Abstract
Transcriptional regulation of the gene encoding brain-derived neurotrophic factor (BDNF) has been widely studied. However, an understanding of mechanisms modifying chromatin, events that are essential for controlling transcription, is rudimentary. We focused on two activation dependent regions of the Bdnf gene physically linked to known transcription sites for exons 1 and 4. Using chromatin immunoprecipitation (ChIP) assays, we determined that N-methyl-D-aspartate (NMDA) receptor activation derepressed promoters 1 and 4-mediated transcription. This derepression correlated with reduced occupancy by histone deacetylase 1 (HDAC1) and methyl cytosine binding protein 2 (MeCP2) of each promoter region near known transcription start sites in cultured hippocampal neurons. These changes did not occur at all sites upstream of transcription initiation. Taken together, these findings suggest that histone and other DNA binding proteins are involved in remodeling of chromatin at some, but not all sites, within Bdnf promoters 1 and 4 and are associated with NMDA receptor-dependent increases in transcription.
Keywords: rain-derived neurotrophic factor, BDNF, chromatin remodeling, epigenetic modification, histone, methylation, HDAC, MeCP2
Introduction
Activity-dependent transcription is thought to be the mechanism by which neurons convert brief cellular changes into stable alterations in brain function. These transcriptional changes constitute a form of “molecular memory” (Hardingham et al., 2001). Changes in intracellular calcium (Ca2+) brought about through L-type voltage-gated Ca2+channels and N-methyl-D-aspartate (NMDA) glutamate receptors play an essential role in neuronal plasticity (Grover and Teyler, 1990; Bliss and Collingridge, 1993). NMDA receptor activation is critical for synaptic plasticity in the hippocampus (Bashir et al., 1991; Nagerl, et al., 2004; Slutsky et al., 2004). One protein that is crucial to these processes is brain-derived neurotrophic factor (BDNF). Among its primary functions, BDNF modulates the strength of existing synaptic connections and acts in the formation of new synaptic contacts (Thoenen, 1995; Katz and Shatz, 1996; Lu and Figurov, 1997; Chao, 2003).
The Bdnf gene has unique structural features. By the most recent report (Pruunsild et al., 2007), the human gene spans more than 70 kb and is composed of ten 5′ exons that are differentially spliced to a single 3′ terminal exon. These ten 5′ terminal exons are transcribed from unique promoters with alternative splicing occurring in at least five of these uniquely initiated transcripts. This complex transcriptional regulation leads to the creation of at least three pre-pro-BDNF isoforms with longer N-termini, whose function is not completely understood (Marini et al., 2004; Liu et al., 2005; Pruusild et al., 2007). This overall gene architecture is highly conserved in mammals. In rat and mouse, there are eight Bdnf exons with separate promoters that have been characterized (Aid et al., 2007).
Activation of Bdnf transcription has been shown to involve changes in chromatin structure through changes in DNA methylation and posttranslational histone modification (Huang et al., 2002; Chen et al., 2003; Martinowich et al., 2003; Tsankova et al., 2006). Epigenetic modifications include DNA methylation and posttranslational modifications (including acetylation, methylation, phosphorylation, and ubiquitination) at the amino terminal “tails” of core histones, which comprise the nucleosome. Generally, histone acetylation, regulated by histone deacetylases (HDACS) and histone acetyltransferases (HATS), is associated with open chromatin and allows for increased transcription, while histone methylations (regulated by histone methyltransfereases (HMTS) and histone demethylases (HDMS)) are more stable and specific modifications of core histones (H2A, H2B, H3, H4), such as H3 methylation at lysines 9 and 27, are associated with a tighter chromatin structure and have been thought to be associated with transcriptional repression. In contrast, specific modifications on H3, for example methylation at lysine 4, means more open chromatin and is associated with transcriptional activation. We sought to characterize changes in chromatin associated with NMDA receptor-dependent Bdnf gene expression for two major activation promoters that respond to NMDA receptor activation: promoters 1 and 4 by rat hippocampal neurons maintained in culture.
Materials and Methods
Preparation of hippocampal neurons
Hippocampal cultures were prepared from embryonic day 19 (E19) Sprague–Dawley embryos essentially as described previously (Cheng et al., 1995 and references therein; Jiang et al., 2005). The hippocampi were dissected and placed into ice-cold Hank’s solution with 1 mM HEPES and 1µL/mL penicillin-streptomycin (10,000 units penicillin and 10,000 µg/mL streptomycin).The tissue was then incubated with 1.25% trypsin (Invitrogen, Carlsbad, CA, USA) in 10 mL Hank’s solution and incubated at 37° C for 15 min and then rinsed with 10 mL Hank’s solution. Soybean trypsin inhibitor (2 mg/mL) in 10 mL Hank’s solution was then added and the mixture incubated at 37° C for 5 min. The cell preparation was then washed with 10 mL of minimum essential medium (MEM, Invitrogen) containing: 10 mM sodium bicarbonate, 1 mM pyruvate (pyruvic acid, sodium salt), 20 mM potassium chloride, 1 mM HEPES plus 10% fetal bovine serum, and 2 mM L-glutamine, pH 7.2. The cell suspension was then diluted with MEM to achieve a seeding density of 2.5 × 105−5.0 × 105 cells/mL and distributed into 10 cm dishes or 6 well plates that were pre-coated with poly D-lysine (50 µg/mL). The cells were incubated for 5 h at 37° C in a humidified incubator containing 95% air/5% CO2. The cell media was then aspirated and replaced with the identical volume of neurobasal media containing 2% B27 supplement, 1 M HEPES, 2 mM Lglutamine, and 50 mg/mL gentamicin, which is preheated to 37° C. On day in vitro (DIV) 3, and every other day thereafter, one-fifth of the culture medium was removed and replaced with fresh media to replenish vital nutrients. Cultures were used between DIV 8–11. Procedures using animals were conducted according to the principles set forth in the National Institutes of Health (NIH) Publication no. 85–23,Guide for the Care & Use of Laboratory Animals & the Animal Welfare Act of 1986, as amended. Every effort was made to minimize the number of animals.
Drug treatments
Trichostatin A (TSA) (Sigma, St. Louis, MO, USA) was prepared in dimethyl sulfoxide (DMSO) (Sigma). Hippocampal neurons were plated into 6 well plates grown in Neurobasal medium with B27 supplement. Neurons were treated with a final concentration of 25, 100, 250, or 500 nM TSA for 8–48 hours or with DMSO (vehicle control). Hippocampal neurons were maintained in six well plates for real time reverse-transcribed PCR (RT-PCR) or 10cm dishes for histone extraction, or chromatin immunoprecipitation (ChIP) assays. Subsequently, neurons were treated with NMDA (50µM) for various times (0–24 h).
Reverse-transcribed PCR
Total RNA was extracted from hippocampal neurons using RNA STAT-60 (Tel-Test, RNeasy mini kit (Qiagen, Valencia, CA, USA) and DNA-free kit (Applied Biosystems/Ambion, Austin, TX). The final RNA preparation was free of DNA contamination, as determined by a reverse transcriptase-less control, was reverse transcribed using a cloned AMV first-strand cDNA synthesis kit (Invitrogen). Real time PCR was performed on an ABI PRISM® 7700 sequence detector with the sequence specific PCR primers and a detection probe designed using Primer Express® software (ABI) and synthesized by the manufacturer.
Real time PCR (RT-PCR) primers and probes
The rat Bdnf exon 1 primers and probe sequences were: Forward primer: 5'-GCGTTGAGAAAGCTGCTTCAG-3'; Reverse primer: 5'-GAATGAGCGAGGTTACCAATGAC-3'. The Bdnf exon 1-specific probe sequence was: 5'-CGCCCGCTATATAGCAGGGCAGT-3'. The amplification primers for rat Bdnf exon 4: 5'-TTCCACTATCAATAATTTAACTTCTTTGC-3' (forward primer) and 5'-CTCTTACTATATATTTCCCCTTCTCTTCAGT-3' (reverse primer).
The exon 4-specific detection probe sequence: 5'-TACATATCGGCCACCAAAGACTCGC-3' Rat Gapdh was used as an internal reference and was purchased from ABI.
Chromatin immunoprecipitation (ChIP) detection primers within Bdnf promoters 1 and 4
Bdnf exons 1 and 4 are separated by approximately 17 kb (Figure 1). To determine sites within the promoter regions that respond to chromatin remodeling following NMDA receptor activation, we designed three sets of PCR primers (two within promoter 1, one within promoter 4) based on sequences for the Bdnf gene. Promoter 1 primers: the site proximal to the transcription start site of exon 1 (pm51): forward, 5'- AACTTTTCTAAGAAGTTTCC TTTTTACCA-3'; reverse, 5'- TGAGCC AGTTACGTGACCAACT-3'; a distal site, approximately 4 kb 5' of the transcription start site (pm9): forward, 5'- AGCTCTTGCAGACTAAATCGTGAGTTT-3'; reverse, 5'- CAAGAACCTGGTGTTGAGCTCATATTT-3'. The PCR primers for the −65 to −152 region from promoter 4 (pmS4): forward; 5′-CTAGGACTGGAAGTGGAAA-3′ and reverse; 5′-ATTTCATGCTAGCTCGCCG-3′ were described by Chen et al., (2003).
Figure 1.
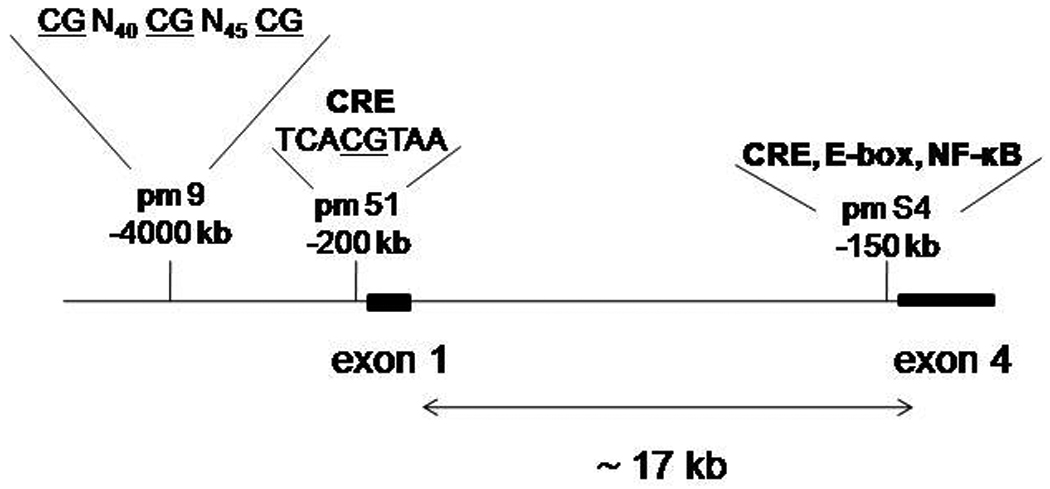
Schematic of genomic region of Bdnf promoters 1 and 4. The DNA sequences from approximately −4000 to −200 relative to exon 1 and approximately −150 relative to exon 4. Location of ChIP PCR amplicons within the promoter regions are: pm 9, the distal site of promoter 1, pm 51, the proximal site in promoter 1, and pmS4, the proximal promoter 4 region. CRE, E-box, and NF-κB elements within each promoter are labled. The CpG dinucleotides within promoter 1 are underlined, separated by nucleotide sequence, indicated in subscript. The promoter 4 elements have been described previously in Jiang et al. (2008). Exons 1 and 4 are separated by approximately 17 kb of genomic DNA. For simplicity, exons 2, 3, 5, and 6 were not included in the map.
ChIP
The ChIP assay was modified from the EZ- ChIP kit protocol (Upstate Biotechnology, Lake Placid, NY, USA). Rat hippocampal neurons in 10 cm dishes (E19, 9 DIV, 0.5 × 107 cells per dish) were treated with 50 µM NMDA or 250 nM TSA for 45 min to 12 hours or treated with vehicle. Crosslinking was performed at room temperature by adding 37% formaldehyde to the cell culture medium at 1% final concentration and incubated for 10 min. The reaction was stopped by the addition of 2.5 M glycine to a final concentration of 125 mM, followed by 5 min incubation at room temperature. Cells were washed twice with 8mL cold phosphate-buffered saline (PBS) containing a protease inhibitor cocktail (Roche, Nutley, NJ), then removed to 15 mL tube in 1 mL PBS with protease inhibitor. Samples were centrifuged at 700 × g for 10 min 4º C. The cell pellet from four dishes was pooled and suspended in 1 mL SDS lysis buffer with protease inhibitor cocktail. The lysate was snap frozen in the ethanol-dry ice bath and then thawed in a 37º C water bath. This process was repeated four times. The final lysate was sonicated on ice using a Misonix sonicator 3000, MicrotipTM 419 (Farmingdale, NY, USA) at a power setting of 6, 12 pulses (12 sec on, 2.5 min off). Samples were centrifuged at 12,000 × g, 4º C for 10 min to remove insoluble material, and the supernatant containing DNA–protein complexes was collected. To test for reproducibility, 5 µL of the sonicated chromatin was retained and 90 µL ddH2O were added, followed by 4 µL 5 M NaCl. The solution was then incubated 4–5 hours at 65° C to reverse the DNA- protein crosslinks. RNase A (1 µL from 1 U stock) was added and the mixture incubated for 30 minutes at 37° C, followed by incubation with 2 µL 0.5M EDTA, 4µL 1M Tris-HCl and 1 µL Proteinase K at 45° C for 1–2 hours. A sample of 5–20 µL was fractionated on a 1.5% native agarose gel with a 100 bp DNA marker. The sonicated DNA produced fragments from 150 to 1000 bp. The cell supernatant from three different cultures was pooled. The pooled cell supernatant was precleared with salmon sperm DNA/protein A or G agarose 50% slurry for 1 h at 4º C with agitation , and centrifuged at 1,500 rpm for 2 minutes to recover the beads. The beads were then resuspended in dilution buffer for subsequent immunoprecipitation. An aliquot (20 µL) of the precleared DNA preparation served as an input control and was reserved for later use. Antibodies for ChIP assays were anti-HDAC1 and anti-MeCP2. The antibodies were added to the precleared chromatin preparation, incubated on a rocking platform 16 h at 4°C. As a negative control, nonimmune rabbit or mouse IgG (Upstate Inc.) were used in place of specific antibodies. Immune complexes were precipitated by the addition of salmon sperm DNA/protein A or G agarose slurry for 1–2 h at 4ºC and pelleted at 700 × g for 1 min. The pellets were washed five times (1 ml each) in a low-salt wash buffer, high-salt wash buffer, and LiCl wash buffer, and twice with TE buffer. Immune complexes were treated twice in 100 µL of 1% SDS and 100 mM NaHCO3 at room temperature for 15 min. Protein–DNA cross-links were reversed by adding 8 µL 5 M NaCl and incubating the mixture at 65ºC for 5 hours, followed by treatment with 1 U of RNase A and 20 µg proteinase K at 37ºC for 1 h. The DNA was then processed using PCR purification columns (Qiagen,Valencia, CA). Levels of specific proteins occupying Bdnf promoter 1 were determined by a semi-quantitative PCR assay. Conditions for PCR were: 1 cycle 94°C, 10 min; 24–40 cycles 94°C, 15 sec, 50–65°C, 20 sec, 72° C, 30 sec, followed by an extension at 72° C, 7 min. PCR products were resolved on TBE polyacrylamide gels and the amplicon band visualized by ethidium bromide staining. Molecular size was determined using appropriate size standards (O’GeneRuler, Fermentas Inc., Hanover, MD). Input and immunoprecipitated DNA reactions were performed in triplicate. Representative ChIP results are presented in the Results section. Buffer-only “no antibody” conditions were used as negative ChIP controls and did not produce a band following PCR. In addition, immunoprecipitated chromatin with nonimmune rabbit IgG produced similar results as “no antibody” controls.
Statistical analysis
Data are presented as mean ± SD or fold change relative to control (no addition). ANOVA with post hoc Tukey were used for analysis. p < 0.05 is considered significant.
Results
Expression of alternative rat Bdnf transcripts containing exon 1 or exon 4 are differentially regulated by NMDA receptor activation
In rat, mouse, and human brain, Bdnf transcripts containing exon 1 or exon 4 are prominently expressed (Liu et al., 2005; Aid et al., 2007; Pruunsild et al., 2007). The promoters initiating transcription of exons 1 and 4 are highly regulated and respond a number of different activators, including the neurotransmitters dopamine (Fang et al., 2003) and glutamate (Timmusk et al., 1993; Marini et al., 1998). Neuronal activity elicits an immediate early gene response in a subset of Bdnf promoters in hippocampus (Lauterborn et al., 1996). Previously, it was known that activity-dependent regulation of BDNF by kainic acid (Timmusk et al., 1994). Tabuchi and coworkers (2000) showed in cortical neurons that Bdnf gene promoters respond differently to calcium signals; NMDA receptor activation predominantly increased exon 4-specific mRNA levels while calcium influx through L-type voltage dependent calcium channels mostly induced exon 1-specific Bdnf mRNA levels.
In cultured hippocampal neurons, NMDA receptor activation elicited a time-dependent increase in exon 1 and exon 4-specific Bdnf mRNA levels. Beginning at 3 h, exon 1-containing transcripts increased 25-fold, increasing to 65-fold, at least up to 12 h (Figure 2). In contrast, exon 4-specific transcripts peaked at 6 h, producing a 19-fold increase relative to the baseline control (Figure 2).
Figure 2.
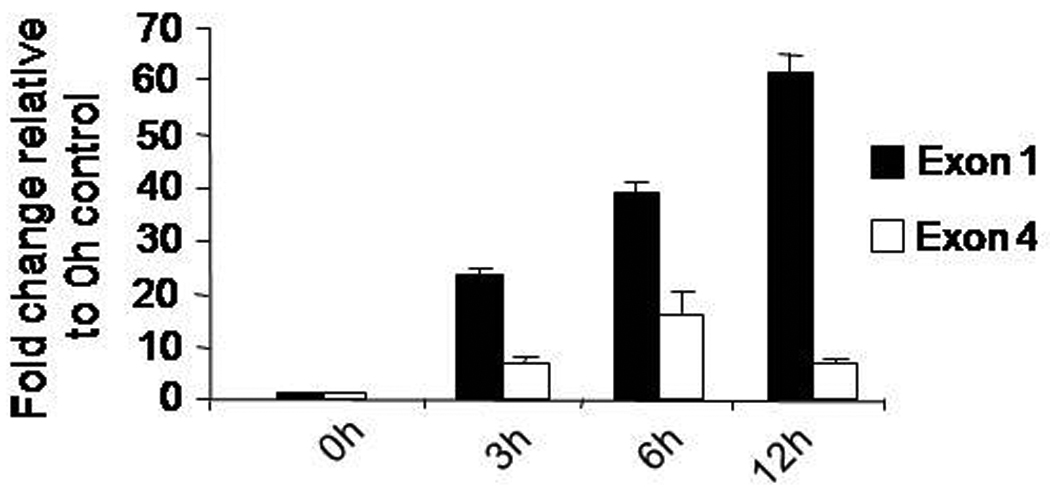
Time course for induction of Bdnf exon 1 and exon 4-specific mRNA levels in NMDA-treated hippocampal neurons. Cultured hippocampal neurons were incubated with a 50 µM for the indicated times on day in vitro eight). Mean expression values for each Bdnf transcript were determined after normalization to the average expression values of the Gapdh gene from each template. Data are presented as fold-change relative to the baseline (0 h) control (means ± SD).
HDAC inhibitor, TSA, enhances Bdnf exon 1-specific mRNA levels but not exon 4-containing transcripts
Histone modification by acetylation is generally regarded as a feature of transcriptionally active genes. An inhibitor of histone deacetylase (HDAC) activity, trichostatin A (TSA), has differential effects on Bdnf promoters 1 and 4 in Neuro2A cells treated with TSA (300 nM) for 48 hrs (Aid et al., 2007). We wanted to determine if TSA caused a dose-dependent difference in Bdnf exon 1 and exon 4-specific mRNA levels in cultured hippocampal neurons. As determined by real-time RT-PCR, levels of exon 1-specific transcripts increased in a dose-dependent manner, producing up to a 20-fold increase at 250 nM over untreated neurons (Figure 3). Levels of Bdnf exon 4 increased only 2-fold at the same concentration. These results showed that the regulatory regions for exons 1 and 4 respond differently to an HDAC inhibitor, suggesting that different mechanisms are in responsible for altering chromatin structure for each of these promoters in response to NMDA.
Figure 3.
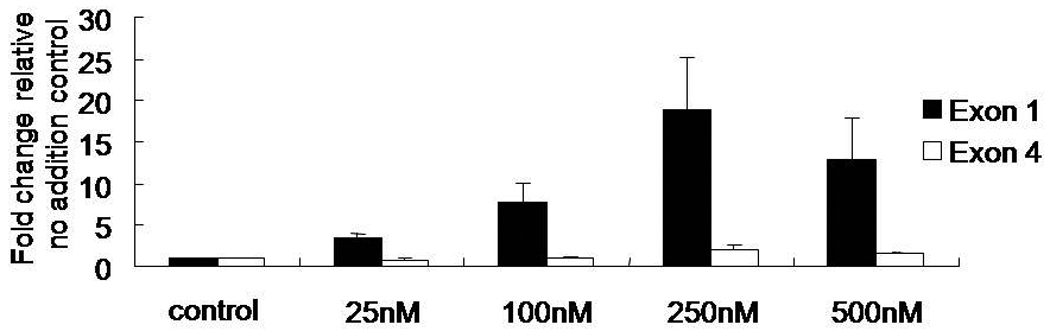
Hippocampal neurons treated with Trichostatin A (TSA) increase levels of Bdnf exon 1 mRNA in a dose-dependent manner. Neuronal cultures were treated with increasing concentrations of TSA (25, 100, 250, or 500 nM) for 24 hr on day in vitro eight. Total RNA was extracted from cells, first strand cDNA synthesized, and real time PCR performed to amplify exons 1 or 4 using intron spanning detection probes. Gapdh mRNA levels were used as the internal reference. Data are presented relative to the vehicle control (means ± SD).
HDAC1 is released from Bdnf promoters 1 and 4 following NMDA receptor activation
HDAC1 is a part of a protein complex involved in transcriptional repression via deacetylation of core histones (Hassig et al., 1998). Bdnf promoters 1 and 4 are activated when histone deacetylation is inhibited in vivo (Tsankova et al., 2006) or conversely, when acetylated histones are enhanced at promoters 1 and 4 in vivo (Bredy et al., 2007). Other investigators have noted differential histone changes associated with promoter 1, being consistently hyperacetylated, while the downstream promoter 4 had a different response in the hippocampus following pilocarpine treatment in vivo (Huang et al 2002). As a first approach to characterizing histone changes taking place at Bdnf promoter 1 following NMDA receptor activation, we analyzed two sites from the 5' flanking sequence of exon 1, which also contains multiple potential transcription factor binding sites and CpGs, for presence of modified HDAC1 in primary hippocampal neurons treated with NMDA (50 µM) or vehicle-treated neurons by ChIP assays. One site analyzed is located 200 bp 5' from a know transcription start site mapped previously by Nakayama and colleagues (1994) and is contained within a ChIP PCR probe that responds in vivo (Bredy et al., 2007), while the second site is located approximately 4 kb 5' from this transcription start site. We compared HDAC1 occupancy of the promoter 1 sites with one critical region from promoter 4 known to respond to NMDA receptor activation (Jiang et al., 2008) at 45min or 3 h after NMDA treatment. The transcription start site proximal region, pm51, showed a rapid time-dependent decrease in HDAC1 binding following NMDA receptor activation (Figure 4A). In addition, there was an increase in NMDA-induced histone modifications (acetylated H3, data not shown). HDAC1 was not associated with the distal promoter 1 region (pm9) (Figure 4A). There were no changes in levels of acetylated H3 for this region (data not shown). For the same neuronal cultures, HDAC1 occupancy of the promoter 4 region, defined by the pmS4 amplicon, also decreased following NMDA receptor activation (Figure 4B), although this was associated with reduced levels of acetylated H3 (data not shown). Take together, these results support the idea that region-specific alterations in histone acetylation at promoter 1 are associated with transcriptional derepression and may explain, at least in part, the differential responsiveness of Bdnf promoters 1 and 4.
Figure 4.
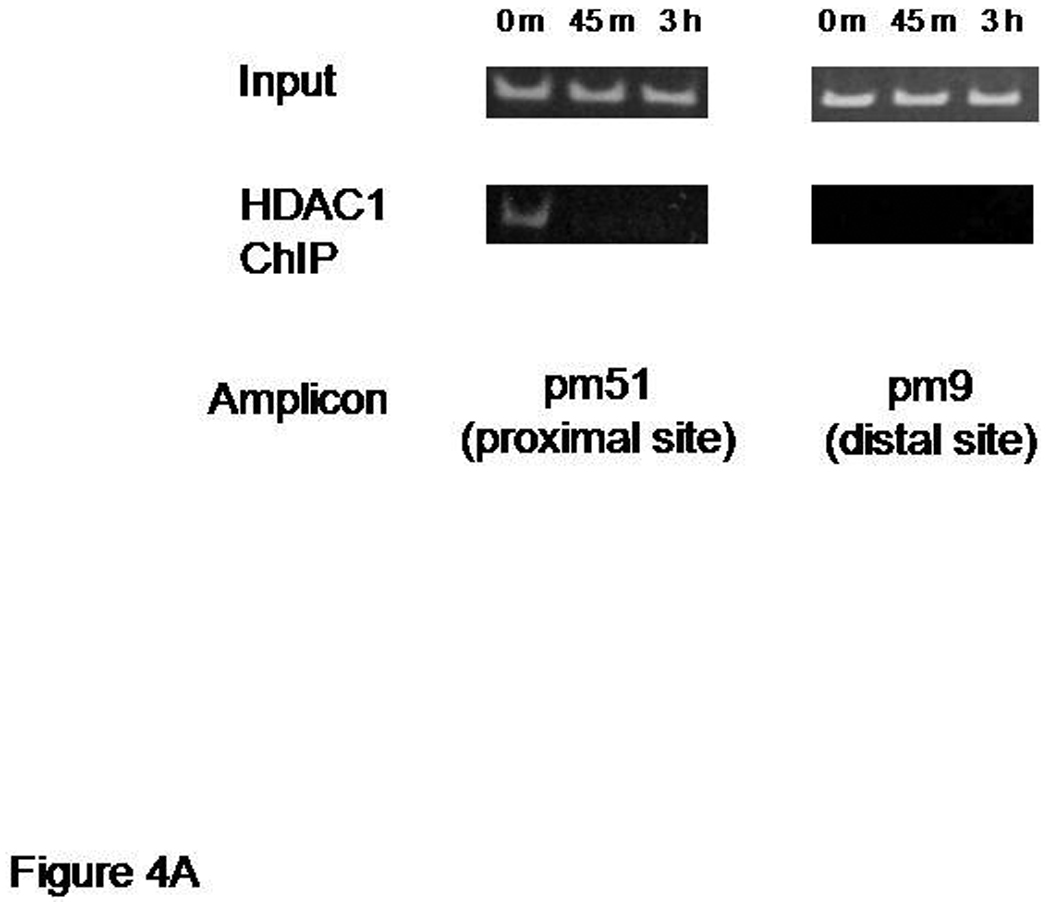

HDAC1 is released from sequences near transcription start sites of Bdnf promoters 1 and 4. Chromatin immunoprecipitation (ChIP) assays showing release of histone deacetylase 1 (HDAC1) from Bdnf promoters 1 and 4 within 45 min of NMDA receptor activation. Panel A: changes in promoter 1 occupancy are shown using a semi-quantitative PCR assay (Materials and Methods). Note that sequences defining the proximal site of promoter 1 (pm51 amplicon), nearest the transcription start site, but not a distal site of promoter 1 (pm9), are associated with HDAC1. Panel B: promoter 4 occupancy by HCAC1 is seen under basal conditions that was lost within 45 min treatment of hippocampal neuron cultures with 50 µM NMDA. The representative semi-quantitative PCR assay also shows preimmunoprecipitated “inputs” from NMDA-treated and untreated neurons (0 h). Buffer-only “no antibody” conditions were used as negative controls and did not produce a band following PCR. In addition, immunoprecipitated chromatin with nonimmune rabbit IgG produced similar results as “no antibody” controls (data not shown).
NMDA receptor activation reduces MeCP2 occupancy of promoters 1 and 4
MeCP2 has been shown to function as a transcriptional repressor at promoter 4 of the Bdnf gene (Chen et al., 2003; Martinowich et al, 2003). To determine if endogenous MeCP2 also acts at promoter 1 and is subject to regulation by NMDA receptors, we performed ChIP assays as in the previous section. Cross-linked chromatin from hippocampal neuron cultures treated with NMDA (50 µM) or vehicle-treated neurons were sheared by sonication and subsequently incubated with antibodies specific to MeCP2 to immunoprecipitate proteins bound to the native promoter region. Immunoprecipitated DNA was released from cross-linked proteins followed by semi-quantitative PCR, using primers that amplified the Bdnf promoter 1 regions located approximately 200 bp or 4000 bp 5' of the exon 1 transcription start site. Immunoprecipitation of chromatin with anti-MeCP2 antibodies resulted in decreased amplification of the −200 Bdnf promoter 1 sequence. Kinetically, this reduction in MeCP2 binding following NMDA receptor activation was slower than the decrease in HDAC1 occupancy of the same site, in vivo. A temporally slower change in MeCP2 occupancy was also seen at the more distal promoter 1 site, mp9, in the same neurons treated with NMDA (Figure 5A) while a more rapid release of MeCP2 occurred at the promoter 4 site, pmS4 (Figure 5B).
Figure 5.
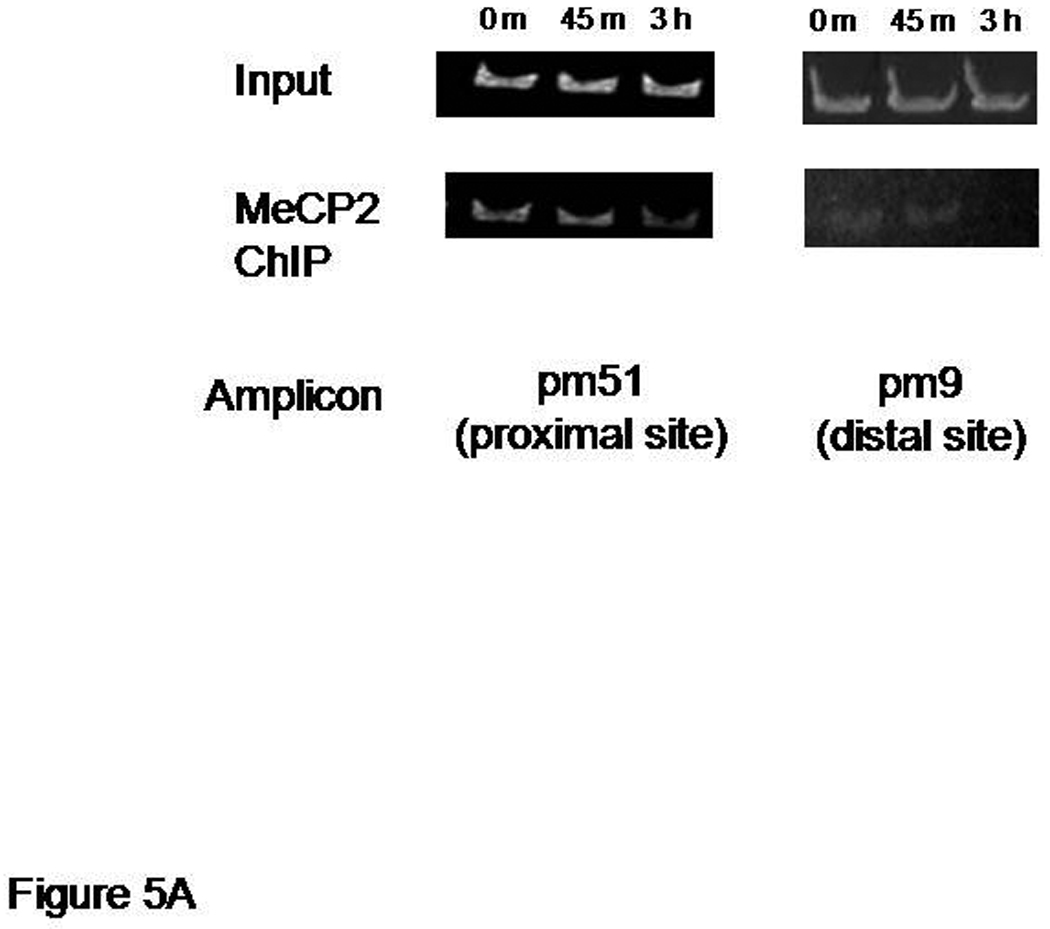

Decreased MeCP2 binding to different sites of Bdnf promoter 1 after NMDA treatment. Panel A: a representative chromatin immunoprecipitation (ChIP) assay showing the time dependent release of MeCP2 from sequences defining the proximal and distal sites in promoter 1 (pm51 and pm9 amplicons, respectively). Panel B: ChIP assay showing the release of MeCP2 from Bdnf promoter 4 (pmS4 amplicon) following NMDA receptor activation. Results shown are from the same chromatin preparation used in Figure 4.
Discussion
In this study, we observed differential effects of NMDA receptor activation on transcription from Bdnf promoters 1 and 4 by rat hippocampal neurons in culture. Following NMDA receptor activation, Bdnf exon 1 mRNA levels showed a rapid, yet more sustained increase compared to exon 4 mRNA levels. To approach this differential response mechanistically, we incubated neurons with different concentrations of TSA, an inhibitor of HDAC activity, and quantified exon 1 and exon 4 Bdnf mRNA levels. Treating the neurons with TSA produced a significant dose –dependent increase in Bdnf exon 1 mRNA levels, while the level of exon 4 transcripts in the same cells was changed only slightly. These results clearly show that promoters 1 and 4 respond differentially to signals affecting chromatin structure. This differential response is bolstered by structural features of the two promoter regions in that each promoter is part a small cluster of genomic sequence of approximately three kb, made up of three exons and their 5' flanking regions. The first cluster contains exons 1–3, while the second cluster contains exons 4–6. The clusters are separated by about 17 kb of genomic DNA. We hypothesized that changes in chromatin structure may account for the transcriptional differences seen in responsiveness to NMDA. As an initial approach, we examined these two Bdnf promoters for endogenous differences in occupancy by HDACs, specifically HDAC1. Promoter occupancy by HDACs has been associated with transcriptional silencing, while enhanced histone acetylation, with loss of HDAC from chromatin, is correlated with transcriptional activation. We noted that in vivo HDAC1 binding of promoter 1 near a know transcription start site was reduced, at least up to 3 h following NMDA receptor activation, consistent with enhanced exon 1-containing transcripts, which were first detected at 3 h, increasing at 12 h to levels greater than 60-fold over basal conditions. In contrast, NMDA receptor activation, produced greater than a 10-fold increase in exon 4-specific transcripts relative to control levels (Figure 2) and showed similar temporal changes in HDAC1 binding to a region of promoter 4 physically close to known transcription start sites in human and rodent brain (Timmusk et al., 1993; Nakayama et al., 1994; Liu et al., 2005; Pruunsild et al., 2007). The reduction in promoter 1 occupancy by HDAC1 and increased modification of core histone H3 near the start of transcription in the presence of NMDA is consistent with histone modifications covering the same transcription initiation region from promoter 1 in mice undergoing an extinction of conditioned fear paradigm (Bredy et al., 2007). Interestingly, the learning-induced histone modification (increased histone H3 acetylation) did not produce an increase in Bdnf exon 1 mRNA expression in the mouse model. Although the ChIP experiments performed by us and those of Bredy et al. (2007) differed greatly by choice of animal model, brain region selected, and the fact that whole tissue was used compared to disassociated neurons maintained in culture, the combined findings show that a common region near a known transcription initiation site within promoter 1 is also a common target for particular activation-dependent histone modifications. Thus, at least some signaling pathways are shared in an early step of chromatin remodeling necessary for recruiting transcription factors to promoters 1 and 4. In addition, our findings and those of Bredy et al. suggest that early remodeling of chromatin is not necessarily sufficient for transcriptional activation.
Because the Bdnf gene is critically involved in CNS development, neuronal survival, and learning and memory, its disrupted expression is thought to contribute to the pathopysiology of Rett syndrome, an X-linked disorder caused by mutations in the gene encoding MeCP2 (reviewed by Francke, 2006). Previously, it was shown that Bdnf promoter 4 had multiple sites for MeCP2 binding and that this was a major site of MeCP2 protein-DNA interactions in cortical neurons (Chen et al., 2003). In particular, a well-described binding site for the transcription factor cAMP response element binding protein (CREB), known to play an important role in neuronal and synaptic plasticity (Frank and Greenberg, 1994), contains a CpG site within the CREB binding site (CRE) that is methylated in vivo (Chen et al., 2003; Martinowich et al., 2003). In addition to binding MeCP2, this region of promoter 4 and flanking sequences are known to play critical roles in activation-dependent transcription with binding sites for regulatory transcription factors including upstream stimulatory factor (USF), nuclear factor kappa B (NF-κB), and the basic helix- loop-helix protein BHLHB2 (Tabuchi et al., 2002, Lipsky et al., 2001, Jiang et al., 2008). Computational analysis of Bdnf promoter 1 predicts binding sites for these transcription factors, although in different sequence contexts. In particular, a CRE site in promoter 1, which was previously shown to play a major role in activation-dependent Bdnf exon 1 mRNA expression (Tabuchi et al., 2002), is also a potential target for MeCP2 binding. Because the CpG sequence found at this site is the only potential methylation target present in the pm51 PCR amplicon used in the ChIP assay, this result strongly suggests that MeCP2 also has a role in regulating Bdnf promoter 1 activity. Although the CRE site in promoter 1 is known to bind CREB in vitro and to activate Bdnf exon1 transcription in transfected cortical neurons (Tabuchi et al., 2002), it is not know if activated CREB occupies this site in vivo. To address this question, we performed preliminary ChIP experiments using NMDA-treated and unstimulated hippocampal neurons. These experiments detected CREB occupying promoter 1 in the region defined by the pm51 PCR amplicon only after 3 h NMDA receptor activation (FT, RHL unpublished data). This finding suggests that activated CREB may not be rapidly recruited to promoter 1 in hippocampal neurons and is in keeping with our observation that NMDA receptor activation is associated with a slow release of MeCP2 from the pm51 site of promoter 1, relative to promoter 4 (Figures 5A and 5B).
What factors could be responsible for the rapid transcriptional responsiveness of Bdnf promoter 1? Another transcription factor, USF, also activates Bdnf promoter 1 transcription (Tabuchi et al., 2002). Thus, it is plausible that USF proteins may occupy the region defined by the pm51 amplicon following NMDA receptor activation. Future experiments will focus on differences in chromatin remodeling and recruitment of specific transcription factors in different brain regions and also address changes in methylation status of promoter 1 CpGs under basal and activation-dependent conditions.
The signal transduction pathways mediating promoter-specific changes in chromatin architecture have yet to be elucidated. However, as additional activation-dependent chromatin-associated targets are characterized for each of these Bdnf regulatory regions, it is anticipated that the pathways activated via Ca2+ signaling, such as the Ca2+-calmodulin kinases, extracellular signal-regulated kinase/mitogen-activated protein kinase, adenylyl cyclase, and IκB kinase, just to name a few, will be defined and integrated into the transcription networks regulating the molecular responses for neuronal survival and synaptic plasticity.
Acknowledgements
This study was supported by NIH intramural grant NIAAA Z01-AA00325 (RHL) and extramural grant F-192EG-C1 from the Defense Brain and Spinal Cord Injury Program (AMM).
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir ZI, Alford S, Davies SN, Randall AD, Collingridge GL. Long-term potentiation of NMDA receptor-mediated synaptic transmission in the hippocampus. Nature. 1991;349:156–158. doi: 10.1038/349156a0. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;36:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem. 2007;14:268–276. doi: 10.1101/lm.500907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MV. Neurotrophins and their receptors: a convergence point for many signaling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheng B, Furukawa K, O’Keefe JA, Goodman Y, Kihiko M, Fabian T, Mattson MP. Basic fibroblast growth factor selectively increases AMPA-receptor subunit GluR1 protein level and differentially modulates Ca2+ responses to AMPA and NMDA in hippocampal neurons. J Neurochem. 1995;65:2525–2536. doi: 10.1046/j.1471-4159.1995.65062525.x. [DOI] [PubMed] [Google Scholar]
- Fang H, Chartier J, Sodja C, Desbois A, Ribecco-Lutkiewicz M, Walker PR, Sikorska M. Transcriptional activation of the human brain-derived neurotrophic factor gene promoter III by dopamine signaling in NT2/N neurons. J Biol Chem. 2003;278:26401–26409. doi: 10.1074/jbc.M211539200. [DOI] [PubMed] [Google Scholar]
- Francke U. Mechanisms of disease: neurogenetics of MeCP2 deficiency. Nat Clin Pract Neurol. 2006;2:212–221. doi: 10.1038/ncpneuro0148. [DOI] [PubMed] [Google Scholar]
- Frank DA, Greenberg ME. CREB: a mediator of long-term memory from mollusks to mammals. Cell. 1994;79:5–8. doi: 10.1016/0092-8674(94)90394-8. [DOI] [PubMed] [Google Scholar]
- Grover LM, Teyler TJ. Two components of long-term potentiation induced by different patterns of afferent activation. Nature. 1990;347:477–479. doi: 10.1038/347477a0. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4:261–267. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleisher TC, Owa T, Grable PG, Ayer DE, Schreiber SL. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci U S A. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J Neurosci. 2002;22:8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Tian F, Mearow K, Okagaki P, Lipsky RH, Marini AM. The excitoprotective effect of N-methyl-D-aspartate is mediated by a BDNF autocrine loop in cultured hippocampal neurons. J Neurochem. 2005;94:713–722. doi: 10.1111/j.1471-4159.2005.03200.x. [DOI] [PubMed] [Google Scholar]
- Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L, Wu X, Pan H, Hu XZ, Xu K, Kenney H, Egan SE, Turley H, Harris AL, Marini AM, Lipsky RH. BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J Neurosci. 2008;28:1118–1130. doi: 10.1523/JNEUROSCI.2262-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Lauterborn JC, Rivera S, Stinis CT, Hayes VY, Isackson PJ, Gall CM. Differential effects of protein synthesis inhibition on the activity-dependent expression of BDNF transcripts: evidence for immediate-early gene responses from specific promoters. J Neurosci. 1996;16:7428–7436. doi: 10.1523/JNEUROSCI.16-23-07428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky RH, Xu K, Zhu D, Kelly C, Terhakopian A, Novelli A, Marini AM. Nuclear factor kappaB is a critical determinant in N-methyl-D-aspartate receptor-mediated neuroprotection. J Neurochem. 2001;78:254–264. doi: 10.1046/j.1471-4159.2001.00386.x. [DOI] [PubMed] [Google Scholar]
- Liu QR, Walther D, Drgon T, Polesskaya O, Lesnick TG, Strain KJ, de Andrade M, Bower JH, Maraganore DM, Uhl GR. Human brain derived neurotrophic factor (BDNF) genes, splicing patterns, and assessments of associations with substance abuse and Parkinson's Disease. Am J Med Genet B Neuropsychiatr Genet. 2005;134:93–103. doi: 10.1002/ajmg.b.30109. [DOI] [PubMed] [Google Scholar]
- Lu B, Figurov A. Role of neurotrophins in synapse development and plasticity. Rev Neurosci. 1997;8:1–12. doi: 10.1515/revneuro.1997.8.1.1. [DOI] [PubMed] [Google Scholar]
- Marini AM, Rabin SJ, Lipsky RH, Mocchetti I. Activity-dependent release of brain-derived neurotrophic factor underlies the neuroprotective effect of N-methyl-D-aspartate. J Biol Chem. 1998;273:29394–29399. doi: 10.1074/jbc.273.45.29394. [DOI] [PubMed] [Google Scholar]
- Marini AM, Jiang X, Wu X, Tian F, Zhu D, Okagaki P, Lipsky RH. Role of brain-derived neurotrophic factor and NF-kappaB in neuronal plasticity and survival: from genes to phenotype. Restor Neurol Neurosci. 2004;22:121–130. [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Nagerl UV, Eberhorn N, Cambridge SB, Bonhoeffer T. Bidirectional activity-dependent morphological plasticity in hippocampal neurons. Neuron. 2004;44:759–767. doi: 10.1016/j.neuron.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Gahara Y, Kitamura T, Ohara O. Distinctive four promoters collectively direct expression of brain-derived neurotrophic factor gene. Brain Res Mol Brain Res. 1994;21:206–218. doi: 10.1016/0169-328x(94)90251-8. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky I, Sadeghpour S, Li B, Liu G. Enhancement of synaptic plasticity through chronically reduced Ca2+ flux during uncorrelated activity. Neuron. 2004;44:835–849. doi: 10.1016/j.neuron.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Nakaoka R, Amano K, Yukimine M, Andoh T, Kuraishi Y, Tsuda M. Differential activation of brain-derived neurotrophic factor gene promoters I and III by Ca2+ signals evoked by L-type voltage-dependent and N-methyl-D-aspartate receptor Ca2+ channels. J Biol Chem. 2000;275:17269–17275. doi: 10.1074/jbc.M909538199. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Sakaya H, Kisukeda T, Fushiki H, Tsuda M. Involvement of an upstream stimulatory factor as well as cAMP-responsive element-binding protein in the activation of brain-derived neurotrophic factor gene promoter I. J Biol Chem. 2002;277:35920–35931. doi: 10.1074/jbc.M204784200. [DOI] [PubMed] [Google Scholar]
- Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–596. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Belluardo N, Persson H, Metsis M. Developmental regulation of brain-derived neurotrophic factor messenger RNAs transcribed from different promoters in the rat brain. Neuroscience. 1994;60:287–291. doi: 10.1016/0306-4522(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]


