Abstract
Liver regeneration triggered by 2/3 partial hepatectomy is accompanied by elevated hepatic levels of endotoxin, which contributes to the regenerative process, but liver inflammation and apoptosis remain paradoxically limited. Here we show that STAT3, an important anti-inflammatory signal, is activated in myeloid cells after partial hepatectomy and its conditional deletion results in an enhanced inflammatory response. Surprisingly, this is accompanied by an improved rather than impaired regenerative response with increased hepatic STAT3 activation, which may contribute to the enhanced liver regeneration. Indeed, conditional deletion of STAT3 in both hepatocytes and myeloid cells results in elevated activation of STAT1 and apoptosis of hepatocytes, and a dramatic reduction in survival after partial hepatectomy, whereas additional global deletion of STAT1 protects against these effects. Conclusions: An interplay of myeloid and hepatic STAT3 signaling is essential to prevent liver failure during liver regeneration through tempering a strong innate inflammatory response mediated by STAT1 signaling.
Keywords: Liver regeneration, STAT3, myeloid cells, inflammatory response
Introduction
The liver has great ability to regenerate after injury or tissue loss, which is tightly controlled by multiple signaling pathways induced by a wide variety of cytokines, growth factors, and hormones.1-4 Liver regeneration triggered by 2/3 partial hepatectomy (PHx), a widely used experimental model, proceeds initially by proliferation of hepatocytes and then by proliferation of nonparenchymal cells, including biliary epithelial, sinusoidal endothelial, and hepatic stellate cells.1-4 Emerging evidence suggests that a variety of factors contribute to the initiation and progression of liver regeneration. These include hepatocyte growth factors, platelet-derived serotonin, stem cell factor, complements, and the innate inflammatory response.1-4 Among these, the role of the innate inflammatory response has been extensively investigated.1-4 It is generally accepted that PHx leads to elevation of serum levels of bacterial endotoxin (LPS),5 which stimulates Kupffer cells to produce TNF-α and IL-6. The latter then targets the IL-6 receptor complex (gp80/gp130) in hepatocytes, triggering the activation of signal transducer and activator of transcription 3 (STAT3), which promotes hepatocyte survival and proliferation.1-4 However, a recent study suggests that MyD88 rather than LPS acting via TLR4 and CD14 contributes to IL-6/STAT3 activation after PHx, and the role of MyD88 in liver regeneration has been controversial.6, 7 IL-6 knockout mice display acute liver failure and impaired liver regeneration after PHx,8 although recent studies suggest that IL-6 may play a more important role in hepatoprotection during liver regeneration.9, 10 IL-6 activation of STAT3 also induces expression of suppressor of cytokine signaling 3 (SOCS3), which in turn terminates STAT3 signaling and negatively regulates liver regeneration.11 Although mice with global knockout of IL-6 develop acute liver failure following PHx, 8, 9 conditional deletion of the IL-6 downstream signaling molecule gp130 or STAT3 in hepatocytes did not result in acute liver failure, resulting in either no effect or only impaired liver regeneration after PHx.10, 12 The more severe liver damage following IL-6 knockout may be due to STAT3-independent signaling of IL-6, to extrahepatic actions of IL-6, or both.
After PHx, portal and systemic plasma concentrations of LPS are significantly elevated with peak levels reaching 140 pg/ml.5 Despite such high circulating levels of LPS, hepatocyte apoptosis and hepatic and systemic inflammation remain minimal.1-3 For example, PHx reportedly induced only slight or no elevation of hepatocyte apoptosis,13 and even made hepatocytes resistant to Fas- and LPS-induced apoptosis.13, 14 PHx is associated with elevated serum levels of proinflammatory cytokines, but except for IL-6, these changes are minimal (supplemental Table S1),15 and there are no obvious inflammatory foci in the liver after PHx.1-3 At present, the mechanisms that temper liver inflammation and apoptosis post PHx remain obscure. STAT3, a key signal for cell survival,16 is activated by PHx in the liver. However, deletion of STAT3 in hepatocytes only moderately impaired PHx-induced liver regeneration without inducing hepatocyte apoptosis.12 Here we demonstrate for the first time that STAT3, an important anti-inflammatory signal,17 is also markedly activated in immune cells by PHx. Conditional deletion of STAT3 in myeloid lineage cells resulted in an enhanced inflammatory response and increased liver regeneration. Combined conditional ablation of STAT3 in both hepatocytes and myeloid cells, but not in either cell type alone, resulted in a dramatic reduction in survival with elevated activation of STAT1 and hepatocyte apoptosis after PHx. These findings suggest that an interplay of STAT3 in myeloid cells and hepatocytes plays an critical role in ensuring normal liver regeneration via tempering systemic and hepatic innate inflammatory responses.
Materials and Methods
Mice
Eight- to 10-week-old male mice were used in this study. Hepatocyte-specific STAT3KO (STAT3Hep−/−) and Myeloid cell-specific STAT3KO (STAT3Mye−/−) mice were described previously.18 Littermate wild-type mice (STAT3flox/flox) were used as controls. STAT3Mye−/− mice have been proved to be a valuable tool in analyzing the physiologic role of STAT3 in monocytes/macrophages and neutrophils.17 Male STAT3Mye−/− mice were bred with female STAT3Hep−/− mice to generate four lines of mice: wild-type littermates (STAT3flox/flox), STAT3Mye−/−, STAT3Hep−/−, STAT3Mye−/−Hep−/− mice in which the STAT3 gene was deleted in both myeloid cells and hepatocytes.
STAT3Hep−/−STAT1−/− and STAT3Mye−/−STAT1−/− mice were developed via several steps of crossing STAT3Hep−/− mice with STAT1−/− mice, and STAT3Mye−/− mice with STAT1−/− mice, repsectively. Male STAT3Mye−/−STAT1−/− mice were bred with female STAT3Hep−/−STAT1−/− mice to generate STAT3Mye−/−Hep−/−STAT1−/− triple KO mice, in which the STAT3 gene was deleted in myeloid cells and hepatocytes while the STAT1 was deleted globally. All knockout strains mentioned above were developmentally normal and have normal life spans. All animal studies were approved by the Institutional Animal Care and Use Committees of the NIAAA, NIH.
Partial hepatectomy (PHx) model
For 2/3 partial hepatectomy (PHx) surgery, mice were anesthetized with sodium pentobarbital, followed by laparotomy, ligation of the median and left lateral lobes of the liver at their stem and excision under aseptic conditions, as described previously.19 For sham operation, mice were anesthetized and then subjected to laparotomy, followed by brief manipulation of the intestines, but not the liver, with cotton swabs before wound closure. The animals were killed by decapitation at the indicated times following surgery.
Statistical Analysis
Data are expressed as mean±SD. To compare values obtained from 2 groups, the Student's t test was performed. To compare values obtained from three or more groups, 1-factor analysis of variance (ANOVA) was used, followed by Tukey's post hoc test. Statistical significance was taken at the P <0.05 level.
Other methods
All other methods are described in the supporting document.
Results
Activation of STAT3 in myeloid cells after PHx
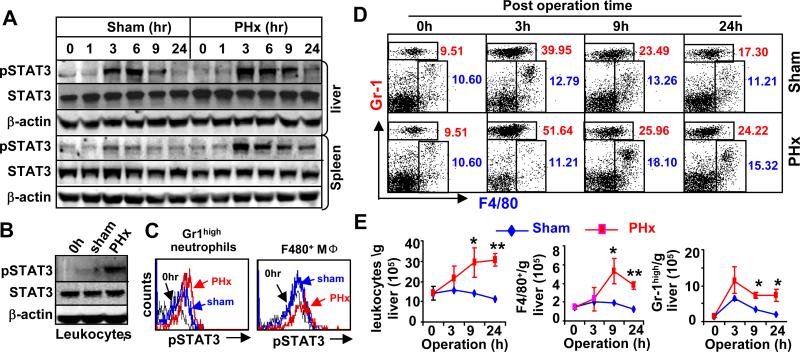
Figs. 1A and 1B show that phospho-STAT3 was markedly elevated by PHx in the liver and spleen tissues (Fig. 1A) as well as in liver leukocytes (Fig. 1B). Flow cytometric analyses show that phospho-STAT3 was elevated in both Gr1high neutrophils and F4/80+ macrophages in the liver post PHx (Fig. 1C and Fig. S1). In addition, flow cytometric analyses reveal that the percentage of Gr1high neutrophils was significantly increased in the liver post PHx while the percentage of F4/80+ macrophages was slightly increased (Fig. 1D). The total number of leukocytes, neutrophils, and macrophages was markedly elevated in the liver post PHx compared to the sham group (Fig. 1E). Increased levels of pSTAT3 were also detected in the liver but not in the spleen after sham operation to a lesser extent (Fig. 1A), which is in agreement with earlier findings.8
Fig. 1. STAT3 activation in liver neutrophils and macrophages after PHx.
A. Western blot analyses of mouse liver and spleen tissues post sham and PHx. B. Western blot analyses of liver leukocytes 3 hrs post sham and PHx. C. STAT3 activation in liver neutrophils and macrophages 3 hrs post PHx as determined by FACS analyses. D. Percentage of Gr1high neutrophils and F4/80+ macrophages analyzed by flow cytometric analyses post sham and PHx. E. Total number of F4/80+ macrophages and Gr1high neutrophils increased after PHx. Values represent means ± SD (n=6). *P<0.05, **P<0.01 in comparison with corresponding sham group.
Deletion of STAT3 in myeloid cells and hepatocytes results in enhanced and reduced liver regeneration, respectively, while deletion of STAT3 in both cell types results in liver failure after PHx
To explore whether STAT3 activation in neutrophils/macrophages plays a role in controlling liver inflammation after PHx, we generated myeloid cell-specific STAT3 knockout mice (STAT3Mye−/−), in which the STAT3 gene had been deleted in myeloid lineage cells, including neutrophils, monocytes and macrophages.17 To further understand the interaction of myeloid and hepatic STAT3 in controlling liver inflammation and regeneration, we also generated hepatocyte-specific STAT3 knockout (STAT3Hep−/−) and hepatocyte/myeloid cell-specific double knockout (STAT3Mye−/−Hep−/−) mice. After PHx, STAT3Mye−/− and STAT3Hep−/− mice showed no obvious adverse phenotype and no mortality, and their liver/body weight ratios were similar to that in wild-type mice (Fig. S2a). In contrast, 75% of STAT3Mye−/−Hep−/− mice died between 24 and 40 hrs post PHx (Fig. 2A), with the remaining 25% surviving for at least 1 month after PHx. Liver histology showed that the number of inflammatory foci was greater in STAT3Mye−/− and STAT3Mye−/−Hep−/− mice compared to wild-type and STAT3Hep−/− mice (Figs. 2B-C). Compared to wild-type mice, hepatocyte proliferation as determined by BrdU incorporation and mitosis was significantly reduced in STAT3Hep−/− mice but was elevated in STAT3Mye−/− mice 40 hrs post PHx. The surviving STAT3Mye−/−Hep−/− mice also had lower BrdU incorporation and mitosis in hepatocytes 40 hrs post PHx compared with wild-type mice (Figs. 2B, 2D, and Fig. S2b) and had reduced serum albumin compared with other groups (Fig. S2c). TUNEL analyses revealed that the number of apoptotic hepatocytes was much greater in STAT3Mye−/−Hep−/− mice than in the other groups (Figs. 2B, 2E).
Fig. 2.
Increased liver regeneration in STAT3Mye−/− mice, reduced liver regeneration in STAT3Hep−/− mice, and liver failure in STAT3Mye−/−Hep−/− mice after PHx. A. Survival rate post PHx. B. Representative photographs of H&E, Brdu, and TUNEL staining of livers 9, 40, and 24 hrs post PHx, respectively. Red arrows indicate TUNEL+ apoptotic hepatocytes. C. The number of inflammatory foci was quantified 9 hrs post PHx. D, E. The percentage of BrdU+ and TUNEL+ hepatocytes were quantified. Values represent means ± SD (n=5-10 mice). ND: not done in STAT3Mye−/−Hep−/− group. *P<0.05, **P<0.01, ***P<0.0001 in comparison with corresponding wild-type littermate controls (WT).
Deletion of STAT3 in myeloid cells enhances inflammatory response after PHx
Flow cytometric analyses show that in wild-type mice, the number of infiltrating Gr1high cells, which represent neutrophils,20 was elevated significantly 3 and 6 hrs post sham operations and maintained at slightly higher levels at 24 hrs, with such elevations being more pronounced and prolonged following PHx (Figs. S3 and Fig. 3A). The number of F4/80+ macrophages was slightly elevated post sham and PHx. Similar elevation of Gr1high and F4/80+ cells was also observed in STAT3Hep−/− mice. In addition, the total number of infiltrating Gr1high and F4/80+ cells was much higher in the livers of STAT3Mye−/− and STAT3Mye−/−Hep−/− mice as compared to wild-type and STAT3Hep−/− mice, both before surgery, as well as post sham and PHx. This suggests that PHx-induced activation of STAT3 in myeloid cells inhibits their infiltration into the liver.
Fig. 3.
Depletion of STAT3 in myeloid cells enhances inflammation after PHx. A, Total number of Gr1high neutrophils and F4/80+ macrophages after sham and PHx. B, Serum cytokines after sham and PHx. Values represent means ± SD (n=5-10). *P<0.05, **P<0.01; ***P<0.001 in comparison with corresponding wild-type littermate controls.
Fig. 3B summarized the serum levels of proinflammatory cytokines such as TNF-α, IL-6, and IFN-γ. In general, after sham operation, serum levels of IL-6 but not other cytokines was elevated in wild-type mice, while both STAT3Mye−/− and STAT3Mye−/−Hep−/− mice had higher serum levels of IL-6 as well as TNF-α and IFN-γ. After PHx, serum levels of IL-6 were markedly elevated in wild-type and STAT3Hep−/− mice, such elevation was prolonged in STAT3Mye−/− and STAT3Mye−/−Hep−/− mice. Serum levels TNF-α and IFN-γ were slightly elevated in wild-type and STAT3Hep−/− mice but were dramatically higher in STAT3Mye−/− and STAT3Mye−/−Hep−/− mice. In addition, serum TNF-α levels were higher in STAT3Mye−/−Hep−/− mice than STAT3Mye−/− mice 6 and 9 hrs post PHx
Deletion of STAT3 results in enhanced STAT1 in myeloid cells and hepatocytes after PHx
Next, we investigated the mechanisms underlying liver failure and impaired liver regeneration in STAT3Mye−/−Hep−/− mice by analyzing the activation of the STAT3 pathway, which promotes hepatocyte survival and liver regeneration,12, 16 as well as STAT1 activation, which induces hepatocyte apoptosis and inhibits liver regeneration.21 As illustrated in Fig. 4A, STAT3 was activated by PHx in wild-type mice, as reflected by elevated levels of pSTAT3 that peaked 3-6 hr post-surgery. Compared to wild-type mice, in STAT3Mye−/− mice the STAT3 pathway was constitutively active, with both the STAT3 protein levels and pSTAT3 being elevated (PHx 0 hr), and increased slightly further following PHx. In contrast, hepatic STAT3 and pSTAT3 were both very low in STAT3Hep−/− and STAT3Mye−/−Hep−/− mice, as expected.
Fig. 4.
STAT1 activation is enhanced the liver and leukocytes in STAT3Hep−/− and STAT3Mye−/− mice, respectively, and is enhanced in both the liver and leuokcytes in STAT3Mye−/−Hep−/− mice after PHx. A, Western blot (upper panel) and RT-PCR (low panel) analyses of liver tissues post PHx. B, Western blot analyses of liver leukocytes 3 hrs post sham (s) and PHx (p). Representative Western blot photography was shown from 2 independent experiments with similar results. “–” represents control group without operation. WT: represent littermate controls.
STAT1 activation (pSTAT1) was not detected in the liver of wild-type and STAT3Mye−/− mice after PHx. However, in both STAT3Hep−/− and STAT3Mye−/−Hep−/− mice hepatic levels of pSTAT1 were greatly increased following PHx, with peaks occurring 3-6 hrs post-PHx. A delayed increase in the expression of the STAT1 protein was observed 24 to 40 hrs post PHx in STAT3Hep−/− mice, whereas in STAT3Mye−/−Hep−/− mice, STAT1 protein levels were constitutively elevated compared to wild-type and STAT3Hep−/− mice. Weak pSTAT3 activation was also detected in the liver after sham operation in wild-type and STAT3Mye−/− mice but not in STAT3Hep−/− and STAT3Mye−/−Hep−/− mice (Fig. S4a). Interestingly, in the spleen STAT3 was activated similarly after PHx in all four lines of mice, while pSTAT1 activation was not detected in any of them (Fig. S4b).
Expression of cyclin D was induced in wild-type mice 24 and 40 hrs post PHx. Such induction was higher in STAT3Mye−/− mice but lower in STAT3Hep−/− and STAT3Mye−/−Hep−/− mice (Fig. 4 and Fig. S5), which is consistent with the grade of liver regeneration in these mice, as illustrated in Fig. 2. In addition, SOCS3 but not SOCS1 was induced after PHx in wild-type mice, consistent with earlier findings.11 Similar induction of SOCS3 was also observed in STAT3Mye−/− mice. Interestingly, SOCS1 but not SOCS3 was significantly induced after PHx in both STAT3Hep−/− and STAT3Mye−/−Hep−/− mice.
pSTAT3 and pSTAT1 activation were also examined in liver leukocytes after sham operation or PHx. As shown in Fig. 4B, pSTAT3 was detected post-PHx in the liver leukocytes from wild-type and STAT3Hep−/− mice but not from STAT3Mye−/− and STAT3Mye−/−Hep−/− mice. Constitutive activation of pSTAT1 was detected in the liver leukocytes of STAT3Mye−/− mice before or after sham or PHx, in agreement with previous reports.17 pSTAT1 was detected in the liver leukocytes 3 hrs post PHx in all groups with the highest levels in STAT3Mye−/−Hep−/− mice.
Deletion of STAT1 restores liver regeneration in STAT3Hep−/− mice, reduces inflammatory responses in STAT3Mye−/− mice, and rescues STAT3Mye−/−Hep−/− mice from post-PHx liver failure
The above data (Fig. 4) indicate increased activation of pSTAT1 in the inflammatory cells of STAT3Mye−/− mice and in the liver of STAT3Hep−/− mice, respectively, and increased activation in both the inflammatory cells and the liver in STAT3Mye−/−Hep−/− mice. Since STAT1 plays a key role in the induction of inflammation, cell apoptosis and cell cycle arrest,21 it is possible that elevation of STAT1 in hepatocytes contributes to reduced liver regeneration in STAT3Hep−/− mice, elevation of STAT1 in inflammatory cells contributes to enhanced inflammation in STAT3Mye−/− mice, while the simultaneous elevation of pSTAT1 in both inflammatory cells and the liver contributes to liver failure and impaired liver regeneration in STAT3Mye−/−Hep−/− mice. To test these possibilities, we generated STAT3Hep−/−STAT1−/−, STAT3Mye−/−STAT1−/−, and STAT3Mye−/−Hep−/−STAT1−/− mice.
As illustrated in Fig. 5A, expression of STAT1 protein in the liver was induced in STAT3Hep−/− mice but not in wild-type mice, which is consistent with previous findings.12 Western blot analyses confirmed the absence of STAT1 and STAT3 protein expression in the liver of STAT3Hep−/−STAT1−/− mice (Fig. 5B). All STAT3Hep−/−STAT1−/− mice survived after PHx (Fig. 5C) and had a greater number of Brdu+ hepatoctyes than STAT3Hep−/− mice after PHx (Fig. 5D), suggesting that deletion of STAT1 in STAT3Hep−/− mice restores the ability of the liver to regenerate. Fig. 5E illustrates that treatment with a low dose of IFN-γ induced stronger pSTAT1 activation in STAT3Hep−/− than in wild-type hepatocytes. As expected, no STAT1 or STAT3 proteins were detected in STAT3Hep−/−STAT1−/− hepatocytes. Furthermore, STAT3Hep−/− hepatocytes were more susceptible to IFN-γ inhibition of cell proliferation, an effect that was abolished in STAT3Hep−/−STAT1−/− hepatocytes.
Fig. 5. Deletion of STAT1 restores liver regeneration in STAT3Hep−/− mice and abolishes IFN-γ inhibition of hepatocyte proliferation.
A, Expression of STAT1 in the liver is enhanced in STAT3Hep−/− mice after PHx. B, Western blot analyses of liver tissues to confirm STAT3Hep−/−STAT1−/− mice. C, STAT3Hep−/−STAT1−/− mice survived after PHx. D, STAT3Hep−/−STAT1−/− mice showed increased liver regeneration compared to STAT3Hep−/− mice. E, Western blotting of primary mouse hepatocytes treated with IFN-γ. F, Primary hepatocytes were treated with IFN-γ for 48 hrs, cell proliferation was then determined by measuring [3H] thymidine uptake. Values represent means ± SD (n=6-8). In panel D, **P<0.01 in comparison with STAT3Hep−/− mice. In panel F, *P<0.05, **P<0.01, in comparison with corresponding wild-type littermate controls.
Western blot analyses (Fig. 6A) confirmed that in STAT3Mye−/−STAT1−/− mice STAT1 protein was undetectable and STAT3 protein was very low in liver leukocytes. All STAT3Mye−/−STAT1−/− mice survived after PHx (data not shown) and had comparable liver regeneration as STAT3Mye−/− mice (Fig. 6B). Infiltration of neutrophils and macrophages was reduced in STAT3Mye−/−STAT1−/− mice compared with STAT3Mye−/− mice (data not shown). Elevation of serum inflammatory cytokines was also diminished in the former relative to the latter group (Fig. 6C).
Fig. 6. Deletion of STAT1 diminishes inflammatory responses in STAT3Mye−/− mice after PHx.
A. Western blot analyses of liver leukocytes to confirm STAT3Mye−/−STAT1−/− mice. Liver leukocytes were isolated 3 hrs post PHx. B. STAT3Mye−/−STAT1−/− mice had comparable liver regeneration as STAT3Mye−/− mice after PHx. C. STAT3Mye−/−STAT1−/− mice showed reduced serum cytokines after PHx compared to STAT3Mye−/− mice. Values represent means ± SD (n=5-9). *P<0.05, **P<0.01, ***P<0.001 in comparison with STAT3Mye−/− mice.
Western blotting (Fig. 7A) confirmed the absence of STAT1 protein expression in hepatocytes and liver leukocytes, and the absence of STAT3 protein expression in hepatocytes and its very low level in liver leukocytes in STAT3Mye−/−Hep−/−STAT1−/− triple KO mice. All STAT3Mye−/−Hep−/−STAT1−/− triple KO mice survived after PHx, in contrast to the 25% survival of STAT3Mye−/−Hep−/− mice (Fig. 7B). Furthermore, as illustrated in Fig. 7C-D, the STAT3Mye−/−Hep−/−STAT1−/− mice had enhanced liver regeneration, reduced hepatocyte apoptosis, and reduced serum cytokines after PHx compared to STAT3Mye−/−Hep−/− mice. These findings suggest that deletion of STAT1 rescues liver function and regeneration, and attenuates the innate inflammatory response as compared to STAT3Mye−/−Hep−/− double KO mice.
Fig. 7. Deletion of STAT1 rescues liver failure in STAT3Mye−/−Hep−/− mice after PHx.
A. Western blot analyses of liver tissues and leukocytes to confirm the STAT3Mye−/−Hep−/−STAT1−/− KO mice. B-D. Deletion of STAT1 increases the survival rate (B), restores liver regeneration and reduces apoptosis (C), and reduces the inflammatory response (D) in STAT3Mye-Hep−/− mice after PHx. Values represent means ± SD (n=4-8). *P<0.05, **P<0.01, ***P<0.001 in comparison with STAT3Mye-Hep−/− mice.
Discussion
In this paper, we demonstrate for the first time that PHx results in STAT3 activation in immune cells, in addition to its activation in the liver, as reported previously.8 Additionally, our results indicate that activation of STAT3 in myeloid lineage cells and hepatocytes act in concert to effectively temper the systemic and hepatic inflammatory responses, ensuring normal liver regeneration, as summarized in a proposed model in Fig. 8. The rationale for this model is presented below.
Fig. 8. An interplay of myeloid and hepatic STAT3 in preventing liver failure during liver regeneration via tempering innate immunity.
Myeloid STAT3 inhibits infiltration of innate immune cells, inhibits STAT1 activation in innate immune cells, and inhibits production of inflammatory cytokines such as TNF-α, IL-6, and IFN-γ; while hepatic STAT3 inhibits inflammatory signaling STAT1 in the liver and promotes hepatocyte survival.
STAT3 is activated in both the liver and myeloid cells after PHx (Fig. 1). Elevation of IL-6 are likely responsible for STAT3 activation in the liver because such activation is markedly diminished in IL-6 knockout mice.8, 11 At present, the mechanisms underlying PHx-induced STAT3 activation in myeloid cells are not clear. Both IL-6 and IL-10 are known to activate STAT3 in myeloid cells and to be elevated in the liver and serum after PHx.8, 22 Thus, both of these cytokines likely contribute to STAT3 activation in myeloid cells.
Deletion of STAT3 in myeloid cells resulted in increased infiltration of macrophages and neutrophils into the remnant liver following PHx. Production of the proinflammatory cytokines TNF-α and IL-6 by these cells leads to activation of STAT3 in the liver. This may be responsible for the enhanced liver regeneration observed in STAT3Mye−/− mice after PHx (Fig. 2), because all of these factors have been shown to promote liver regeneration.8, 23, 24 Deletion of STAT3 in myeloid cells also resulted in elevated serum levels of IFN-γ, a cytokine known to induce hepatocyte apoptosis and cell cycle arrest via activation of the STAT1 signaling pathway.21 However, despite the high serum levels of IFN-γ, activation of STAT1 was not detected in the liver of STAT3Mye−/− mice after PHx (Fig. 4A). This may be due to suppression of STAT1 signaling by hepatic STAT3 in STAT3Mye−/− mice. Indeed, deletion of hepatic STAT3 resulted in enhanced hepatic pSTAT1 in both STAT3Hep−/−Mye−/− and STAT3Hep−/− mice. In addition, the strong inflammatory response in STAT3Mye−/− mice after PHx may be partly due to enhanced STAT1 activation in leukocytes (Fig. 4B), as deletion of STAT1 markedly reduced cytokine production (Fig. 6).
Disruption of STAT3 in hepatocytes resulted in decreased liver regeneration without mortality after PHx, consistent with previous reports.12 Interestingly, we have previously shown that mortality rate was significantly higher in mice with STAT3 deficiency in hepatocytes and digestive tissues than wild-ype controls.25 These findings suggest that STAT3 in digestive tissues may play a hepatoprotective role while STAT3 in hepatocyte stimulates hepatocyte proliferation during liver regeneration. It is believed that the stimulatory effect of STAT3 on liver regeneration is mediated via induction of several immediate early genes.12 Here we demonstrated that deletion of STAT1 restored liver regeneration in STAT3Hep−/− and STAT3Hep−/−Mye−/− mice (Figs. 5 and 7), suggesting that inhibition of STAT1 signaling is one of the mechanisms through which STAT3 activation promotes liver regeneration. However, the mechanism by which STAT3 suppresses STAT1 signaling is not well understood. STAT signaling pathways can be negatively regulated by several mechanisms, including induction of SOCSs, tyrosine phosphatases, PIAS etc.26 Fig. 4 shows that induction of SOCS3 and SOCS1 correlates with activation of STAT3 and STAT1, respectively, suggesting that STAT3 activation is responsible for SOCS3 induction while SOCS1 induction is dependent on STAT1 activation after PHx. It is probable that STAT3 inhibits STAT1 signaling via at least in part induction of SOCS3 expression because SOCS3 has been shown to inhibit STAT1 signaling.27
No mortality and no obvious hepatocyte apoptosis were observed in STAT3Mye−/− mice after PHx despite high levels of inflammatory cytokines such as TNF-α and IFN-γ. This is probably due to prolonged STAT3 activation in the liver that protects against hepatocyte death, STAT3 being a survival signal for hepatocytes.16 Indeed, deletion of hepatic STAT3 both in hepatocytes and myeloid cells caused massive apoptosis after PHx. Interestingly, deletion of the IL-6 signaling molecule gp130 in both hepatocytes and bone marrow cells did not result in liver failure after PHx.10 This suggests that the critical role of myeloid STAT3 activation in liver regeneration is mediated by a mediator other than IL-6. In addition, deletion of hepatic STAT3 also resulted in further increases in serum levels of TNF-α in STAT3Mye−/− mice after PHx (Fig. 3B). This may be due to increased hepatocyte apoptosis in STAT3Mye−/−Hep−/− mice that can stimulate macrophages/Kupffer cells to produce inflammatory cytokines.28 Such high levels of TNF-α may also contribute to the massive apoptosis and liver failure in STAT3Mye−/−Hep−/− mice after PHx because abnormally high levels of TNF-α were shown to contribute to liver failure after PHx in Timp3−/− mice.29 Although STAT3 is a survival signal for hepatocytes, selective deletion of STAT3 in hepatocytes did not induce apoptosis and mortality. This may be due to maintained STAT3 activation in myeloid cells that limits inflammatory responses such as TNF-α and IFN-γ production. Deletion of STAT3 in both myeloid cells and hepatocytes in STAT3Hep−/−Mye−/− mice resulted in high levels of serum TNF-α and IFN-γ and hepatic STAT1 activation, which resulted in massive apoptosis of hepatocytes and high mortality. It has been recently proposed that STAT3 inhibitors may be used in the treatment of hepatocellular carcinoma (HCC).30 The present findings advocate caution with such an approach, because global inhibition of STAT3 may result in a strong innate inflammatory response and liver failure, especially in the remnant liver of HCC patients following liver resection. Indeed, liver failure after resection was often seen in HCC patients with elevated inflammatory responses due to sepsis.31 Additionally, elevated STAT1 expression and activation in the liver were found in patients with chronic liver disease,32 which may impair liver regeneration. Thus, a strategy to increase STAT3/STAT1 ratio in both hepatocytes and leukocytes may have a beneficial effect in preventing liver failure in HCC patients with elevated inflammatory responses after liver resection.
Supplementary Material
Fig. S1. Mean fluorescence intensity (MFI) of pSTAT3 in F4/80+ cells and Gr1bright cells qualified from the figures in Fig. 1E. Left panel shows that STAT3 activation (pSTAT3) in F4/80+ macrophages from the liver 3 hrs post sham and PHx. Right panel shows STAT3 activation in Gr1bright neutrophils from the liver 3 hrs post sham or PHx. Values represent means ± SD (n=4).
Fig. S2a. The ratio of liver/body weight after PHx. ND. Not done in STAT3Mye−/−Hep−/− group.
Fig. S2b. The percentage of mitotic Hepatocytes post PHx
Fig. S2c: Serum levels of albumin post PHx
Fig. S3: Depletion of STAT3 in myeloid cells enhances inflammation after PHx. Representative flow cytometric analyses of liver leukocytes with anti-Gr1 and anti-F4/80 antibodies post sham or PHx. Gr1high cells in red gates represent neutrophils. F4/80+ cells represent macrophages.
Fig. S4a. Activation of STAT1 and STAT3 in the liver from sham groups.
Fig. S4b. Activation of STAT1 and STAT3 in the spleen tissues from partially hepateomized mice.
Fig. S5: Cycline D protein levels 40 h post PHx from 3 independent experiments were Quantified. Values represent means ± SD. P<0.05, **P<0.01
Supporting Materials
Interplay of hepatic and myeloid STAT3 in facilitating liver regeneration via tempering innate immunity
Hua Wang, Ogyi Park, Fouad Lafdil, Kezhen Shen, Norio Horiguchi, Shi Yin, Xin-Yuan Fu, George Kunos, Bin Gao
Materials and Methods:
Materials. Anti-STAT3, anti-phospho-STAT3 (Tyr705), anti-pSTAT1, anti-STAT1, anti-cyclin D antibodies were purchased from Cell Signaling Technology (Beverly, MA).
Isolation of liver leukocytes. Liver leukocytes were isolated as described previously 1. Briefly, mouse livers were pressed through a 200-gauge stainless steel mesh. The liver cell suspension was suspended in RPMI 1640 media (Invitrogen, Carlsbad, CA) and centrifuged at 50×g for 5 minutes. Supernatants containing leukocytes were collected, washed in PBS, and resuspended in 35% Percoll (Sigma) containing 100 U/mL heparin. The cell suspension was centrifuged for 15 minutes at 500×g. The cell pellet containing leukocytes was collected and re-suspended in 5 mL red blood cell lysis solution (155 mmol/L NH4CL, 10 mmol/L KHCO3, 1 mmol/L EDTA, 170 mmol/L Tris, pH 7.3). After incubation for 5 minutes on ice, cells were washed twice in RPMI 1640 containing 5% fetal bovine serum.
Flow cytometry analysis of neutrophils and macrophages in the liver. Neutrophils and macrophages in the livers were determined by anti-Gr1 and anti-F4/80 antibodies (PharMingen, San Diego, CA) through a fluorescent-activated cell sorter (FACScalibur, Becton Dickinson, Mountain View, CA). Gr1high cells mainly represent neutrophils, where Gr1intermediate cells represent monocytes and eosinophils 2.
Determination of the rate of liver regeneration. Liver regeneration rate was determined by the BrdU incorporation assay. Briefly, partially hepatectomized mice were injected intraperitoneally with BrdU (50 μg/g body weight) at various time points post PHx. Mice were euthanized 2 hours later and the livers were harvested and fixed in 10% neutral buffered formalin for 24 hours. Fixed livers were embedded in paraffin, cut into 5-μm tissue sections by a microtome, and then adhered to poly-L-lysine-coated glass slides. The slides were dried overnight at 37°C. BrdU incorporation was visualized immunohistochemically, using a BrdU immunostaining kit (BD Biosciences) according to the manufacturer's instructions. BrdU-labeled hepatocytes were quantified by counting positively stained hepatocyte nuclei in 3 to 6 low-power (100×) microscope fields, and the mean was calculated.
Histology and immunohistochemistry. Formalin-fixed liver samples were processed, and paraffin sections of 5-μm thickness were stained with hematoxylin and eosin (H&E). Inflammatory foci were counted in 10 randomly selected fields (×100 magnification).
TUNEL assay. Apoptotic cells in sections of mouse livers were detected using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Chemicon International Inc., Temecula, CA) according to the manufacturer's instructions. Numbers of TUNEL+ hepatocytes were determined by counting positively stained hepatocyte nuclei in 5 high-power (200×) microscope fields, and the mean was calculated.
Serum cytokine levels. Serum cytokine levels were measured using cytometry bead array (BD Bioscience) according to the manufacturer's protocol.
Serum albumin levels. Serum levels of albumin were measured using a kit from Drew scientific Ltd (Barrow-in-Furness, UK).
Western blotting. Western blot analyses were performed with proteins from liver homogenates (80 μg) using various primary antibodies. Briefly, 50-80 μg of proteins were loaded. Immunoreactive bands were visualized on nitrocellulose membranes using alkaline-phosphotase-linked anti-mouse or rabbit antibody and the ECF detection system with a PhosphorImager (GE Healthcare, Piscataway, NJ).
RT-PCR: RT-PCR was used to determine the expression of SOCS1 and SOCS3 in the liver. The primer sequences include SOCS1 (forward): GAA GCC ATC TTC ACG CTG, (reverse): ACA CTC ACT TCC GCA CCT TC (PCR product 210 bp); SOCS3 (forward): GGG TGG CAA AGA AAA GGA G, (reverse) GTT GAG CGT CAA GAC CCA GT (PCR product 450bp).
Statistical Analysis. Data are expressed as means ± SEM. To compare values obtained from 2 groups, the Student t test was performed. To compare values obtained from three or more groups, 1-factor analysis of variance (ANOVA) was used, followed by Tukey's post hoc test. Statistical significance was taken at the P <0.05 level.
References:
1. Liu ZX, Han D, Gunawan B, Kaplowitz N. Neutrophil depletion protects against murine acetaminophen hepatotoxicity. Hepatology 2006;43:1220-1230.
2. Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol 2008;83:64-70.
3. Fujita J, Marino MW, Wada H, Jungbluth AA, Mackrell PJ, Rivadeneira DE, et al. Effect of TNF gene depletion on liver regeneration after partial hepatectomy in mice. Surgery 2001;129:48-54.
Supplemental TABEL S1:
Serum levels of cytokines after PHx. Values represent means ± SEM (ng/ml) (n=3-6)
Acknowledgments
This work was supported by the intramural program of NIAAA, NIH. No conflicts of interest exist for all authors.
Abbreviations
- KO
knockout
- PHx
2/3 partial hepatectomy
- STAT
signal transducer and activator of transcription
- STAT3Hep−/−
hepatocyte-specific STAT3 knockout mice
- STAT3Mye−/−
myeloid cell-specific STAT3 knockout mice
- STAT3Hep−/−Mye−/−
hepatocyte and myeloid cell-specific STAT3 double knockout mice
- STAT3Hep−/−Mye−/−STAT1−/−
hepatocyte and myeloid cell-specific STAT3 and STAT1 triple knockout mice
References
- 1.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 3.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 4.Koniaris LG, McKillop IH, Schwartz SI, Zimmers TA. Liver regeneration. J Am Coll Surg. 2003;197:634–659. doi: 10.1016/S1072-7515(03)00374-0. [DOI] [PubMed] [Google Scholar]
- 5.Cornell RP. Gut-derived endotoxin elicits hepatotrophic factor secretion for liver regeneration. Am J Physiol. 1985;249:R551–562. doi: 10.1152/ajpregu.1985.249.5.R551. [DOI] [PubMed] [Google Scholar]
- 6.Campbell JS, Riehle KJ, Brooling JT, Bauer RL, Mitchell C, Fausto N. Proinflammatory cytokine production in liver regeneration is Myd88-dependent, but independent of Cd14, Tlr2, and Tlr4. J Immunol. 2006;176:2522–2528. doi: 10.4049/jimmunol.176.4.2522. [DOI] [PubMed] [Google Scholar]
- 7.Seki E, Tsutsui H, Iimuro Y, Naka T, Son G, Akira S, et al. Contribution of Toll-like receptor/myeloid differentiation factor 88 signaling to murine liver regeneration. Hepatology. 2005;41:443–450. doi: 10.1002/hep.20603. [DOI] [PubMed] [Google Scholar]
- 8.Cressman DE, Greenbaum LE, DeAngelis RA, Ciliberto G, Furth EE, Poli V, et al. Liver failure and defective hepatocyte regeneration in interleukin-6- deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 9.Blindenbacher A, Wang X, Langer I, Savino R, Terracciano L, Heim MH. Interleukin 6 is important for survival after partial hepatectomy in mice. Hepatology. 2003;38:674–682. doi: 10.1053/jhep.2003.50378. [DOI] [PubMed] [Google Scholar]
- 10.Wuestefeld T, Klein C, Streetz KL, Betz U, Lauber J, Buer J, et al. Interleukin-6/glycoprotein 130-dependent pathways are protective during liver regeneration. J Biol Chem. 2003;278:11281–11288. doi: 10.1074/jbc.M208470200. [DOI] [PubMed] [Google Scholar]
- 11.Campbell JS, Prichard L, Schaper F, Schmitz J, Stephenson-Famy A, Rosenfeld ME, et al. Expression of suppressors of cytokine signaling during liver regeneration. J Clin Invest. 2001;107:1285–1292. doi: 10.1172/JCI11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Liang X, Kellendonk C, Poli V, Taub R. STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem. 2002;277:28411–28417. doi: 10.1074/jbc.M202807200. [DOI] [PubMed] [Google Scholar]
- 13.Helling TS, Dhar A, Helling TS, Jr., Moore BT, VanWay CW. Partial hepatectomy with or without endotoxin does not promote apoptosis in the rat liver. J Surg Res. 2004;116:1–10. doi: 10.1016/s0022-4804(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 14.Desbarats J, Newell MK. Fas engagement accelerates liver regeneration after partial hepatectomy. Nat Med. 2000;6:920–923. doi: 10.1038/78688. [DOI] [PubMed] [Google Scholar]
- 15.Bergheim I, Luyendyk JP, Steele C, Russell GK, Guo L, Roth RA, et al. Metformin prevents endotoxin-induced liver injury after partial hepatectomy. J Pharmacol Exp Ther. 2006;316:1053–1061. doi: 10.1124/jpet.105.092122. [DOI] [PubMed] [Google Scholar]
- 16.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest. 2003;112:978–980. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 18.Horiguchi N, Wang L, Mukhopadhyay P, Park O, Jeong WI, Lafdil F, et al. Cell type-dependent pro- and anti-inflammatory role of signal transducer and activator of transcription 3 in alcoholic liver injury. Gastroenterology. 2008;134:1148–1158. doi: 10.1053/j.gastro.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 20.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 21.Sun R, Park O, Horiguchi N, Kulkarni S, Jeong WI, Sun HY, et al. STAT1 contributes to dsRNA inhibition of liver regeneration after partial hepatectomy in mice. Hepatology. 2006;44:955–966. doi: 10.1002/hep.21344. [DOI] [PubMed] [Google Scholar]
- 22.Rai RM, Loffreda S, Karp CL, Yang SQ, Lin HZ, Diehl AM. Kupffer cell depletion abolishes induction of interleukin-10 and permits sustained overexpression of tumor necrosis factor alpha messenger RNA in the regenerating rat liver. Hepatology. 1997;25:889–895. doi: 10.1002/hep.510250417. [DOI] [PubMed] [Google Scholar]
- 23.Selzner N, Selzner M, Odermatt B, Tian Y, Van Rooijen N, Clavien PA. ICAM-1 triggers liver regeneration through leukocyte recruitment and Kupffer cell-dependent release of TNF-alpha/IL-6 in mice. Gastroenterology. 2003;124:692–700. doi: 10.1053/gast.2003.50098. [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Kirillova I, Peschon JJ, Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci U S A. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moh A, Iwamoto Y, Chai GX, Zhang SS, Kano A, Yang DD, et al. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest. 2007;87:1018–1028. doi: 10.1038/labinvest.3700630. [DOI] [PubMed] [Google Scholar]
- 26.Greenhalgh CJ, Hilton DJ. Negative regulation of cytokine signaling. J Leukoc Biol. 2001;70:348–356. [PubMed] [Google Scholar]
- 27.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273:35056–35062. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 28.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 29.Mohammed FF, Smookler DS, Taylor SE, Fingleton B, Kassiri Z, Sanchez OH, et al. Abnormal TNF activity in Timp3-/- mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat Genet. 2004;36:969–977. doi: 10.1038/ng1413. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Amin R, Gallicano GI, Glasgow E, Jogunoori W, Jessup JM, et al. The STAT3 inhibitor NSC 74859 is effective in hepatocellular cancers with disrupted TGF-beta signaling. Oncogene. 2009;28:961–972. doi: 10.1038/onc.2008.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capussotti L, Vigano L, Giuliante F, Ferrero A, Giovannini I, Nuzzo G. Liver dysfunction and sepsis determine operative mortality after liver resection. Br J Surg. 2009;96:88–94. doi: 10.1002/bjs.6429. [DOI] [PubMed] [Google Scholar]
- 32.Duong FH, Filipowicz M, Tripodi M, La Monica N, Heim MH. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology. 2004;126:263–277. doi: 10.1053/j.gastro.2003.10.076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Mean fluorescence intensity (MFI) of pSTAT3 in F4/80+ cells and Gr1bright cells qualified from the figures in Fig. 1E. Left panel shows that STAT3 activation (pSTAT3) in F4/80+ macrophages from the liver 3 hrs post sham and PHx. Right panel shows STAT3 activation in Gr1bright neutrophils from the liver 3 hrs post sham or PHx. Values represent means ± SD (n=4).
Fig. S2a. The ratio of liver/body weight after PHx. ND. Not done in STAT3Mye−/−Hep−/− group.
Fig. S2b. The percentage of mitotic Hepatocytes post PHx
Fig. S2c: Serum levels of albumin post PHx
Fig. S3: Depletion of STAT3 in myeloid cells enhances inflammation after PHx. Representative flow cytometric analyses of liver leukocytes with anti-Gr1 and anti-F4/80 antibodies post sham or PHx. Gr1high cells in red gates represent neutrophils. F4/80+ cells represent macrophages.
Fig. S4a. Activation of STAT1 and STAT3 in the liver from sham groups.
Fig. S4b. Activation of STAT1 and STAT3 in the spleen tissues from partially hepateomized mice.
Fig. S5: Cycline D protein levels 40 h post PHx from 3 independent experiments were Quantified. Values represent means ± SD. P<0.05, **P<0.01
Supporting Materials
Interplay of hepatic and myeloid STAT3 in facilitating liver regeneration via tempering innate immunity
Hua Wang, Ogyi Park, Fouad Lafdil, Kezhen Shen, Norio Horiguchi, Shi Yin, Xin-Yuan Fu, George Kunos, Bin Gao
Materials and Methods:
Materials. Anti-STAT3, anti-phospho-STAT3 (Tyr705), anti-pSTAT1, anti-STAT1, anti-cyclin D antibodies were purchased from Cell Signaling Technology (Beverly, MA).
Isolation of liver leukocytes. Liver leukocytes were isolated as described previously 1. Briefly, mouse livers were pressed through a 200-gauge stainless steel mesh. The liver cell suspension was suspended in RPMI 1640 media (Invitrogen, Carlsbad, CA) and centrifuged at 50×g for 5 minutes. Supernatants containing leukocytes were collected, washed in PBS, and resuspended in 35% Percoll (Sigma) containing 100 U/mL heparin. The cell suspension was centrifuged for 15 minutes at 500×g. The cell pellet containing leukocytes was collected and re-suspended in 5 mL red blood cell lysis solution (155 mmol/L NH4CL, 10 mmol/L KHCO3, 1 mmol/L EDTA, 170 mmol/L Tris, pH 7.3). After incubation for 5 minutes on ice, cells were washed twice in RPMI 1640 containing 5% fetal bovine serum.
Flow cytometry analysis of neutrophils and macrophages in the liver. Neutrophils and macrophages in the livers were determined by anti-Gr1 and anti-F4/80 antibodies (PharMingen, San Diego, CA) through a fluorescent-activated cell sorter (FACScalibur, Becton Dickinson, Mountain View, CA). Gr1high cells mainly represent neutrophils, where Gr1intermediate cells represent monocytes and eosinophils 2.
Determination of the rate of liver regeneration. Liver regeneration rate was determined by the BrdU incorporation assay. Briefly, partially hepatectomized mice were injected intraperitoneally with BrdU (50 μg/g body weight) at various time points post PHx. Mice were euthanized 2 hours later and the livers were harvested and fixed in 10% neutral buffered formalin for 24 hours. Fixed livers were embedded in paraffin, cut into 5-μm tissue sections by a microtome, and then adhered to poly-L-lysine-coated glass slides. The slides were dried overnight at 37°C. BrdU incorporation was visualized immunohistochemically, using a BrdU immunostaining kit (BD Biosciences) according to the manufacturer's instructions. BrdU-labeled hepatocytes were quantified by counting positively stained hepatocyte nuclei in 3 to 6 low-power (100×) microscope fields, and the mean was calculated.
Histology and immunohistochemistry. Formalin-fixed liver samples were processed, and paraffin sections of 5-μm thickness were stained with hematoxylin and eosin (H&E). Inflammatory foci were counted in 10 randomly selected fields (×100 magnification).
TUNEL assay. Apoptotic cells in sections of mouse livers were detected using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Chemicon International Inc., Temecula, CA) according to the manufacturer's instructions. Numbers of TUNEL+ hepatocytes were determined by counting positively stained hepatocyte nuclei in 5 high-power (200×) microscope fields, and the mean was calculated.
Serum cytokine levels. Serum cytokine levels were measured using cytometry bead array (BD Bioscience) according to the manufacturer's protocol.
Serum albumin levels. Serum levels of albumin were measured using a kit from Drew scientific Ltd (Barrow-in-Furness, UK).
Western blotting. Western blot analyses were performed with proteins from liver homogenates (80 μg) using various primary antibodies. Briefly, 50-80 μg of proteins were loaded. Immunoreactive bands were visualized on nitrocellulose membranes using alkaline-phosphotase-linked anti-mouse or rabbit antibody and the ECF detection system with a PhosphorImager (GE Healthcare, Piscataway, NJ).
RT-PCR: RT-PCR was used to determine the expression of SOCS1 and SOCS3 in the liver. The primer sequences include SOCS1 (forward): GAA GCC ATC TTC ACG CTG, (reverse): ACA CTC ACT TCC GCA CCT TC (PCR product 210 bp); SOCS3 (forward): GGG TGG CAA AGA AAA GGA G, (reverse) GTT GAG CGT CAA GAC CCA GT (PCR product 450bp).
Statistical Analysis. Data are expressed as means ± SEM. To compare values obtained from 2 groups, the Student t test was performed. To compare values obtained from three or more groups, 1-factor analysis of variance (ANOVA) was used, followed by Tukey's post hoc test. Statistical significance was taken at the P <0.05 level.
References:
1. Liu ZX, Han D, Gunawan B, Kaplowitz N. Neutrophil depletion protects against murine acetaminophen hepatotoxicity. Hepatology 2006;43:1220-1230.
2. Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol 2008;83:64-70.
3. Fujita J, Marino MW, Wada H, Jungbluth AA, Mackrell PJ, Rivadeneira DE, et al. Effect of TNF gene depletion on liver regeneration after partial hepatectomy in mice. Surgery 2001;129:48-54.
Supplemental TABEL S1:
Serum levels of cytokines after PHx. Values represent means ± SEM (ng/ml) (n=3-6)