Abstract
Background
A hallmark of prostate cancer (PCa) is that distinct tumor foci may arise independently, which has important biologic and clinical implications. Recent studies characterizing ERG rearranged PCa possessing intrafocal homogeneity but interfocal heterogeneity support this hypothesis.
Objective
Using ERG rearrangement as marker of clonality, we interrogated multifocal PCa to determine its predilection for metastasis.
Design, Setting, and Participants
We studied 26 patients who underwent prostatectomy and lymphadenectomy with at least two distinct PCa foci and one lymph node (LN) metastasis.
Measurement
Each focus was assessed for size, Gleason score, ERG rearrangement, and TMPRSS2-ERG transcript.
Results
15/26 cases exhibited interfocal homogeneity with regard to ERG rearrangement (i.e., presence versus absence of ERG rearrangement). ERG rearrangement was present in all foci for 6 and absent in all foci for 9 cases. Two cases revealed interfocal heterogeneity with regard to rearrangement mechanism (i.e., rearrangement through insertion or deletion). 8/26 cases revealed interfocal heterogeneity with regard to rearrangement status. In all cases with at least one ERG rearranged focus, we found the corresponding LN metastasis harboring an ERG rearrangement. Interestingly, in a subset of cases the rearrangement status in the LN did not correspond to size or Gleason score. All but two ERG rearranged foci had detectable TMPRSS2-ERG transcript levels.
Conclusions
When multifocal PCa demonstrates both ERG positive and negative foci, the positive foci have a greater predilection for metastasis. Larger studies are needed to confirm the potential additional risk an ERG rearranged focus confers on the likelihood of disease progression.
Keywords: prostate cancer, metastatic, ERG rearrangements, TMPRSS2-ERG
Introduction
Rearrangement of the ETS gene ERG (21q22.2), most commonly with the androgen-regulated gene TMPRSS2 (21q22.3), results in the most frequent recurrent genomic rearrangement event in human prostate cancer (PCa) (1-3). Gene rearrangements involving TMPRSS2 and the other ETS family members (e.g., ETV1 and ETV5) occur less frequently (4-6). Fusion of the 5 prime (5’)-untranslated region of TMPRSS2 with the 3 prime (3’) end of ERG leads to the over-expression of an androgen- and estrogen-responsive mRNA fusion product encoding a truncated form of ERG. Although still controversial, two studies following the natural history of PCa in the Watchful Waiting setting have demonstrated a significant association between ERG rearrangement and cancer specific death (7, 8). However, there has not been a consistent correlation between clinical outcome and ERG rearrangement status observed after treating men with clinically localized PCa with radical prostatectomy (3, 9).
Following the initial discovery of recurrent gene fusions in PCa, we demonstrated evidence that ERG gene rearrangement occurs as an early clonal event (2). A few small studies on matched primary and metastatic tumors report concordance in the fusion status, thus supporting the argument for clonal expansion of PCa (2, 10). In these studies, however, multifocal occurrence of localized PCa was not taken into account. At the time of diagnosis, more than 80% of PCa cases harbor multiple tumor foci (11). It is well known that PCa displays histologic and molecular interfocal heterogeneity, suggesting that the different cancer foci arise independently of one another (12-14). In addition to our previous report of clonal intrafocal homogeneity for ERG rearrangement (2), we and others subsequently demonstrated interfocal heterogeneity with regard to ERG rearrangement status in multifocal PCa (14-16). In contrast, various other cancer types have been shown to have clonal multifocal tumors. For example, a recent study reported that a great majority (77%) of multifocal lung cancer cases harbor common clonal origin (17).
The aim of this study was to investigate the ERG rearrangement status in multifocal localized PCa and corresponding lymph node(s) (LN) metastasis to explore if ERG rearrangement could be used as a marker of clonal expansion for aggressive disease.
Materials and methods
The study cohort was derived from patients who underwent radical prostatectomy and lymphadenectomy in Ulm, Germany between 1986 and 2002 (17). We identified 26 cases that had at least two distinct cancer foci in the prostate gland and one LN metastasis and for which formalin-fixed, paraffin-embedded (FFPE) material was available for tissue microarray (TMA) construction. Patient demographics are described elsewhere (17). In brief, 14 patients (54%) were diagnosed with stage pT3 disease, 2 patients (8%) with pT2, and 10 patients (39%) with stage pT4. Four of the patients received hormonal treatment and one had radiation therapy. In this cohort, all Gleason scores were above 6. Slightly over half of the PCa tumors were Gleason score 7 (14/26 (54%)), 1/26 (4%) Gleason score 8, and 11/26 cancers (42%) Gleason score 9.
A PCa sample was considered multifocal if tumor nodules were identified on contralateral sides of the gland. Ipsilateral tumor nodules were defined as separated by a minimum of 3 mm from the nearest tumor nodule in any single section or by a minimum of 4 mm from the closest nodule on the adjacent section above or below (13, 18). Tumor maps were generated by tracking each section according to the pathology report and reconstructing them in a whole-mount manner. Primary, secondary, tertiary and quaternary tumor foci from both the prostate and metastatic LN were designated in decreasing order based on tumor diameter (cm). Although not part of standard clinical routine, we assigned a primary and secondary Gleason score for each PCa focus and metastatic LN focus based on a prior exploratory study (17).
Tissue micro array (TMA) construction
Two TMAs were constructed from 26 patients comprising all identified PCa and LN foci from the prostatectomy and LN specimens. Three randomly selected, representative 0.6 mm cores were taken from each possible cancer focus (i.e., from up to four foci in the prostate and the lymph nodes).
Assessment of gene rearrangement status by dual-color interphase fluorescence in-situ hybridization (FISH)
Four micron thick TMA sections were used for interphase FISH. ERG rearrangement was determined using a dual-color break-apart interphase FISH assay as described previously (1, 2, 19). In brief, two differentially labeled probes were designed to span the telomeric and centromeric neighboring regions of each locus. The following telomeric/centromeric BAC clones were selected to design break-apart assays to assess for rearrangement status (i.e., rearrangement versus no rearrangement): ERG (RP11-372O17 and RP11-24A11), TMPRSS2 (RP11-120C17 and RP11-35C4), SLC45A3 (RP11-131E5 and RP11-249H15), and NDRG1 (RP11-1145H17 and RP11-185E14). In cases of ERG rearrangement, the assay was also capable of differentiating between two different mechanisms of ERG rearrangement as previously described by our group (19). These two mechanisms are ERG rearrangement through insertion and ERG rearrangement through deletion of DNA between TMPRSS2 and ERG. A nucleus without an ERG rearrangement demonstrates two pairs of juxtaposed red and green signals (mostly forming 2 yellow signals). A nucleus with an ERG insertion shows the split of one red-green (yellow) signal pair, resulting in a single red and green signal for the rearranged ERG allele, and a still combined (yellow) signal for the non-rearranged ERG allele in each nucleus. Finally, a nucleus with an ERG rearrangement through deletion shows one juxtaposed red-green signal pair (yellow) for the non-rearranged allele, and a single red signal for the allele involved in the rearrangement. Slides were analyzed under a x60 oil immersion objective using an Olympus (Center Valley, PA) BX-51 fluorescence microscope equipped with appropriate filters and a charge-coupled device camera, and the CytoVision FISH imaging and capturing software (Applied Imaging, San Jose, CA). All foci were assessed for ERG rearrangement. Only cases that revealed an ERG rearrangement but no TMPRSS2-ERG mRNA expression were assessed for rearrangement of TMPRSS2 and other potential 5’ fusion partners of ERG (i.e. SLC45A3 and NDRG1).
Assessment of TMPRSS2-ERG mRNA expression
Total RNA was extracted from a 1.5 mm FFPE tissue core as previously described (20). Nine complete cases out of the 26 cases with valid FISH results had sufficient FFPE material for RNA extraction. One 1.5 mm core biopsy per tumor focus was taken and placed in one well of a 96-well plate for high-throughput RNA extraction. The CyBio-Well liquid handling system (CyBio AG, Jenna, Germany) was used for high-throughput extraction. Cores were first deparaffinized by incubating them with 800 μL Citrisolv (Fisher Scientific, Pittsburgh, PA) at 60°C for 20 minutes and then with 1.2 mL Citrisolv:absolute alcohol (2:1) at room temperature for 10 minutes. Subsequently, cores were washed with absolute alcohol, dried at 55°C, and incubated overnight at 45°C in 300 μL lysis buffer (10 mM NaCl, 500 mM Tris [pH 7.6], 20 mM EDTA, 1% sodium dodecyl sulfate) containing 1 mg/mL proteinase K (Ambion, Austin, TX). RNA was extracted from the lysate using the TRIzol LS reagent (Invitrogen, Carlsbad, CA) and quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE). TMPRSS2-ERG mRNA and PSA mRNA were quantified using magnetic target capture (purification of target mRNA by hybridization to magnetic particles via target-specific oligonucleotides), transcription-mediated amplification (amplification of target RNA sequences), and a hybridization protection assay (specific detection of amplification products by use of target-specific acridinium ester (AE)-labeled probes) as described in Groskopf et al. (21). The most prevalent TMPRSS2-ERG mRNA isoform, TMPRSS2-ERGa (1), also known as TMPRSS2-ERG type III (22) was targeted. Amplification primers for TMPRSS2-ERG mRNA were located in TMPRSS2 exon 1 and ERG exon 4 yielding an amplification product of 86 nucleotides. The AE probe for TMPRSS2-ERG spanned the junction between the two exons. Primers for PSA targeted exons 2 and 3 of the PSA mRNA, and the AE probe spanned the exon 2/3 junction. Calibrators and controls consisted of TMPRSS2-ERG or PSA in vitro transcripts (IVTs) in detergent solution. The TMPRSS2-ERG IVT was prepared from a plasmid provided by Arul Chinnaiyan at the University of Michigan (1). IVT copy levels were determined by A260. Assays were performed using Gen-Probe DTS 400 Systems; the assay protocol utilizes the reagent addition volumes, incubation times and temperatures specified in the APTIMA COMBO2 package insert and as described in Groskopf et al. (21). PSA mRNA was used to normalize for the total amount of prostate-specific mRNA in each sample. The dynamic range of transcript copy levels for TMPRSS2-ERG and PSA were 100-1,000 and 100-39,400, respectively. Samples yielding less than 100 copies of PSA mRNA per 10 ng total RNA were considered invalid due to insufficient mRNA for analysis. Data from samples yielding copies higher than the upper limit (>39,400 for PSA and/or >1,000 for TMPRSS2-ERG) were included in the analysis but used to calculate relative abundance. Data within the indicated ranges were presented as relative abundance by dividing the copies per reaction obtained for TMPRSS2-ERG by copies per reaction obtained for PSA.
Data analysis and interpretation
This unique clinical cohort enabled us to study ERG rearrangement dependent metastatic spread in multifocal PCa. Statistical analysis was performed, but was constrained to primarily a descriptive analysis due to the limited number of cases (n=26) evaluated.
Statistical Analysis
Within the set of individuals with interfocal homogeneity of ERG rearrangement status, we applied Fisher Exact test (two-tailed test) to investigate for significant associations between ERG rearrangement status of PCa cases and ERG rearrangement status of the corresponding LN metastases. We then focused on the set of individuals with interfocal heterogeneity of ERG rearrangement status. To answer the question whether the rearrangement status of the LN metastases is determined by the rearrangement positive focus in the corresponding prostate, we evaluated the joint probability as the product of the single individual binomial probabilities. This provides an estimate of the probability of obtaining the observed result by chance.
Results
We obtained valid ERG break apart (b/a) FISH results from all tumor foci for 26 patients with multifocal PCa and corresponding LN metastasis (see Figure 1A). Clinical information was available for all patients. The mean age was 64 years (range 51-74). The vast majority (21/26 (81%)) of patients had been treated with neoadjuvant hormonal ablation therapy.
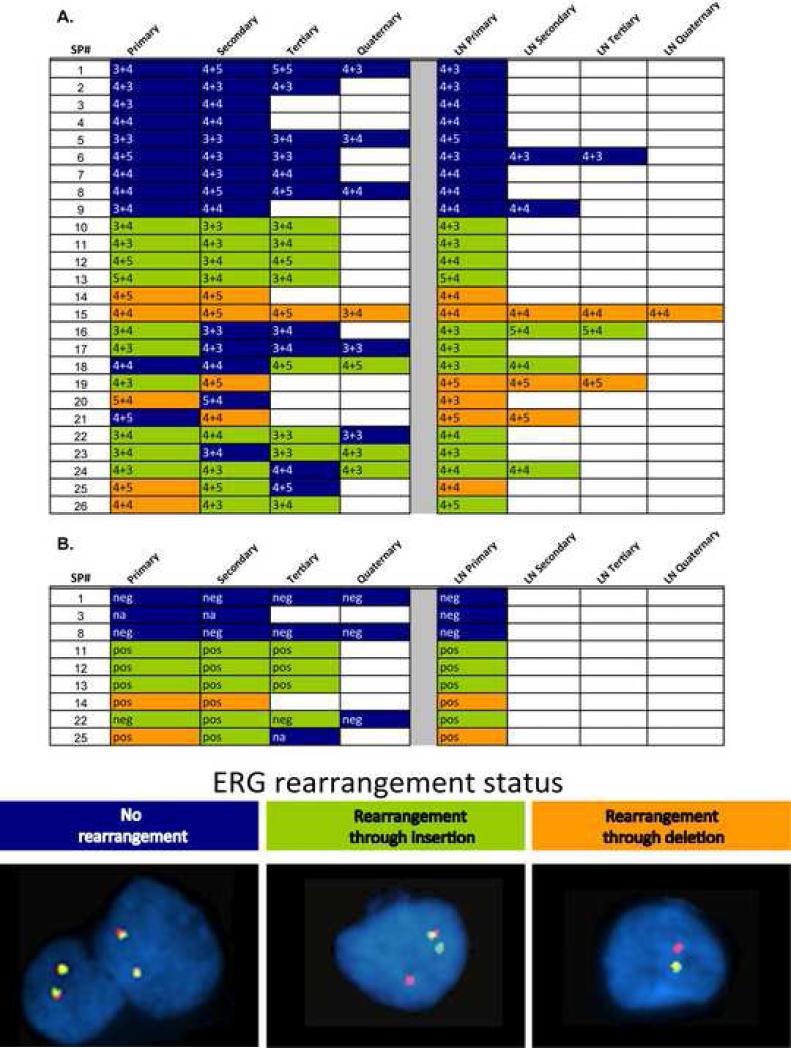
Figure 1. Matrix representation of ERG rearrangement and TMPRSS2-ERG mRNA expression status in multifocal PCa and corresponding LN mets.
The numbers in the cells indicated primary and secondary Gleason grade of each focus. Green highlighted cells indicate ERG rearrangement through insertion, orange indicates rearrangement accompanied by a deletion, and blue indicates no ERG rearrangement, as assessed by FISH seen in the representative pictures for this study. B. PCa and accompanying LN foci from 9 samples that have bold labels in A were analyzed for TMPRSS2 (exon 1)-ERG (exon 4) mRNA expression. The assay that we used provides transcript copy number detected per reaction. Copy numbers for TMPRSS2-ERG mRNA were standardized using PSA mRNA copy numbers from the same sample. “neg” indicates the sample yielded copy numbers within the range of quantification for PSA but yielded no detected copy numbers for TMPRSS2-ERG mRNA. “pos” indicates that we obtained copy numbers within or above the quantifiable range for both PSA and TMPRSS2-ERG mRNAs.
Fifteen out of the 26 cases (58%) (designated case #1 – 15) exhibited interfocal homogeneity with regard to ERG rearrangement status (9 cases did not display an ERG rearrangement in any focus (cases #1 – 9) and 6 cases had ERG rearrangement in all foci (cases #10 – 15)). Two out of the 26 cases (8%) (cases #19 and #26) revealed interfocal heterogeneity with regard to rearrangement mechanism. Eight out of the 26 cases (30%) (cases #16 – 18 and #20 – 24, and #26) revealed interfocal heterogeneity with regard to rearrangement status. One case (#25) was characterized by interfocal heterogeneity with regard to rearrangement status and interfocal heterogeneity with regard to rearrangement mechanism. The 15 cases with interfocal homogeneous ERG rearrangement status showed concordant ERG rearrangement status in the corresponding LN metastasis (p = 0.0002). In the cases that exhibited interfocal heterogeneity, the rearrangement status of the corresponding LN metastasis was defined by the rearrangement positive focus in the prostate gland (p = 0.003). In cases #19 and #25, the rearrangement status of the LN metastases were defined by the focus with rearrangement accompanied by deletion, whereas in case #26, the ERG rearrangement status of the LN metastases was defined by the focus with rearrangement through insertion.
In 3 out of 11 cases (cases #21, #24 and #26) that exhibited interfocal rearrangement status heterogeneity, the LN metastases Gleason score did not correspond to the highest Gleason score observed in the foci. In two of these cases (#21 and #26) and two other cases (#18 and #19), the LN metastasis did not resemble the rearrangement status or mechanism of the largest tumor focus. Taking these two observations together, we found that in 2 cases (#21 and #26), the ERG rearrangement status (and not the highest Gleason score or the largest diameter) was more prone to lead to metastatic dissemination to the LNs (Figure 2). Interestingly, when looking at the results on a focus by focus basis, there is no overall association between the presence of the rearrangement and Gleason score. However, 10 out of 11 foci in the prostate gland (cases #14-15, 19-21 and 25-26) with rearrangement through deletion have Gleason score 8 or above. None of the cases studied had evidence of polysomy of chromosome 21/polyploidy/amplification of the ERG locus, as far as this is assessable by the FISH assay applied.
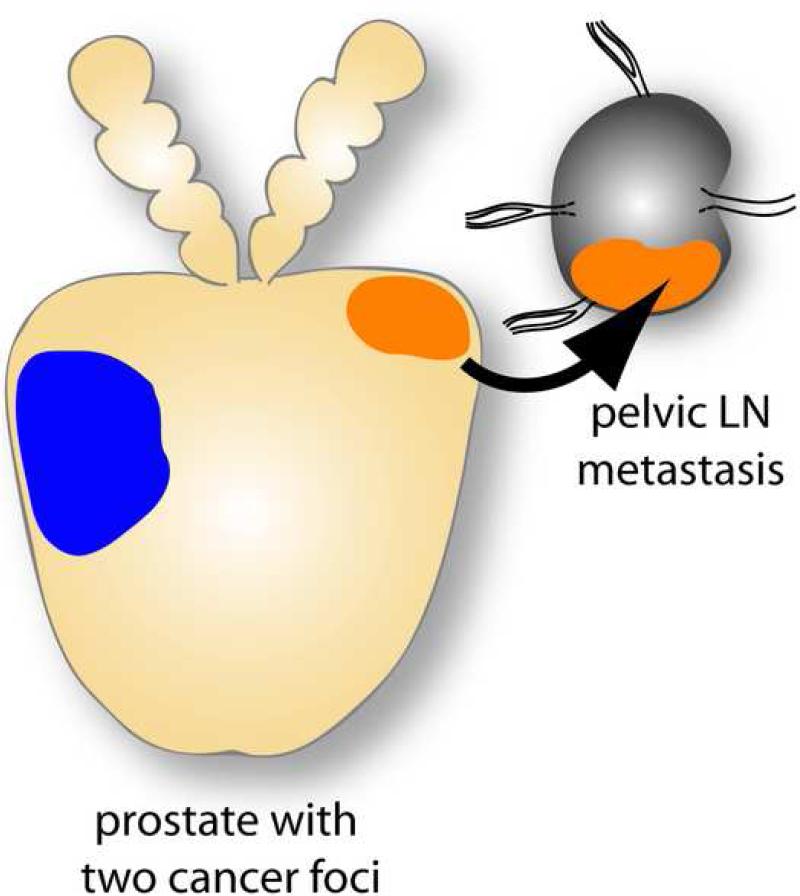
Figure 2. Illustration of how metastatic dissemination of multifocal PCa depends on ERG rearrangement status rather than tumor size or Gleason grade.
The blue focus represents the dominant focus with regard to size and Gleason grade but is negative for ERG rearrangement, while the secondary focus (with regard to size and Gleason grade) is positive for ERG rearrangement and drives regional metastatic dissemination.
When assessing for the agreement between TMPRSS2-ERG mRNA expression in the different PCa and metastatic LN foci, we were able to evaluate transcript copy numbers for PSA and TMPRSS2-ERGa (TMPRSS2-ERG type III isoform) in all the foci from 9 cases (Figure 1B) All foci that were negative for rearrangement based on the ERG b/a FISH assay were also negative for TMPRSS2-ERG mRNA expression. Two foci from case #22 (the primary and tertiary foci) showed ERG b/a rearrangement by the FISH b/a assay but did not yield detectable levels of TMPRSS2-ERG mRNA expression. To investigate if these two foci are characterized by rarer TMRPSS2-ERG isoforms, we first assessed if TMPRSS2 was rearranged by FISH. Both foci were negative for TMPRSS2 rearrangement, which suggests another fusion partner for ERG and not some other less prevalent isoform. We also failed to identify rearrangements for two other known 5’ fusion partners of ERG, SLC45A3 (6) and NDRG1 (23). Therefore the 5’ prime fusion partner of this ERG rearranged case remains unknown.
Discussion
The clinical practice of pathology has widely assumed that in PCa with multiple tumor nodules, the tumors with the largest diameter nodules and/ or the highest Gleason score represent the dominant nodule. The dominant nodule would also be typically used in molecular studies for the development of biomarkers to predict outcome. This current study, using ERG rearrangement as a clonal marker, suggests that the dominant nodule is not determinable by these two clinical parameters. In fact, we did not observe a consistent association between the largest tumor diameter or the tumor nodule with the highest Gleason score and metastasis. Although ERG negative tumors are capable of metastasis we found that cases exhibiting interfocal rearrangement heterogeneity the nodule with the ERG rearrangement is more prone to clonal metastatic dissemination suggesting that these cancer cells may represent a more aggressive subgroup (Figure 1A).
In addition to assessing the genomic ERG rearrangement we were also able to assess most prevalent TMPRSS2-ERG mRNA variant. Frozen material is preferred for TMPRSS2-ERG transcripts, no such material was available for this cohort. Due to this limitation we were used the available FFPE material and could confirm, in a subset of cases, that the majority of ERG rearranged cases results in the expression of TMPRSS2-ERG mRNA (Figure 1B). Interestingly, we found that all foci that were negative for rearrangement based on the ERG b/a FISH assay were also negative for the most frequent TMPRSS2-ERG isoform. However, we found 2 foci that were positive for ERG rearrangement but negative for TMPRSS2-ERG mRNA. After further investigating these two foci, we assessed that they were negative for TMPRSS2 rearrangement, thus unlikely to express a less frequent type of TMRPSS2-ERG isoform, and negative for SLC45A3-ERG and NDRG1-ERG rearrangement – two recently identified, less prevalent 5’ fusion partners of ERG. This suggests that ERG has other, yet to be determined, fusion partners. When assessing PCa for the TMPRSS2-ERG gene fusion, we believe that a combination of assessment on the genomic as well as the transcriptional level might be ideal in order to test for other 5’ partners of ERG or for rare TMPRSS2-ERG fusion transcripts. However, the impact of other, rarer gene fusions or TMPRSS2-ERG fusion transcripts still needs to be elucidated.
Although the limited number of samples that were assessed for TMPRSS2-ERG mRNA did not allow for a statistical evaluation, we found that the LN foci yielded higher relative amounts of TMPRSS2-ERG mRNA than the PCa foci (data not shown). While we could show that the ERG rearrangement status on the DNA level is a marker of clonal expansion, it might be that the increment of TMPRSS2-ERG mRNA expression in the LN metastasis compared to the primary PCa may be an indicator of disease progression. Paris et al. reported a similar finding in PCa where matched primaries and LN metastases showed similar copy number profiles that are distinct from primary tumors that fail to metastasize (24). We hypothesize that alterations at the DNA level are markers of clonal expansion, while changes on the transcript level may reflect disease progression.
This study confirms our earlier observation that there is concordance between rearrangement status in matched primary PCa and hormone-naïve LN metastasis (19). Specifically, it links the existence of interfocal fusion status heterogeneity in multifocal PCa (10, 15, 16) and the observation that multiple androgen-independent metastatic PCa sites from an individual case all harbor the same gene fusion status (10). In addition, this study confirms the early observation that the ERG rearrangement is an early clonal event (2).
Our current study is the first to assess for ERG rearrangement status in multifocal primary PCa and matched hormone-naïve LN metastasis. Although we had a limited sample size, our findings clearly suggest that localized PCa harboring the ERG rearrangement can result in metastatic spread to regional lymph nodes. This has potential clinical impact on disease progression but will need to be confirmed in other larger studies. The mechanism behind the selective metastatic potential associated with ERG rearrangement needs to be further investigated and is out of the scope of this study, however several reports have demonstrated a critical role of ERG over expression in cell migration and invasion (25-28). Another intriguing finding that needs confirmation is that 10/11 foci in the prostate that harbor ERG rearrangement through deletion had a Gleason score of 8 or above. This could be an indicative of a more aggressive subtype of disease. In agreement with a recent report by Attard et al. (29), this study provides clear evidence that ERG rearrangement is a marker of clonal expansion and does not reflect an increase in genetic alterations in metastatic disease (30).
PCa is a clinically heterogeneous disease. The presence of multiple discrete tumor clones in each gland harboring PCa may be an explanation for this well known clinical observation. Due to current biopsy sampling approaches, it is possible that the molecularly dominant nodule may be missed. Emerging approaches using urine to screen for molecular alterations in PCa promise to help ameliorate this process. Assays to detect TMPRSS2-ERG fusion transcripts can reliably determine if one harbors a gene fusion within the gland (31). Multiplex-PCR screening urine assays to detect gene fusions and other biomarkers may complement current biopsy strategies to assess the risk of PCa disease progression.
Acknowledgement
We thank Christopher LaFargue for his excellent technical support with the FISH experiments and Himisha Beltran for helpful comments.
Funding:
Department of Defense Grant to S.P. (PC61474) and NCI Grant to FD and MAR (R01CA125612).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–8. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 3.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–44. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 4.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG Fusion by Translocation or Interstitial Deletion Is Highly Relevant in Androgen-Dependent Prostate Cancer, But Is Bypassed in Late-Stage Androgen Receptor-Negative Prostate Cancer. Cancer Res. 2006;66:10658–63. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 6.Han B, Mehra R, Dhanasekaran SM, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–37. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 8.Attard G, Clark J, Ambroisine L, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–63. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–6. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–90. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greene DR, Wheeler TM, Egawa S, Dunn JK, Scardino PT. A comparison of the morphological features of cancer arising in the transition zone and in the peripheral zone of the prostate. J Urol. 1991;146:1069–76. doi: 10.1016/s0022-5347(17)38003-5. [DOI] [PubMed] [Google Scholar]
- 12.Ruijter ET, van de Kaa CA, Schalken JA, Debruyne FM, Ruiter DJ. Histological grade heterogeneity in multifocal prostate cancer. Biological and clinical implications. J Pathol. 1996;180:295–9. doi: 10.1002/(SICI)1096-9896(199611)180:3<295::AID-PATH663>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 13.Arora R, Koch MO, Eble JN, Ulbright TM, Li L, Cheng L. Heterogeneity of Gleason grade in multifocal adenocarcinoma of the prostate. Cancer. 2004;100:2362–6. doi: 10.1002/cncr.20243. [DOI] [PubMed] [Google Scholar]
- 14.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 Gene Rearrangements in Multifocal Prostate Adenocarcinoma: Molecular Evidence for an Independent Group of Diseases. Cancer Res. 2007;67:7991–5. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 15.Clark J, Attard G, Jhavar S, et al. Complex patterns of ETS gene alteration arise during cancer development in the human prostate. Oncogene. 2008;27:1993–2003. doi: 10.1038/sj.onc.1210843. [DOI] [PubMed] [Google Scholar]
- 16.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70:630–3. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofer MD, Kuefer R, Huang W, et al. Prognostic factors in lymph node-positive prostate cancer. Urology. 2006;67:1016–21. doi: 10.1016/j.urology.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 18.Cheng L, Song SY, Pretlow TG, et al. Evidence of independent origin of multiple tumors from patients with prostate cancer. J Natl Cancer Inst. 1998;90:233–7. doi: 10.1093/jnci/90.3.233. [DOI] [PubMed] [Google Scholar]
- 19.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 20.Setlur SR, Mertz KD, Hoshida Y, et al. Estrogen-Dependent Signaling in a Molecularly Distinct Subclass of Aggressive Prostate Cancer. J Natl Cancer Inst. 2008;100:815–25. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groskopf J, Aubin SM, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006;52:1089–95. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Cai Y, Ren C, Ittmann M. Expression of Variant TMPRSS2/ERG Fusion Messenger RNAs Is Associated with Aggressive Prostate Cancer. Cancer Res. 2006;66:8347–51. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- 23.Pflueger D, Rickman DS, Sboner A, et al. Transcriptome Sequencing Identifies NDRG1-ERG Fusion Prostate Cancer. 2009. Submitted.
- 24.Paris PL, Hofer MD, Albo G, et al. Genomic profiling of hormone-naive lymph node metastases in patients with prostate cancer. Neoplasia. 2006;8:1083–9. doi: 10.1593/neo.06421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klezovitch O, Risk M, Coleman I, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–10. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attard G, Swennenhuis JF, Olmos D, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69:2912–8. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 30.Rubin MA, Gerstein A, Reid K, et al. 10q23.3 loss of heterozygosity is higher in lymph node-positive (pT2-3,N+) versus lymph node-negative (pT2-3,N0) prostate cancer. Hum Pathol. 2000;31:504–8. doi: 10.1053/hp.2000.6713. [DOI] [PubMed] [Google Scholar]
- 31.Laxman B, Morris DS, Yu J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–9. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]