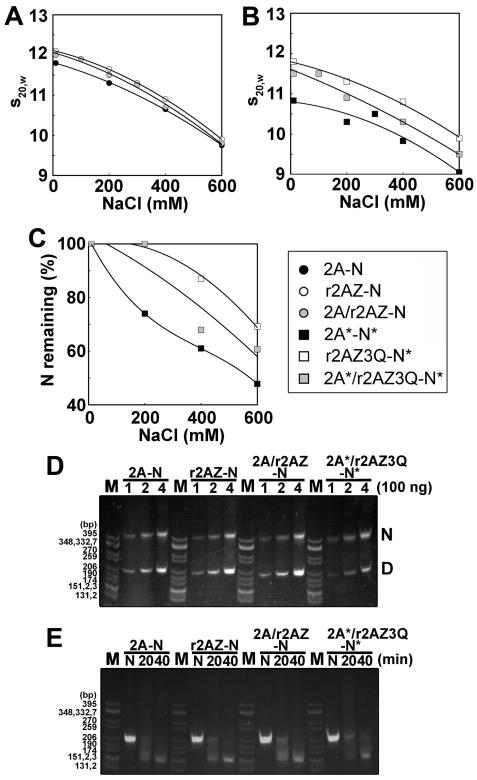
On the basis of the observations of the existence of an altered interface between the H2A.Z–H2B dimers from the crystal structure (36), we hypothesized next that the H2A.Z–H2A heteromorphic NCP would be less stable than the H2A.Z NCP. To test this hypothesis, we analyzed the hybrid H2A- and H2A.Z-containing nucleosomes in the analytical ultracentrifuge. However, we had previously demonstrated that the presence of His-tagged H2A.Z affects the conformation and decreases the stability of the NCP (39) compared to untagged native H2A.Z (18). Therefore, the approach used in Figure 6 to generate a homogeneous population of His-tagged H2A.Z–H2A hybrid nucleosomes could not be used to generate particles for AUC analysis. Since we were able to generate approximately 50% heteromorphic nucleosomes using the His-tagged H2A.Z (Figure 7C), we conducted the reconstitution using an equimolar mixture of recombinant untagged H2A.Z and H2A and a full (H2B, H3, and H4) histone complement. The NCP mixture thus obtained was analyzed in the analytical ultracentrifuge. The salt dependence of the sedimentation coefficient (Figure 8A) shows no noticeable difference with that observed with homomorphic NCPs. This indicates that the heteromorphic NCPs present in the reconstitution mixture must have a conformation very similar (Figure 8A) to those of the H2A and H2A.Z NCPs. However, acetylation of the histones resulted in heteromorphic particles with an intermediate conformation and stability (Figure 8B,C). Finally, we reconstituted untagged heteromorphic nucleosomes onto a sea urchin 5S rRNA 208 bp DNA template (Figure 8D) and performed a time course digestion using micrococcal nuclease to determine if the hybrid NCP organizes the same amount of DNA as homomorphic nucleosomes. As shown in Figure 8E, all the NCPs tested exhibited an almost identical pattern of digestion to approximately 146 bp after digestion for 40 min. This suggests that neither H2A.Z nor the presence of H2A/H2A.Z mixtures used in the NCP reconstitutions has any significant effect on the amount of DNA protected by their corresponding histone core in the reconstituted NCPs.
Figure 8.
Heteromorphic H2A–H2A.Z nucleosome with a conformation and stability intermediate between those of H2A and H2A.Z homomorphous nucleosomes. (A and B) Ionic strength dependence of the sedimentation coefficient (s20,w) of nucleosome core particles reconstituted with several constructs. Reconstituted nucleosome core particles were dialyzed against 20 mM Tris-HCl, 0.1 mM EDTA (pH 7.5) buffer containing different NaCl concentrations and analyzed by analytical ultracentrifugation at 44000 rpm and 20 °C. (C) Nucleosome stability analysis of acetylated H2A (black square), H2A.Z 3Q (white square), or an acetylated H2A/H2A.Z 3Q mixture (gray square) with acetylated complement. Results show the percentage of nucleosome core particles containing a full histone octamer. (D) Reconstituted nucleosomes run on a 4% native gel. M: CfoI-digested pBR322 plasmid DNA as a marker. H2A; nucleosome containing H2A. H2A.Z: nucleosome containing H2A.Z. Mix: heteromorphic nucleosome containing H2A and H2A.Z. AcMix: heteromorphic nucleosome containing acetylated H2A and H2A.Z 3Q with acetylated complement. Numbers are the amounts loaded, and N and D mark the nucleosome and free DNA, respectively. (E) All nucleosomes have the same sensitivity to MNase. Lanes are the same as in panel D, and the numbers are the time of MNase treatment in minutes.
DISCUSSION
Vertebrate Histone H2A.Z Is Mainly Acetylated at Lysines 4, 7, and 11, and the Chromatin Distribution of the Acetylated Forms Is Altered by HDAC Inhibition upon Treatment with Sodium Butyrate
Using an antibody against a synthetic peptide corresponding to the N-terminal tail of chicken H2A.Z acetylated at residues K4, K7, and K11, it was shown that hyperacetylated H2A.Z is a feature of active genes where it is concentrated at the 5′ end in the enhancers of some actively transcribed genes (10, 11). The acetylation signature of this replacement variant is erased at mitosis (10). Selection of the peptide for the generation of this antibody was based on information from htz1 acetylation in yeast since the sites of vertebrate H2A.Z acetylation remained undetermined at that time.
The MS analyses reported here provide the first quantitative determination of the acetylation sites in vertebrate H2A.Z and underscore the notion that a significant portion of this acetylation occurs as dimers and trimers of K4, K7, and K11 (Table 2) in contrast to yeast Htz1, where K14 represents the most abundant acetylated site (40). In Tetrahymena, the retention of one acetylatable lysine is sufficient to provide the essential function of H2A.Z acetylation (17). The fact that an important part of the acetylation pattern in vertebrate H2A.Z involves the presence of doubly acetylated forms (Table 2) indicates that the function of H2A.Z acetylation in vertebrates may involve the neutralization of more than one charge in the N-terminal region.
The finding that treatment with sodium butyrate leads to a redistribution of acetylated H2A.Z from the P fraction to the S1 fraction could be a result of the chromatin rearrangement that has been observed upon exposure of cells to HDAC inhibitors such as sodium butyrate (32, 33) and could probably be one of the reasons why H2A.Z has a lower chromatin binding affinity upon sodium butyrate treatment when analyzed by HAP chromatography (18) (see below).
Acetylation of H2A and H2A.Z Works Synergistically with Acetylation of the Rest of the Core Histone Tails To Alter the Conformation and Stability of the NCP
In a previous paper, we demonstrated that histone H2A.Z produces a small but noticeable stabilization of the native NCP against salt dissociation. A greater effect was observed for the salt-dependent dissociation of H2A.Z from chromatin using hydroxyapatite-immobilized fractions. It was observed that this salt-dependent stability could be reversed in part by histone acetylation (18). It was not clear, however, whether this was the result of histone H2A.Z acetylation itself or if it was a global effect resulting from the overall acetylation of all core histones, including H2A.Z. To address this question, we have reconstructed an extensive repertoire of NCPs (Table 1) and performed an analysis of their stability and conformation using the analytical ultracentrifuge.
The results from the sedimentation velocity analyses show that the salt-dependent loss of stability is due in part to the well-known loss of stability imparted on the NCP by the overall acetylation of core histones (41-43). However, acetylated NCPs consisting of acetylated H2A.Z are still slightly more stabilized than those consisting only of acetylated canonical H2A. Hence, the salt-mediated destabilization of interactions within the particles that contain acetylated H2A.Z must involve other factors and may be related to the DNA sequences or the nature of the chromatin regions with which the acetylated H2A.Z NCPs are associated (see below).
An unexpected result of our structural analysis was the previously undisclosed critical role of H2A and H2A.Z acetylation in the loss of stability and conformational transition undergone by acetylated NCPs (Figures 3-6). As well as decreasing its salt-dependent stability, global core histone acetylation has also been previously shown to alter the conformation of the NCP (34, 41, 44, 45) and to unfold the chromatin fiber in the absence of linker histones (35, 46). The conformational transition associated with the decrease in the sedimentation coefficient (Figure 3C) of the acetylated NCP (34) is still controversial and has been ascribed to partial release of the DNA flanking ends at its entry and exit in the NCP (35, 43) or to a distortion of its overall shape that decreases the number of times the DNA winds around the central axis of the NCP (47). Regardless of its nature, the fact that H2A and H2A.Z can critically affect this conformational transition is intriguing and suggests that these histones operate as a regulatory structural “switch”.
In terms of the unique role of H2A and H2A.Z acetylation in the decreased stability of the acetylated NCP, interestingly it has been shown by others that acetylation of H2A and H2B plays a critical role in weakening the histone–DNA off axis interactions in the acetylated NCP (48). Also, FRAP analysis has shown that histone H2A mobility increases upon acetylation of core histones induced by HDAC inhibitor treatment of living cells (49). The fact that the glutamine mutants of H2A were unable to trigger the conformational change while they could mimic the loss of stability (Figure 6) supports the notion that such loss is the result of a charge neutralization effect, whereas the change in conformation probably involves a change in the structural organization of the H2A.Z and H2A N-terminal tails which cannot be mimicked by the replacement of lysines with glutamines at the acetylation sites. In fact, we have already presented evidence that acetylation of the N-terminal tail of H4 increases the level of the α-helical conformation, shortening the span of interaction of this region with the nucleosomal DNA (20). Although it remains to be shown, it is unlikely that the K to Q replacement mimics produce a similar effect. The inability of the K to Q mutants to completely mimic the structural effects of acetylation is reminiscent of the more physiological observation made in Tetrahymena that revealed that glutamine can only partially mimic the growth effects resulting from H2A.Z acetylation (50).
The possibility also exists that acetylation of H2A and H2A.Z at other K residues (i.e., C-terminal domain) or other PTMs or a combination of these things could be responsible for the generation of the open conformation. However, this is unlikely as acetylation of H2A.Z, which has a very different C-terminal primary structure, is also able to impart (albeit to a slightly lower extent) a more open conformation (Figure 3C) and loss of stability (Figure 4B) of the NCP. Furthermore, the acetylation-essential N-terminal tail of H2A.Z (17) can be replaced by the N-terminal tail of canonical H2A (14).
A similar synergistic structural effect between acetylation of H2A and H2A.Z and that of the other core histones has also been described to operate at the level of the internucleosomal association between chromatin fibers (51). As surprising as the synergistic structural activity of acetylation of core histones may be, it is worth noting that, perhaps not so coincidentally, in vivo some of these tails share common acetyltransferase complexes as exemplified by NuA4 which can acetylate both H4 and H2A (52), including H2A.Z (53).
Heteromorphic H2A–H2A.Z NCPs Have a Conformation and Stability Similar to Those of Homomorphic H2A and H2A.Z NCPs
Whether H2A.Z is a heritable epigenetic mark is still as controversial as its functional and structural implications. The ability of certain sequence-specific transcription factors to preferentially position histone H2A.Z within chromatin has been viewed as part of an epigenetic process for the modulation of gene expression (54). However, it has been argued that should histone H2A.Z have an epigenetic role, H2A.Z should be uniquely present in a homotypic NCP or spread in neighboring NCPs (37). Whereas other histone variants such as replacement histone H3.3 and CenpA have been shown to exist as homotypic NCPs in vivo (55, 56), H2A.Z appears to be present mainly in heterotypic NCPs (37). Thus, regardless of its epigenetic role, there is experimental evidence to suggest that the heteromorphic H2A.Z–H2A NCPs are commonplace within the cell.
On the basis of some of the distinctive structural features of the canonical NCP when compared to the H2A.Z-containing NCP (36), it had been hypothesized that heteromorphic nucleosome constructs consisting of a molecule of H2A and a molecule of H2A.Z would be unstable (36). Our results with NCP mixtures containing a proportion of heterotypic H2A–H2A.Z NCPs indicate that the salt-dependent acetylation-mediated destabilization of chromatin (18) could be affected by the presence these heterotypic H2A–H2A.Z NCPs.
In conclusion, it seems that the butyrate-induced loss of the enhanced stability which H2A.Z appears to confer on the NCP within the chromatin setting (18) may be more complex than initially anticipated. From this study, it appears to be the result of a generic H2A–H2A.Z acetylation-dependent destabilization of the NCP as well as a butyrate-induced chromatin redistribution of acetylated H2A.Z to regions of the chromatin that are more accessible to nucleases (fraction S1). In this regard, there is evidence that indicates that NCPs consisting of canonical H2A and H2A.Z associate with different DNA sequences in chromatin (7, 57, 58). It had been proposed on the basis of this that as stated above, acetylation may operate as a switch that could alter the properties of both H2A and H2A.Z NCPs in a similar way but whose effect would depend on the underlying DNA sequence (14). Indeed, it has been shown that the alterations imparted by histone acetylation on NCPs are DNA sequence-dependent (42). Therefore, it is possible that a full understanding of the differential conformational and stability roles imparted by acetylated H2A.Z to the NCP will not be obtained until they are studied in this context.
Footnotes
This work was supported in part by Canadian Institutes of Health Research Grant MOP 57718 (to J.A.), National Institutes of Health Grant GM37537 (to D.F.H.), and a Michael Smith Health Research Foundation postdoctoral fellowship to T.I.
- AU
- acetic acid-urea
- AUC
- analytical ultracentrifuge
- FRAP
- fluorescence recovery after photobleaching
- HDAC
- histone deacetylase
- MS
- mass spectrometry
- NCP
- nucleosome core particle
- PAGE
- polyacrylamide gel electrophoresis
- PCR
- polymerase chain reaction
- PTM
- post-translational modification
- SDS
- sodium dodecyl sulfate.
REFERENCES
- 1.van Holde KE. Chromatin. Springer-Verlag; New York: 1988. [Google Scholar]
- 2.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol. Cell. Proteomics. 2006;5:541–552. doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Zlatanova J, Thakar A. H2A.Z: View from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Ausió J. Histone variants: The structure behind the function. Briefings Funct. Genomics Proteomics. 2006;5:228–243. doi: 10.1093/bfgp/ell020. [DOI] [PubMed] [Google Scholar]
- 5.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 6.Arimbasseri AG, Bhargava P. Chromatin structure and expression of a gene transcribed by RNA polymerase III are independent of H2A.Z deposition. Mol. Cell. Biol. 2008;28:2598–2607. doi: 10.1128/MCB.01953-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 2005;33:5633–5639. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers FA, Lefevre P, Mantouvalou E, Bruce K, Lacroix C, Bonifer C, Thorne AW, Crane-Robinson C. Developmental activation of the lysozyme gene in chicken macrophage cells is linked to core histone acetylation at its enhancer elements. Nucleic Acids Res. 2006;34:4025–4035. doi: 10.1093/nar/gkl543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20(6):700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pantazis P, Bonner WM. Quantitative determination of histone modification. H2A acetylation and phosphorylation. J. Biol. Chem. 1981;256:4669–4675. [PubMed] [Google Scholar]
- 14.Ren Q, Gorovsky MA. The nonessential H2A N-terminal tail can function as an essential charge patch on the H2A.Z variant N-terminal tail. Mol. Cell. Biol. 2003;23:2778–2789. doi: 10.1128/MCB.23.8.2778-2789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Beck HC, Nielsen EC, Matthiesen R, Jensen LH, Sehested M, Finn P, Grauslund M, Hansen AM, Jensen ON. Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics. 2006;5:1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 18.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausió J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 19.Ausió J, Moore SC. Reconstitution of chromatin complexes from high-performance liquid chromatography-purified histones. Methods. 1998;15:333–342. doi: 10.1006/meth.1998.0637. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Moore SC, Laszckzak M, Ausió J. Acetylation increases the α-helical content of the histone tails of the nucleosome. J. Biol. Chem. 2000;275:35013–35020. doi: 10.1074/jbc.M004998200. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Ausió J. Histones are the major chromosomal protein components of the sperm of the nemerteans Cerebratulus californiensis and Cerebratulus lacteus. J. Exp. Zool. 2001;290:431–436. doi: 10.1002/jez.1085. [DOI] [PubMed] [Google Scholar]
- 22.Ausió J, Dong F, van Holde KE. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 23.Adroer R, Oliva R. Nucleosome positioning in the rat protamine 1 gene in vivo and in vitro. Biochim. Biophys. Acta. 1998;1442:252–260. doi: 10.1016/s0167-4781(98)00184-5. [DOI] [PubMed] [Google Scholar]
- 24.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin SE, Shabanowitz J, Hunt DF, Marto JA. Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 2000;72:4266–4274. doi: 10.1021/ac000497v. [DOI] [PubMed] [Google Scholar]
- 26.Huang SY, Barnard MB, Xu M, Matsui S, Rose SM, Garrard WT. The active immunoglobulin κ chain gene is packaged by non-ubiquitin-conjugated nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 1986;83:3738–3742. doi: 10.1073/pnas.83.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose SM, Garrard WT. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- 28.Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidali G, Boffa LC, Bradbury EM, Allfrey VG. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: A working model of potentiation. J. Biol. Chem. 2004;279:51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- 32.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rada-Iglesias A, Enroth S, Ameur A, Koch CM, Clelland GK, Respuela-Alonso P, Wilcox S, Dovey OM, Ellis PD, Langford CF, Dunham I, Komorowski J, Wadelius C. Butyrate mediates decrease of histone acetylation centered on transcription start sites and down-regulation of associated genes. Genome Res. 2007;17:708–719. doi: 10.1101/gr.5540007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ausió J, van Holde KE. Histone hyperacetylation: Its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 35.Garcia Ramirez M, Rocchini C, Ausió J. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 36.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 37.Viens A, Mechold U, Brouillard F, Gilbert C, Leclerc P, Ogryzko V. Analysis of human histone H2AZ deposition in vivo argues against its direct role in epigenetic templating mechanisms. Mol. Cell. Biol. 2006;26:5325–5335. doi: 10.1128/MCB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakravarthy S, Bao Y, Roberts VA, Tremethick D, Luger K. Structural characterization of histone H2A variants. Cold Spring Harbor Symp. Quant. Biol. 2004;69:227–234. doi: 10.1101/sqb.2004.69.227. [DOI] [PubMed] [Google Scholar]
- 39.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausió J. Characterization of the stability and folding of H2A.Z chromatin particles: Implications for transcriptional activation. J. Biol. Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 40.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oliva R, Bazett-Jones DP, Locklear L, Dixon GH. Histone hyperacetylation can induce unfolding of the nucleosome core particle. Nucleic Acids Res. 1990;18:2739–2747. doi: 10.1093/nar/18.9.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Solis FJ, Bash R, Wang H, Yodh J, Lindsay SA, Lohr D. Properties of nucleosomes in acetylated mouse mammary tumor virus versus 5S arrays. Biochemistry. 2007;46:5623–5634. doi: 10.1021/bi062116z. [DOI] [PubMed] [Google Scholar]
- 43.Gansen A, Toth K, Schwarz N, Langowski J. Structural Variability of Nucleosomes Detected by Single-Pair Forster Resonance Energy Transfer: Histone Acetylation, Sequence Variation, and Salt Effects. J. Phys. Chem. B. 2009;113:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 44.Simpson RT. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- 45.Libertini LJ, Ausió J, van Holde KE, Small EW. Histone hyperacetylation. Its effects on nucleosome core particle transitions. Biophys. J. 1988;53:477–487. doi: 10.1016/S0006-3495(88)83126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norton VG, Imai BS, Yau P, Bradbury EM. Histone acetylation reduces nucleosome core particle linking number change. Cell. 1989;57:449–457. doi: 10.1016/0092-8674(89)90920-3. [DOI] [PubMed] [Google Scholar]
- 47.Bauer WR, Hayes JJ, White JH, Wolffe AP. Nucleosome structural changes due to acetylation. J. Mol. Biol. 1994;236:685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 48.Brower-Toland B, Wacker DA, Fulbright RM, Lis JT, Kraus WL, Wang MD. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 49.Higashi T, Matsunaga S, Isobe K, Morimoto A, Shimada T, Kataoka S, Watanabe W, Uchiyama S, Itoh K, Fukui K. Histone H2A mobility is regulated by its tails and acetylation of core histone tails. Biochem. Biophys. Res. Commun. 2007;357:627–632. doi: 10.1016/j.bbrc.2007.03.203. [DOI] [PubMed] [Google Scholar]
- 50.Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell. 2001;7:1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol. Cell. Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohba R, Steger DJ, Brownell JE, Mizzen CA, Cook RG, Cote J, Workman JL, Allis CD. A novel H2A/H4 nucleosomal histone acetyltransferase in Tetrahymena thermophila. Mol. Cell. Biol. 1999;19:2061–2068. doi: 10.1128/mcb.19.3.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 57.Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, Glaser RL. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem. 2000;275:23267–23272. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- 58.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]