Summary
The specific molecular events that characterize the intrinsic apoptosis pathway have been the subject of intense research due to its fundamental role in development, homeostasis, and cancer. This pathway is defined by the release of cytochrome c from mitochondria into the cytosol, and subsequent binding of cytochrome c to the caspase activator Apaf-1. Here we report that both mitochondrial and cytosolic transfer RNA (tRNA) bind to cytochrome c. This binding prevents cytochrome c interaction with Apaf-1, blocking Apaf-1 oligomerization and caspase activation. tRNA hydrolysis in living cells and cell lysates enhances apoptosis and caspase activation, while microinjection of tRNA into living cells blocks apoptosis. These findings suggest that tRNA, in addition to its well-established role in gene expression, may determine cellular responsiveness to apoptotic stimuli.
Introduction
Apoptosis is essential to development, homeostasis, and the prevention of viral infection and cancer (Thompson, 1995; Vaux and Korsmeyer, 1999). This form of programmed cell death is executed by caspases and characterized by sterotypic morphological and biochemical changes (Chang and Yang, 2000; Li and Yuan, 2008; Riedl and Shi, 2004). Two major apoptosis pathways exist in mammalian cells, one triggered by cell-intrinsic stimuli and the other by extrinsic stimuli. The intrinsic pathway is activated by many cues, including developmental lineage information, oncogene activation, DNA damage, and nutrient deprivation. This pathway is defined by the release of mitochondrial cytochrome c into the cytosol. Once in the cytosol, cytochrome c binds to Apaf-1, which enables Apaf-1 to assemble into the oligomeric apoptosome complex. The apoptosome then recruits and oligomerizes the precursor of an initiator caspase, caspase-9, leading to its auto-proteolytic activation. Caspase-9 activates effector caspases such as caspase-3 and −7, which cleave various cellular proteins leading to cell death (Riedl and Salvesen, 2007; Wang, 2001) (Figure S1). Cytochrome c-induced caspase-9 activation is regulated at multiple levels involving various protein factors, cations such as potassium and calcium, and perhaps most notably, nucleotides (Riedl and Salvesen, 2007; Schafer and Kornbluth, 2006). ATP or dATP is required for the assembly of the apoptosome (Chandra et al., 2006; Kim et al., 2005; Liu et al., 1996; Riedl et al., 2005). However, high levels of nucleotides can also block caspase-9 activation by inhibiting cytochrome c (Chandra et al., 2006). While the involvement of nucleotides in cytochrome c-induced caspase activation is well established, there has been no reported role of polymer ribonucleotide (RNA) in this process.
Transfer RNA (tRNA) is a major RNA in cells, and it has a fundamental role in protein synthesis by linking a genetic codon with an amino acid (Hopper and Shaheen, 2008; Ibba et al., 2000; Weisblum, 1999). In eukaryotes, tRNA is encoded in nuclear, mitochondrial, and plastidial genomes. Each tRNA of a given specificity is bound or charged to its amino acid by a specific aminoacyl-tRNA transferase. A charged tRNA delivers an activated amino acid to the ribosome for polypeptide synthesis. tRNA also serves as a primer for the reverse transcription of RNA genomes, including those from HIV. Furthermore, uncharged tRNA, which is accumulated upon amino acid deficiency, induces expression of genes for amino acid synthesis (Wek et al., 1989). In mammalian cells, tRNA and 5S rRNA, with a few other small RNAs, are transcribed by RNA polymerase (pol) III. tRNA expression is enhanced in tumor cells and rapidly dividing normal cells (Ruggero and Pandolfi, 2003; White, 2005). A recent study showed that an elevation in the level of a specific tRNA could lead to cellular transformation, suggesting a causal effect of tRNA expression on tumorigenesis (Marshall et al., 2008). Despite recent advances, it is still unclear whether high levels of tRNA provide advantages to tumor cells beyond meeting the demand for protein synthesis.
In this current study, we show that tRNA binds to cytochrome c and impairs the association of cytochrome c with Apaf-1, blocking the formation of the apoptosome and the subsequent activation of caspase-9. These findings reveal a direct role for tRNA in promoting cell survival and demonstrate a function beyond tRNA’s well-established role in gene expression. The findings also suggest that tumor cells may rely on high levels of tRNA for apoptosis resistance and support the notion that tRNA is a valuable target for tumor therapy.
Results
RNA hydrolysis enhances cytochrome c-induced caspase-9 activation
Cytochrome c-induced caspase-9 activation is regulated by nucleotides (Chandra et al., 2006; Kim et al., 2005; Liu et al., 1996; Riedl et al., 2005). This observation led us to explore the possibility that RNA could have a role in caspase-9 activation. We first tested the effect of RNA hydrolysis on caspase-9 activation in cell extracts. The addition of cytochrome c to S100 extracts from cell lines such as HeLa and Jurkat results in the auto-activation of procaspase-9, generating the mature p37/p35 and p10 subunits. The effector procaspase-3 is then processed to the mature p20/p17 and p12 subunits (Figures 1A and S2). Interestingly, when HeLa S100 extracts were pre-treated with increasing amounts of RNase A, cytochrome c-induced caspase-9 activation was progressively enhanced (Figures 1A). This stimulatory effect of RNase A correlated with the degradation of cellular RNA in the S100 extracts (Figure 1B) and was abolished by an RNase inhibitor (Figure 1C), confirming that the enhanced caspase-9 activation was due to the catalytic activity of RNase A. In an analogous system, where in vitro-translated procaspase-9 was activated by cytochrome c (Liu et al., 2005), treatment with RNase A enhanced caspase-9 activation (Figure 1D).
Figure 1. RNA hydrolysis enhances cytochrome c-induced caspase activation.
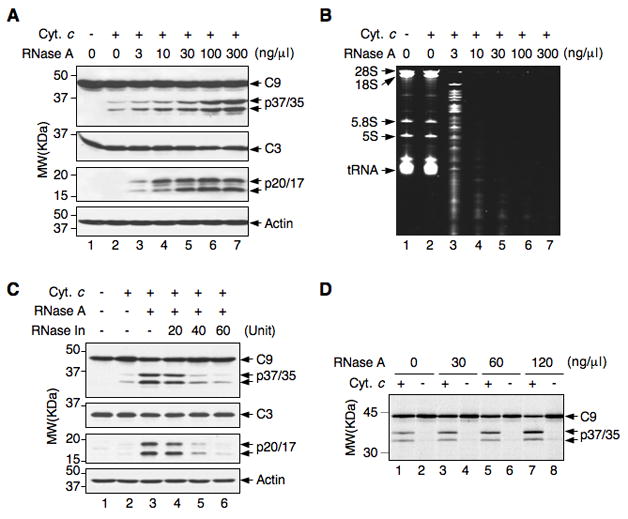
(A and B) HeLa S100 extracts were incubated with cytochrome c (20 μg/ml) in the presence and absence (−) of increasing amounts of RNase A. (A) Extracts were analyzed by Western blot. Molecular weight (MW) markers (in kilo Daltons) are indicated on the left. C9, procaspase-9; C3, procaspase-3. The levels of actin are shown as a loading control. (B) RNA was separated via PAGE under denaturing conditions and stained with ethidium bromide.
(C) Activation of caspase-9 and −3 in HeLa S100 extracts treated with cytochrome c (20 μg/ml), RNase A (30 ng/ml), and RNase inhibitor (In) as indicated.
(D) Reticulocyte lysates containing in vitro-translated, 35S-labeled procaspase-9 were treated with and without cytochrome c (200 μg/ml) in the presence of the indicated concentration of RNase A. Caspase-9 was detected by autoradiography.
Conversely, when exogenous cellular RNA was added to Jurkat S100 extracts, it reduced cytochrome c-induced caspase activation in a dose-dependent manner (Figure 2A). To assess whether cellular RNA directly inhibits caspase-9 activation or indirectly via other cellular factors, we used a reconstituted system. When binding to cytochrome c in the presence of dATP, purified full-length Apaf-1 forms the apoptosome and activates caspase-9 (Zou et al., 1999) (Figure 2B, lane 5). When exogenous total RNA was included in this system, it strongly inhibited Apaf-1-induced caspase-9 processing at low doses, and completely blocked caspase-9 processing at a higher dose (lanes 6–8). This result suggests that RNA may exert its inhibitory effect directly on cytochrome c, Apaf-1, and/or caspase-9.
Figure 2. Total cellular RNA inhibits caspase activation in cell extracts and in a reconstituted system.
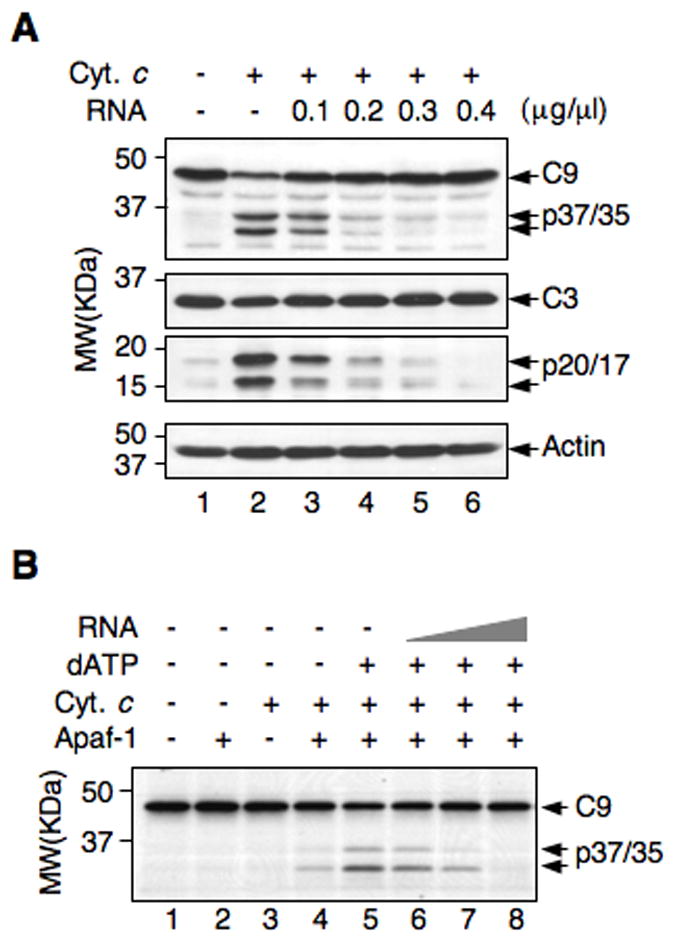
(A) Activation of caspase-9 and −3 in Jurkat S100 extracts treated with cytochrome c (20 μg/ml) alone and in combination with the indicated amounts of RNA.
(B) In vitro-translated, 35S-labeled procaspase-9 was incubated with purified full-length Apaf-1 (10 nM), dATP (1mM), cytochrome c (20 μg/ml), and increasing amounts of total cellular RNA (0.1, 0.2, and 0.4 μg/μl). Caspase-9 was detected by autoradiography.
RNA impairs the cytochrome c:Apaf-1 interaction and prevents the formation of the apoptosome
To investigate the mechanism by which cellular RNA inhibits caspase-9 activation, we first analyzed the effect of RNA on Apaf-1 oligomerization using gel filtration chromatography. In un-stimulated Jurkat S100 extracts, Apaf-1 was eluted as a monomer (~160 kDa), and caspase-9 was not activated (Figure 3A, top two panels). As expected, in extracts treated with cytochrome c, a large portion of the Apaf-1 protein became part of the oligomeric apoptosome (~700 kDa). At the same time, processed procaspase-9 was present in both the apoptosome-bound form and the released form (Figure 3A, middle two panels). In contrast, when purified total cellular RNA was added with cytochrome c to the Jurkat S100 extracts, Apaf-1 could no longer oligomerize, and caspase-9 activation was completely blocked (Figure 3A, bottom two panels). Therefore, RNA prevents the oligomerization of Apaf-1.
Figure 3. RNA interferes with cytochrome c:Apaf-1 interaction and inhibits apoptosome formation.
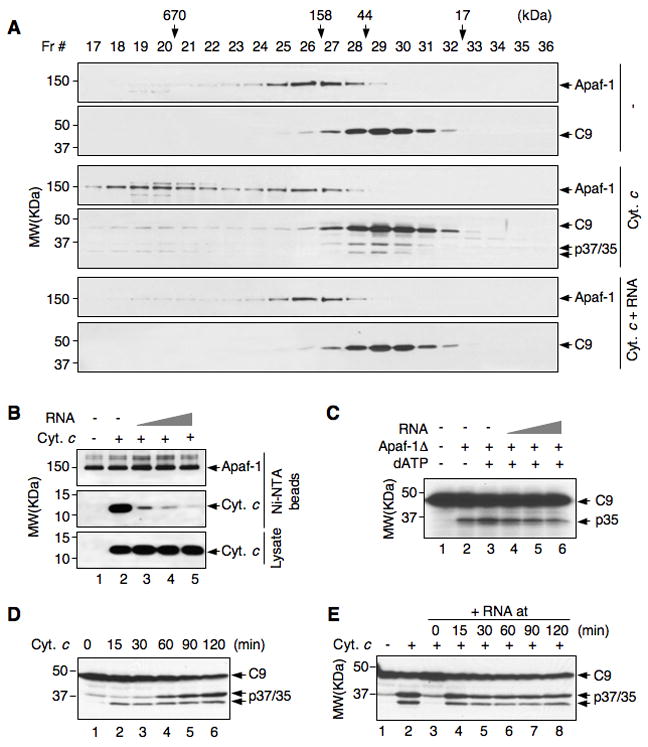
(A) Jurkat S100 extracts were incubated alone (−), with cytochrome c, and with cytochrome c plus RNA at 25 °C for 15 min. Extracts were fractionated on a Superose 6 gel filtration column. The fractions were analyzed by Western blot. The positions of molecular weight standards for the column are marked at the top.
(B) His-tagged Apaf-1 bound to Ni-NTA beads was incubated with and without cytochrome c, and with cytochrome c plus increasing amounts of RNA (0.1, 0.2, and 0.4 μg/μl). Bead-bound proteins and one percent of the input were analyzed by Western blot.
(C) In vitro-translated, 35S-labeled procaspase-9 was incubated with purified Apaf-1Δ, dATP, and increasing amount of RNA as indicated, at 30 °C for 1 h.
(D) Jurkat S100 extracts were incubated with cytochrome c for the indicated durations before being analyzed for caspase-9 activation.
(E) Jurkat S100 extracts were incubated with and without cytochrome c (lanes 1 and 2), and were pre-incubated with cytochrome c before the addition of tRNA at the indicated time (lanes 3–8). The total reaction time for each sample was 2 h. Activation of caspase-9 was analyzed by Western blot.
Next, we determined whether total RNA interferes with the binding of cytochrome c to Apaf-1. Immobilized recombinant full-length Apaf-1 readily pulled down cytochrome c in the solution. However, in the presence of total RNA, the amount of cytochrome c that bound to Apaf-1 was drastically decreased (Figure 3B), suggesting that RNA prevents the binding of cytochrome c to Apaf-1. We also examined whether RNA has any additional effect on Apaf-1 once Apaf-1 is oligomerized. To this end, we used the recombinant Apaf-1Δ protein (containing amino acids 1–591), which retains the caspase-9-binding and oligomerization domains, but lacks the negative regulatory WD40 repeats that bind to cytochrome c (Riedl et al., 2005). Apaf-1Δ spontaneously forms homo-oligomers and activates caspase-9 independently of cytochrome c (Riedl et al., 2005; Srinivasula et al., 1998). As shown in Figure 3C, total RNA had a minimal effect on Apaf-1Δ-induced caspase-9 activation. To confirm this observation, we treated Jurkat S100 extracts with cytochrome c for different lengths of time and then added RNA. Treatment with cytochrome c for as little as 15 minutes rendered RNA entirely ineffective in preventing caspase-9 activation (Figures 3D and 3E). Therefore, RNA inhibits the interaction of cytochrome c with Apaf-1, but not the subsequent Apaf-1-involved events in caspase-9 activation.
Cytochrome c binds to tRNA both in vivo and in vitro and inhibits caspase-9 activation
The above observation suggests that the target of RNA may be cytochrome c. To test this possibility, we used a gel filtration assay to compare the size of cytochrome c in the presence and absence of total cellular RNA. Cytochrome c alone was eluted as a monomer. However, when incubated with total cellular RNA, it formed higher molecular weight complexes (Figure 4A), suggesting that cytochrome c can directly associate with one or more cellular RNA.
Figure 4. Interaction of cytochrome c with tRNA both in vivo and in vitro and the effect of tRNA on caspase-9 activation.
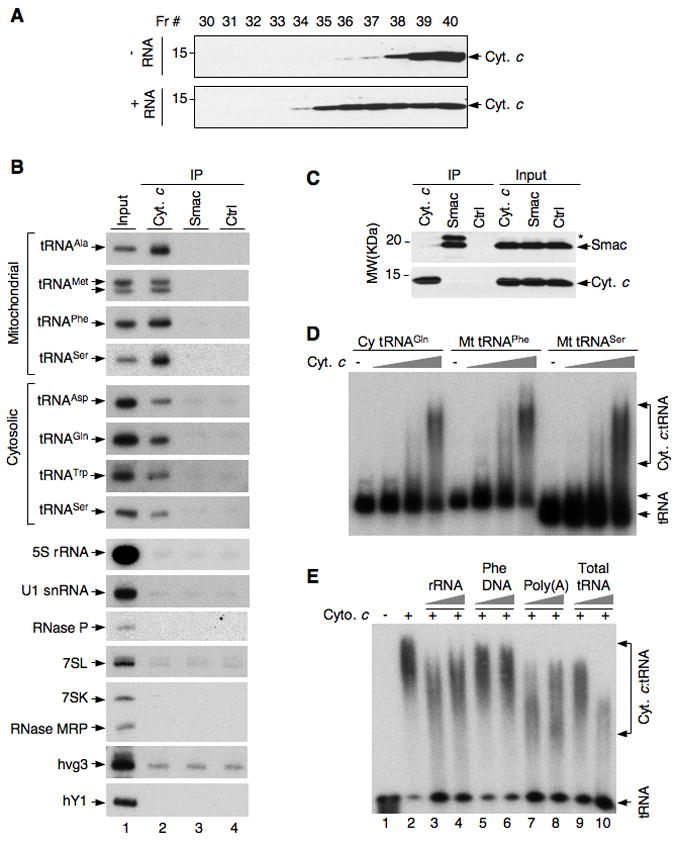
(A) Recombinant cytochrome c was incubated with and without total RNA at 25 °C for 15 min. The reaction mixture was loaded on a Superose 6 column and fractions were analyzed by Western blot.
(B and C) HeLa cells were treated with formaldehyde, and cell lysates were made in Empigen BB-containing buffer and immunoprecipitated with the indicated antibodies.
(B) RNA in the immunoprecipitates was analyzed by Northern blotting using radiolabeled probes for the indicated RNAs. hvg3, human vault RNA; hY1, human Y1 RNA. Input samples contained ~ 1% of the RNA used for IP. (C) Cytochrome c and Smac in immunoprecipitates and ~1.5% of the input were analyzed by Western blot. *, Smac precursor.
(D) In vitro synthesized, [32P]UTP-labeled cytosolic (Cy) and mitochondrial (Mt) tRNAs were incubated with increasing amounts (0.5, 2.5, 12.5 μM) of cytochrome c for 45 min at 30 °C. Mixtures were then incubated with 0.5 M Urea (final concentration) for 10 min and analyzed by 6% native gel electrophoresis and autoradiography. Cytochrome c:tRNA complexes are indicated.
(E) Radiolabeled total cellular tRNA labeled (12 fmole) was incubated with and without cytochrome c (100 pmol), and with cytochrome c in the presence of non-labeled rRNA, poly(A), total tRNA (50 and 250 ng), and mitochondrial Phe DNA (130 and 520 ng). The mixtures were analyzed by native gel electrophoresis and autoradiography.
To determine whether cytochrome c specifically interacts with RNA in cells, and which RNA species it possibly interacts with, we used a stringent immunoprecipitation assay. HeLa cells were first treated with low concentrations of formaldehyde (0.2%) to cross-link specific RNA-protein complexes while avoiding non-specific linkage (Niranjanakumari et al., 2002). Cells were then lysed in a buffer containing the zwitterionic detergent Empigen BB (Choi and Dreyfuss, 1984). The lysates were subjected to immunoprecipitation with an anti-cytochrome c antibody and, as controls, with an isotype-matching unrelated antibody, an anti-Apaf-1 antibody, and an antibody against Smac/DIABLO. The latter, like cytochrome c, resides in the inter-membrane space of mitochondria and is released during apoptosis to participate in caspase activation (Riedl and Salvesen, 2007; Wang, 2001). Initial analyses demonstrated that RNAs around 70–90 nucleotides were enriched in anti-cytochrome c immunoprecipitates but not in anti-Apaf-1, anti-Smac, and control antibody immunoprecipitates (data not shown). Because the size of the RNA species corresponded to that of tRNA, we performed Northern blot analyses using specific probes for various tRNAs, including four mitochondrial and four cytosolic tRNAs. As controls, probes detecting an additional eight RNA species, including several small and structured RNAs (e.g. 5S rRNA and U1snRNA), were also used. Cytochrome c was associated with all four mitochondrial tRNAs in vivo. It was also associated with the four cytosolic tRNAs, although to a lesser extent (Figures 4B and 4C). The specificity of tRNA:cytochrome c interaction is underscored by the absence of tRNA in anti-Smac and control antibody immunoprecipitates and by the lack of binding of cytochrome c to any of the eight control RNAs (Figure 4B).
To determine whether cytochrome c directly binds to tRNA, mitochondrial tRNAPhe and tRNASer and cytosolic tRNAGln were in vitro-transcribed and -radiolabeled. These tRNAs were incubated with and without cytochrome c, and then analyzed by an electrophoretic mobility shift assay (EMSA). The mobility of each of these RNAs was slowed in the presence of cytochrome c (Figure 4D), indicating that cytochrome c directly binds to all of the RNAs. To further characterize the cytochrome c:tRNA interaction, we analyzed the binding of cytochrome c to total tRNA isolated from cells. The mobility of the vast majority of the labeled tRNAs was slowed by cytochrome c (Figure 4E), suggests that cytochrome c may bind to virtually all tRNAs. The binding of cytochrome c to radiolabeled tRNA could be inhibited almost completely by non-labeled total tRNA (lane 10). In contrast, this binding was only partially impeded by non-labeled rRNA and poly(A), and was not affected at all by DNA (lanes 3–8). These results suggest that in vitro cytochrome c can directly binds to RNA but not DNA, and it preferentially binds to tRNA.
To determine the effect of tRNA on caspase-9 activation, we added purified cellular tRNA with cytochrome c to Jurkat S100 extracts. tRNA blocked cytochrome c-induced caspase-9 activation as potently as total RNA. In contrast, neither mRNA nor rRNA exhibited significant inhibitory effect (Figure 5A). Moreover, tRNA inhibited cytochrome c-induced apoptosome formation (Figure 5B). We also compared the minimal concentrations of cytochrome c that could initiate caspase-9 activation in the absence of tRNA and in the presence of two different amounts of tRNA. In the absence of tRNA, cytochrome c induced activation of caspase-9 at a concentration as low as 2 μg/ml. In the presence of 0.1 and 0.2 μg/μl tRNA, the minimal amount of cytochrome c required to induce caspase-9 activation increased to 10 and 20 μg/ml, respectively (Figure 5C). Thus, tRNA sets a threshold for cytochrome c-induced caspase activation.
Figure 5. tRNA block apoptosome formation and caspase-9 activation.
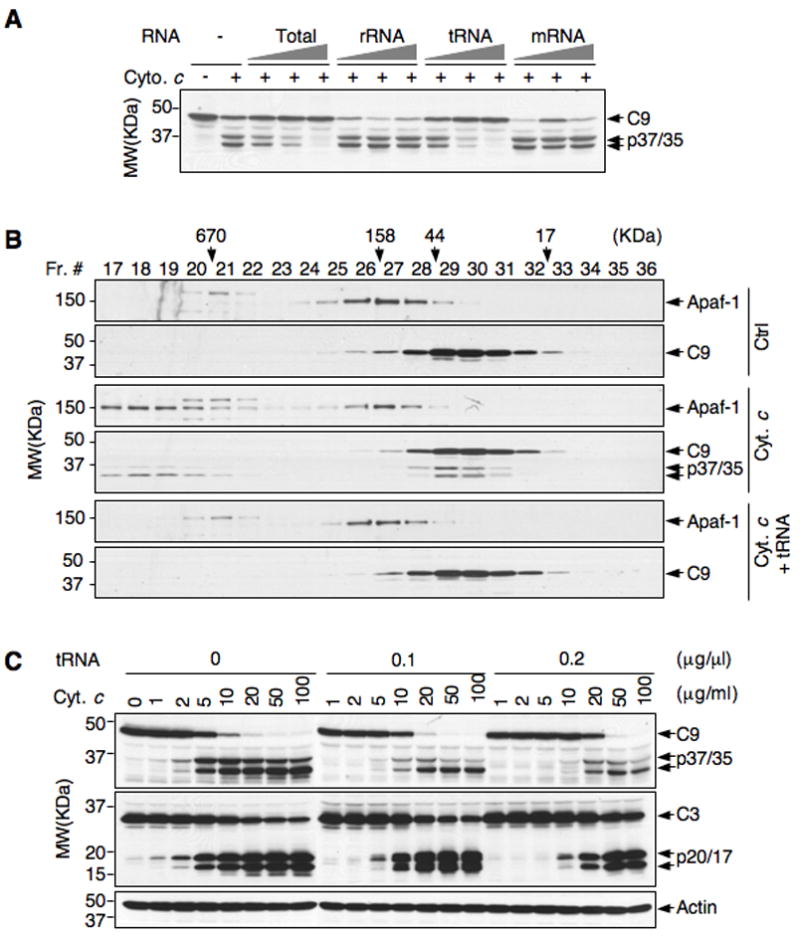
(A) Jurkat S100 extracts were incubated with cytochrome c and increasing amounts of total RNA, rRNA, tRNA (0.1, 0.2, and 0.4 μg/μl), and mRNA (0.02, 0.04, 0.08 μg/μl) at 37 °C for 1 h. Less amounts of mRNA were used because in cells it is expressed at lower levels compared with either tRNA or rRNA. Activation of caspase-9 was analyzed by Western blot.
(B) Jurkat S100 extracts were incubated with and without (ctrl) cytochrome c, and with cytochrome c plus tRNA. The extracts were resolved on Superose 6 gel filtration column, and fractions were analyzed by Western blot.
(C) Jurkat S100 extracts were incubated with increasing concentrations of cytochrome c in the absence and presence of different concentrations of tRNA for 1 h. Activation of caspase-9 and −3 was analyzed by Western blot.
Microinjection of tRNA into cells inhibits cytochrome c-induced apoptosis
To assess the role of tRNA in cytochrome c-induced apoptosis, cytochrome c was microinjected into HEK293 cells alone and together with tRNA. Microinjection of cytochrome c in HEK293 cells led to apoptosis in a significant number of injected cells, as determined by apoptosis-associated morphological changes, such as membrane blebbing and nuclear fragmentation (Figures 6A, panels d-f and Figure 6B, column 2). Pre-treatment of cells with the pan-caspase inhibitor zVAD-fmk nearly blocked apoptosis completely, indicating that cell death was mediated by caspases. Microinjection of tRNA alone did not induce apoptosis. However, when tRNA was co-injected with cytochrome c, apoptosis among injected cells noticeably decreased. Furthermore, in a similar co-injection experiment, rRNA had a minimal effect on cytochrome c-induced apoptosis (Figures 6A and 6B). Therefore, cytochrome c-induced apoptosis is specifically inhibited by tRNA.
Figure 6. Microinjection of tRNA blocks cytochrome c-induced apoptosis.
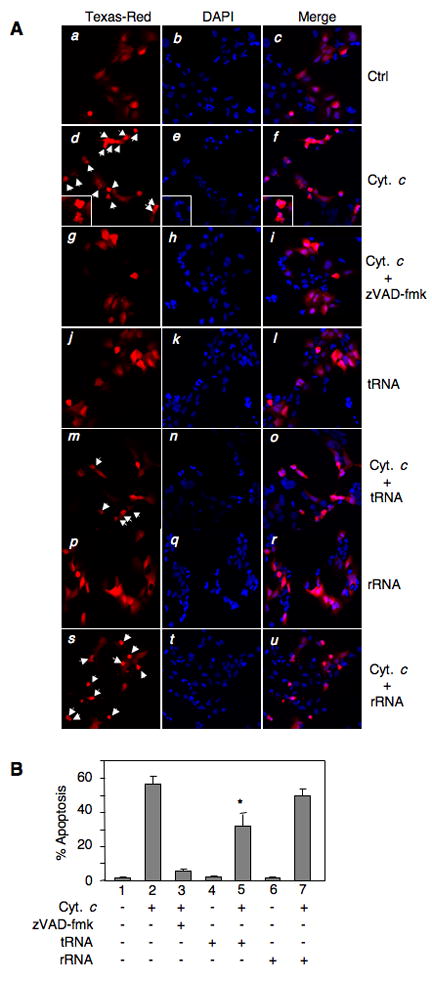
HEK 293 cells were injected with buffer (ctrl) and the indicated combinations of cytochrome c, zVAD-fmk, tRNA, and rRNA. Dextran-Texas Red (Dex-TR) was included in each injection to label the injected cells. Cells were fixed and stained with DAPI 2 h after injection. (A) Representative images of injected cells. Arrowheads indicate apoptotic cells. Larger images of apoptotic cells are shown in panels d–f. (B) Percentage of apoptosis among injected cells. Data presented are the mean ± the standard deviation (SD) of three independent experiments.
Degradation of cellular tRNA enhances apoptosis via the intrinsic pathway
To investigate the effect of tRNA hydrolysis on caspase-9 activation and apoptosis in cells, we used the tRNA-specific ribonuclease onconase (also known as ranpirnase) (Ardelt et al., 2008; Costanzi et al., 2005). Consistent with previous observations (Iordanov et al., 2000; Saxena et al., 2002), transfection of onconase into HeLa cells resulted in the degradation of tRNA, but not various rRNAs (Figure 7A, left panel). The degradation of tRNA was closely followed by the activation of caspase-9 and −3 and the cleavage of the apoptotic substrate PARP (Figure 7A, right panels). This suggests that tRNA degradation could determine onconase-induced apoptosis. In HeLa S100 extracts, treatment with onconase also enhanced cytochrome c-induced caspase activation in a dosage-dependent manner (Figure S3A). To confirm that onconase-induced apoptosis is dependent on Apaf-1, we treated wild type and Apaf-1-decifient (Apaf-1−/−) mouse embryonic fibroblasts (MEFs). Indeed, onconase killed wild type but not Apaf-1−/− MEFs (Figure 7B).
Figure 7. Degradation of tRNA enhances apoptosis via the intrinsic pathway.
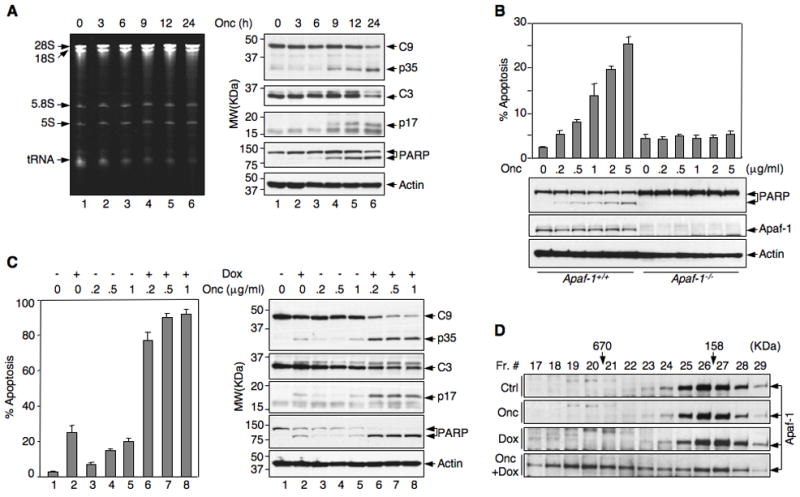
(A) HeLa cells were transfected with 1 μg/ml onconase and cultured for the indicated periods of time. Left: total RNA was separated by 8% urea-containing PAGE and visualized by ethidium bromide staining. Right: activation of caspase-9 and caspase-3, and the cleavage of PARP were analyzed by Western blot.
(B) Apaf-1+/+ and Apaf-1−/− MEF cells were treated with indicated amounts of onconase for 24 h. Top: percentages of apoptosis are shown as mean ± SD of three independent experiments. Bottom: Apaf-1 expression and PARP cleavage in cell lysates.
(C) HeLa cells were transfected with an indicated amount of onconase. 3 h post-transfection, the cells were incubated with and without doxorubicin (Dox, 1μg/ml) for an additional 12 h. Left: percentages of apoptosis. The data represent mean ± SD of three independent experiments. Right: the activation of caspase-9 and −3 and the cleavage of PARP.
(D) HeLa cells were transfected with and without onconase (1 μg/ml). 3 h after transfection, cells were treated with Dox (1 μg/ml) for another 12 h. S100 extracts were fractionated on a Superose 6 gel filtration column, and fractions were analyzed by Western blot.
If onconase promotes apoptosis by reversing the inhibitory effect of tRNA on cytochrome c, it should enhance cell death stimulated by agents that invoke the release of cytochrome c. We tested this possibility by examining the individual and combined effect of onconase and the genotoxic drug doxorubicin on apoptosis induction. At dosages that alone caused substantial tRNA degradation but only minimal levels of caspase activation and apoptosis, onconase promoted doxorubicin-induced caspase activation and enhanced apoptosis from ~30% to over 95% (Figures 7C and S3B). A gel-filtration analysis showed that onconase and doxorubicin together, but not individually, triggered the assembly of Apaf-1 into the apoptosome (Figure 7D). These results indicate that degradation of cellular tRNA enhances apoptosis via the intrinsic pathway.
Discussion
For the past 50 years, tRNA has been studied almost exclusively in the context of the flow of genetic information as the adaptor between genetic codons and amino acids in protein translation and a primer for reverse transcription (Hopper and Shaheen, 2008; Ibba et al., 2000; Weisblum, 1999). Our data provides evidence that tRNA has a previously unexpected role beyond gene expression. By binding to cytochrome c and preventing its association with Apaf-1, tRNA directly inhibits apoptosis. This finding reveals an intimate connection between tRNA and cytochrome c, linking the protein translation pathway to cell death, and may have direct implications in cancer therapy.
Mitochondria are known to contain several other apoptosis inducers in addition to cytochrome c, such as Smac/Diablo, EndoG, and AIF (Riedl and Salvesen, 2007; Wang, 2001). The interaction of mitochondrial tRNA with cytochrome c (Figure 4) suggests that mitochondria harbor at least one antidote of apoptosis, and it might have reflected the need to tightly regulate cytochrome c when it first acquired such a destructive power. Cytosolic tRNA is also capable of associating with cytochrome c in healthy cells. This surprising finding is nevertheless consistent with a recent report that mammalian cytosolic tRNA can be efficiently imported into mitochondria (Rubio et al., 2008). Unlike mitochondria in some other organisms that rely on at least some cytosolic tRNAs for protein synthesis, mammalian mitochondrial genome encodes a complete set of tRNAs. The function of cytosolic tRNA in mitochondria is thus unclear. An intriguing possibility is that cytosolic tRNA transferred into the mitochondria may help coordinate the rate of protein synthesis in the cytosol with cellular resistance to apoptosis stimuli targeted to mitochondria. tRNA in the cytosol could then further block apoptosis once cytochrome c is released from mitochondria. This notion is supported by the effect of tRNA microinjection and hydrolysis on cytochrome c-induced apoptosis (Figures 6 and 7). tRNA in the cytosol may also be important in neutralizing small amounts of cytochrome c that are accidentally released into the cytosol in a healthy cell.
Cytochrome c appears to interact with tRNA but not other RNAs in vivo. This specific binding is likely a combination of certain intrinsic preference of cytochrome c to tRNA (Figure 4) and the relative accessibility of tRNA compared with other RNAs in cells. Notably, cytochrome c has a high content of lysines and arginines, which is somewhat similar to ancillary RNA-binding domains present in the N- and C-termini of certain eukaryotic amino acid-tRNA synthetases (Cahuzac et al., 2000; Kaminska et al., 2001). Because both cytochrome c and tRNA are highly conserved molecules, their interaction may be ancient and could impact other basic functions of these molecules. For example, tRNA may affect the role of cytochrome c in oxidative phosphorylation; vice versa, cytochrome c may affect the availability of tRNA for protein translation. The cytochrome c:tRNA interaction could therefore reflect an innate coordination among protein synthesis, energy production, and cellular survival that might have arisen early in evolution. We speculate that the modulation of protein function by tRNAs and other RNAs may be more prevalent than previously anticipated.
In mammalian cells, transcription of tRNA by Pol III is stimulated by oncogenic proteins Myc and Ras and inhibited by tumor suppressors RB and p53 (Ruggero and Pandolfi, 2003; White, 2005). Tumor cells synthesize tRNA at highly enhanced rates due to the deregulation of these tumor suppressors and oncoproteins, and in some cases, also due to direct elevation in the expression of the pol III factor TFIIIC (White, 2004; White, 2005). Overexpression of tRNA can lead to cellular transformation (Marshall et al., 2008), and we speculate that suppression of apoptosis may be an important part of this transforming activity. Notably, the tRNA-specific RNase onconase kills tumor cells with relatively low systemic toxicity (Ardelt et al., 2008; Costanzi et al., 2005). Onconase also has the advantage of p53-independent killing (Costanzi et al., 2005; Schein, 1997), an important characteristic as many tumors lack p53 activity. It is in phase III clinical trials for the treatment of mesothelioma, a specific lung cancer, and phase II clinical trials for other cancers (Ardelt et al., 2008; Costanzi et al., 2005). The reason for tumor-specific toxicity of onconase is unclear, and onconase could have functions in addition to tRNA degradation. Nevertheless, the effect of onconase on a critical inhibitor of apoptosis may afford an explanation for its toxicity in tumor cells; it also indicates a valuable target for therapeutic intervention. Furthermore, the synergistic action of onconase with agents that elicit the intrinsic apoptosis pathway (Figure 7C) provides a biologic rationale for development of combined cancer therapies.
Experimental Procedures
Reagents and plasmids
The following reagents were obtained from the indicated sources: bovine cytochrome c, yeast rRNA and tRNA, doxorubicin, proteinase K, formaldehyde, RNase A, Empigen BB, zVAD-FMK, and anti-Flag M2 beads (Sigma); protein A agarose and Texas Red-labeled dextran (Invitrogen); RNase Inhibitor (Promega); antibodies against caspase-9 (MBL International Corporation), caspase-3 (Santa Cruz Biotechnology), Smac (Cell Signaling), and actin (Sigma). Anti-cytochrome c antibodies for immunoblotting and immunoprecipitation were purchased from R&D systems and BD Pharmingen, respectively. Anti-Apaf-1 antibody was kindly provided by Dr. X. Wang (Zou et al., 1997). Onconase was kindly provided by the Alfacell Corporation (Somerset, NJ). Flag-caspase-9-pRK5 was previously described (Chang et al., 2003).
Protein preparation
Recombinant full-length Apaf-1 (amino acids 1–1248) was expressed in Hi-5 insect cells and purified as described (Bao et al., 2007; Jiang and Wang, 2000). Apaf-1Δ was expressed in BL21 (DE3) Escherichia coli strain and purified as described (Riedl et al., 2005).
Caspase activation assay
S100 cell extracts of Jurkat and HeLa cells were prepared as described (Liu et al., 1996). Activation of caspase-9 and caspase-3 in the S100 extracts was induced by the addition of 20 μg/ml cytochrome c and subsequent incubation at 37 °C for 1 h. Activation of in vitro-translated, 35S-labeled caspase-9 was generated using Flag-caspase-9-pRK5 and a coupled in vitro-transcription and translation system (Promega) in the presence of 35S-methionine. Activation of [35S]caspase-9 was induced by incubation with purified Apaf-1 (10 nM) or Apaf-1Δ (20 nM), cytochrome c (20 μg/ml), and dATP (1 mM) for 1 h at 30 °C in oligomerization buffer (20 mM Hepes/pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 2 mM EDTA, and 1 mM DTT).
Gel filtration assay
Gel filtration analysis was performed on a Superose 6 HR 10/30 column driven by an Akta FPLC system (GE Healthcare). The column was calibrated with molecular weight standards from Bio-Rad. The buffer contained 20 mM Hepes (pH 7.0), 0.1 % CHAPS, 5mM DTT, 5% Sucrose, and 50 mM NaCl. 500 μg of S100 extracts were injected into the column, and 500 μl was collected for each fraction. To analyze interaction of total RNA with cytochrome c, cytochrome c was incubated with total RNA at 25°C for 15 min before being analyzed by gel filtration.
RNA preparation and cytochrome c:tRNA interaction
Total RNA was prepared using Trizol reagents (Invitrogen) following manufacturer’s instructions. mRNA was further purified using Oligo(dT)-Cellulose (GE Healthcare). RNAs were dissolved in TE buffer (10 mM Tris-HCl, pH7.5, 1 mM EDTA), and the concentration was quantified by NanoVue spectrophotometer (GE Healthcare). To assess the effect of RNA on the binding of cytochrome c to Apaf-1, recombinant full-length Apaf-1 immobilized on Ni-NTA beads was incubated with and without cytochrome c, and with cytochrome c plus total cellular RNA at 4 °C for 1 h. To examine the effect of RNA on caspase-9 activation, 2 μl of RNAs were added to a total of 20 μl of Jurkat S100 extracts.
To examine the tRNA:cytochrome c interaction in vivo, HeLa cells grown in DMEM supplemented with 10% FBS were harvested and washed twice with ice-cold PBS. Cell pellets were re-suspended in PBS containing 0.2% formaldehyde and incubated at room temperature for 10 min. The cross-linking reaction was quenched with 0.15 M (final concentration) of glycine, pH 7.4. Cell extracts were prepared in the immunoprecipitation buffer (20 mM Tris, pH 7.8, 500 mM NaCl, and 2.5 mM MgCl2) containing 1% Empigen BB, and incubated with antibodies 4 °C for 1 h, followed by incubation with protein G agarose beads for an additional 2 h. Immunoprecipitates were treated with proteinase K and RNA was extracted by phenol/chloroform. Purified RNA was separated by 8% denaturing polyacrylamide gel electrophoresis and analyzed by Northern blotting. Oligonucleotide sequences used to probe various RNAs are as follows: Mitochondrial (Mt) tRNAAla: 5′-TGCAAAACCCCACTCTGCATCAACTGAACGCAAATCAGCCACTTTAATTAAG CTAAGCCC-3′; Mt tRNAMet: 5′-TACGGGAAGGGTATAACCAACATTTTCGGGGTATGGGCCCGATAGCTTATTT AGCTGACC-3′; Mt tRNAPhe: 5′-GGGGTGATGTGAGCCCGTCTAAACATTTTCAGTGTATTGCTTTGAGGAGGTAA GCTACAT-3′; Mt tRNASer: 5′-AGCTGGTTTCAAGCCAACCCCATGGCCTCC-3′; Cytosolic (Cy) tRNAAsp: 5′-CGGGGAATCGACCCCCTCTCTCACGCATGACAGGCAGAGATACTCACCACTA TAGTAACA-3′; Cy tRNAGln: 5′-TTCAGAGTCCAGAGTGCTTACCATTACACC-3′; Cy tRNASer: 5′-ACCTGCGCGGGGAGACCCCAATGGATTTCTAGTCCATCGCCTTAACCACTCG GCCACGAC-3′; Cy tRNATrp: 5′-CGACGTGATTTGAACACGCAACCTTCTGATCTGGAGTCAGACGCGCTACCGTT GCGCCAC; U1 snRNA: 5′-CACCTTCGTGATCATGGTATCTCCC-3′. 5S RNA: 5′-CCTACAG CACCCGGTATTCCCAGGC-3′; RNase MRP: 5′-TGCACGTGGCACTCTCTGCCCG AGG-3′; RNase P: 5′-TCCTGCCCAGTCTGACCTCGCGCGG-3′; 7SK: 5′-CGCCTAGCCAGCCAGATCAGCCGAATCAAC-3′; 7SL: 5′-ACCCCTCCTTAGGCAACCTGGTGGTCCCCC-3′; Hvg3: 5′-AGAGGTGGTTTGATGACACG-3′; and hY1: 5′-TGAACAATCAATTGAGATAACTCACTACCT-3′.
To analyze the interaction of cytochrome c with individual tRNAs in vitro, each tRNA was transcribed in vitro in the presence of [32P-UTP]. 20,000 cpm tRNA was incubated with different amounts (0.5, 2.5 and 12.5 μM) of recombinant cytochrome c for 45 min at 30 °C in the buffer containing 20 mM HEPES, pH 7.5, 35 mM KCl, 2.5 mM MgCl2, 0.5 mM EDTA, and 1 mM DTT supplemented with salmon sperm DNA (final concentration 50 ng/μl). The reaction mixture was incubated with sample loading buffer containing urea (final concentration 0.5 M) for 10 min at room temperature. The assembled tRNA:cytochrome c mixture was analyzed by 6% native polyacrylamide (acrylamide:bis=79:1) gel electorphoresis followed by autoradiography.
To analyze the interaction of cytochrome c with total tRNA in vitro, total RNA was isolated from HeLa cells using Trizol method and separated on 6% denaturing PAGE. Total tRNA bands were isolated, and 50 ng of total tRNA was used for labeling in the presence of [32P-pCp]. 20,000 cpm tRNA (approximately 12 fmole with a labeling efficiency of ~50%) was incubated with 100 pmole of cytochrome c in the presence and absence of unlabeled rRNA (28S and 18S), poly(A), total tRNA (50 and 250 ng), or DNA oligonucleotide corresponding to mitochondrial tRNAPhe (5 pmole/132 ng and 20 pmole/528 ng). The binding reaction was done in buffer containing 20 mM HEPES, pH 7.5, 35 mM KCl, 2.5 mM MgCl2, 0.5 mM EDTA, and 1 mM DTT and the reaction product was analyzed on 6% native polyacrylamide gel electrophoresis using buffer containing 25 mM Tris/7.5 and 25 mM Glycine.
Microinjection
Microinjection was performed on an Eppendorf micromanipulator/microinjector. Concentrations of the solutions were: dextran-Texas Red (0.3%, in PBS), cytochrome c (0.5 μg/μl), and tRNA/rRNA (7.5 μg/μl). Based on an injection volume of 0.1pl, the total amount of RNA and cytochrome c injected in each cell was 700 fg and 50 fg, respectively. 2 h after injection, cells were fixed with 4% paraformaldehyde and mounted with a DAPI-containing medium (Vector Laboratories). For each condition, one hundred to one hundred fifty injected cells were counted, and apoptotic cells were determined by the presence of membrane blebbing and nuclear fragmentation.
Treatment with onconase and doxorubicin
Cells were transfected with onconase using lipofectamine 2000 (Invitrogen) similar to that previously described (Iordanov et al., 2000). 3 h after transfection, the cells were incubated with and without doxorubicin (1 μg/ml) for another 12 h. Cells were stained with Hoechst 33342, and apoptotic cells were scored based on nuclear condensation and fragmentation.
Supplementary Material
Acknowledgments
We thank Drs. K. Shogen and W. Ardelt for providing onconase, Dr. X. Wang for providing Apaf-1−/− and wild type MEFs and anti-Apaf-1 antibody, Dr. K. Dittmar for advice on tRNA work, and A. Stonestrom and S. Slattery for help with manuscript preparation. Supported by NIH grants GM060911 and CA108872, and a Leukemia and Lymphoma Society Scholar Award (to X.Y.). G.D. is funded by the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardelt W, Shogen K, Darzynkiewicz Z. Onconase and amphinase, the antitumor ribonucleases from Rana pipiens oocytes. Curr Pharm Biotechnol. 2008;9:215–225. doi: 10.2174/138920108784567245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Q, Lu W, Rabinowitz JD, Shi Y. Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol Cell. 2007;25:181–192. doi: 10.1016/j.molcel.2006.12.013. [DOI] [PubMed] [Google Scholar]
- Cahuzac B, Berthonneau E, Birlirakis N, Guittet E, Mirande M. A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases. Embo J. 2000;19:445–452. doi: 10.1093/emboj/19.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra D, Bratton SB, Person MD, Tian Y, Martin AG, Ayres M, Fearnhead HO, Gandhi V, Tang DG. Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome C and inhibiting apoptosome. Cell. 2006;125:1333–1346. doi: 10.1016/j.cell.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Chang DW, Ditsworth D, Liu H, Srinivasula SM, Alnemri ES, Yang X. Oligomerization is a general mechanism for the activation of apoptosis initiator and inflammatory procaspases. J Biol Chem. 2003;278:16466–16469. doi: 10.1074/jbc.C300089200. [DOI] [PubMed] [Google Scholar]
- Chang HY, Yang X. Proteases for Cell Suicide: Functions and Regulation of Caspases. Microbiol Mol Biol Rev. 2000;64:821–846. doi: 10.1128/mmbr.64.4.821-846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YD, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984;99:1997–1204. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro-apoptotic anticancer strategy: review of the preclinical and clinical data for ranpirnase. Cancer Invest. 2005;23:643–650. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Shaheen HH. A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol. 2008;18:98–104. doi: 10.1016/j.tcb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Ibba M, Becker HD, Stathopoulos C, Tumbula DL, Soll D. The adaptor hypothesis revisited. Trends Biochem Sci. 2000;25:311–316. doi: 10.1016/s0968-0004(00)01600-5. [DOI] [PubMed] [Google Scholar]
- Iordanov MS, Ryabinina OP, Wong J, Dinh TH, Newton DL, Rybak SM, Magun BE. Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis. Cancer Res. 2000;60:1983–1994. [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Kaminska M, Shalak V, Mirande M. The appended C-domain of human methionyl-tRNA synthetase has a tRNA-sequestering function. Biochemistry. 2001;40:14309–14316. doi: 10.1021/bi015670b. [DOI] [PubMed] [Google Scholar]
- Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci U S A. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- Liu H, Chang DW, Yang X. Interdimer processing and linearity of procaspase-3 activation. A unifying mechanism for the activation of initiator and effector caspases. J Biol Chem. 2005;280:11578–11582. doi: 10.1074/jbc.M414385200. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, Alfonzo JD. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci U S A. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- Saxena SK, Sirdeshmukh R, Ardelt W, Mikulski SM, Shogen K, Youle RJ. Entry into cells and selective degradation of tRNAs by a cytotoxic member of the RNase A family. J Biol Chem. 2002;277:15142–15146. doi: 10.1074/jbc.M108115200. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Kornbluth S. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev Cell. 2006;10:549–561. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Schein CH. From housekeeper to microsurgeon: the diagnostic and therapeutic potential of ribonucleases. Nat Biotechnol. 1997;15:529–536. doi: 10.1038/nbt0697-529. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Weisblum B. Back to Camelot: defining the specific role of tRNA in protein synthesis. Trends Biochem Sci. 1999;24:247–250. doi: 10.1016/s0968-0004(99)01396-1. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jackson BM, Hinnebusch AG. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989;86:4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


