Abstract
The social amoeba Dictyostelium discoideum has become established as a simple model for the examination of cell-cell interactions and early studies suggested that shifts in glycosylation profiles take place during its life cycle. In the present study, we have applied HPLC and mass spectrometric methods to show that the major N-glycans in axenic cultures of the AX3 strain are oligomannosidic forms, most of which carry core fucose and/or intersecting and bisecting N-acetylglucosamine residues, including the major structure with the composition Man8GlcNAc4Fuc1. The postulated α1,3-linkage of the core fucose which correlates with the cross-reactivity of Dictyostelium glycoproteins with an anti-horseradish peroxidase antiserum; a corresponding core α1,3-fucosyltransferase activity capable of modifying oligomannosidic N-glycans was detected in axenic Dictyostelium extracts. The presence of fucose on the N-glycans and the reactivity to the antiserum, but not the fucosyltransferase activity, are abolished in the fucose-deficient HL250 strain. In later stages of development, N-glycans at the mound and culmination stages show a reduction in both the size and the degree of modification by intersecting/bisecting residues as compared to mid-log phase cultures, consistent with the hypothesis that glycosidase and glycosyltransferase expression levels are altered during the slime mould life cycle.
Keywords: Dictyostelium, fucose, fucosyltransferase, glycan, mass spectrometry
Cell-cell interactions often rely on the recognition of carbohydrate chains; such interactions are key to the development of multicellular organisms, but are often difficult to understand due to the inherent complexity of such processes, which involve not just carbohydrates, but also the relevant lectin receptors. One of the simplest systems for studying the multicellular state is the cellular slime mould or social amoeba Dictyostelium discoideum. When food is plentiful, it is a unicellular amoeba; however, upon starvation, signals between individual cells result in formation of multicellular aggregates, a process which culminates in the formation of a fruiting body. The spores can then survive until conditions for the organism are more optimal. Thus, a number of processes analogous to those in ‘true’ multicellular organisms, such as cell-cell communication and adhesion, occur [1]. Due to the involvement of carbohydrates in such phenomena in general, it is not surprising that, in the case of Dictyostelium, stage-specifically expressed cell surface oligosaccharides were found to be involved in cell adhesion [2] and that lectins, such as discoidins I and II [3], are produced during the aggregation stage. Over the past thirty years there have been a number of other studies directed at examining the glycosylation of Dictyostelium; however, methodological constraints, as well as the lack until recently of a full genomic sequence, have meant that a full understanding and general consensus about glycan structures, their biosynthesis and their lectin partners in the slime mould is still to be achieved.
In previous studies on the N-linked oligosaccharides of Dictyostelium, endoglycosidases were frequently used to release radiolabelled glycans, which were then primarily analysed by gel filtration chromatography [4-6]; in only two studies were mass spectrometric data obtained for two novel endoglycosidase-released structures containing methylphosphodiester-modified mannose residues [7, 8]. The accumulated data, thereby, lacked specific information about modification of the innermost N-acetylglucosamine residue; the presence of fucose and/or sulphate on the core or peripheral regions of the N-glycans was inferred solely by resistance to endoglycosidase digestion [4, 6, 9]. Indications of shifts in the glycosylation of Dictyostelium during development were also inferred from gel filtration chromatography and glycosidase digestion data [4-6, 10, 11] as well as by alterations in the recognition of glycoproteins by various antibodies, raised against Dictyostelium itself or against horseradish peroxidase [12, 13]. The origin, though, of such epitopes remained unresolved.
Indeed, a general problem in understanding the glycomic potential of Dictyostelium is the limited knowledge of glycosylation-relevant genes and the uncertain glycostructural repercussions of those glycomutants available, some of which affect processing glycosidases, glycosyltransferases or enzymes required for nucleotide sugar biosynthesis [14-21]. The recent sequencing of the Dictyostelium genome opens up the possibility of identifying genes displaying homology to those encoding glycosylation-relevant enzymes in other organisms [22]; however, homology to a glycosyltransferase or glycosidase in itself has poor predictive power as to the exact biochemical function of a gene. Therefore, glycostructural determination, enzymological and molecular glycobiological studies must run in parallel in order to optimally increase our understanding of Dictyostelium oligosaccharide biosynthesis and function. Considering the recent advances in mass spectrometry of glycans, we believe that the application of glycomic profiling methodology to the slime mould is now appropriate.
In this study, we have examined the N-glycans of one standard laboratory and one wild-type strain of Dictyostelium as well as one glycomutant, HL250 (modC), previously shown to have a defect in the biosynthesis of GDP-Fuc, which is the donor substrate required for formation of fucosylated glycans [17]. In the HL250 mutant, we show that the binding of Dictyostelium proteins to anti-horseradish peroxidase, an antiserum raised against a plant glycoprotein is abolished. This correlates with an absence of all core α1,3-fucosylated N-glycans in this strain, which, in the pseudo-‘wild-type’ AX3 strain, display unique structures also determined during the present study for the first time. Furthermore, the structure of these fucosylated glycans also varies during the development of Dictyostelium and a reduced glycan size is observed with similar profiles for the glycans from fruiting bodies of both the axenic AX3 and non-axenic NC4 strains. The correlation of these structures with the multicellular state in Dictyostelium suggests that they have a role during wild-type development.
Experimental Procedures
Dictyostelium cultivation
AX3 (a standard axenic strain; axeA1, axeB1, axeC1), HL250 (axeA1, axeB1, axeC1, modC) and NC4 (wild-type) strains of D. discoideum were obtained from the Dictyostelium Stock Centre and maintained on either E. coli OP50 (for AX3 and HL250) or Klebsiella aerogenes (for NC4); axenic cultures of AX3 and HL250 were grown in HL-5 medium. In one experiment, HL250 cells were also grown in HL-5 medium supplemented with 10 mM fucose. Development of AX3 was initiated on Whatman 50 filter paper from axenic cultures washed in a development buffer containing 5 mM Na2HPO4, 5 mM KH2PO4, 1 mM CaCl2 and 2 mM MgCl2, pH 6.5 [23], followed by exposure to light for between 10 and 24 hours.
Western blotting
Cells (1.25 × 106/ml) were washed, centrifuged and resuspended in SDS-PAGE buffer and incubated at 95 °C for 10 minutes. The supernatant from this extract was then subject to SDS-PAGE and the proteins were transferred to nitrocellulose using a semi-dry blotting apparatus. After blocking with 0.5% (w/v) bovine serum albumin, the membranes were incubated with rabbit anti-horseradish peroxidase (anti-HRP; 1:10000); after washing, alkaline phosphatase conjugated goat anti-rabbit (1:2000) was used with subsequent colour detection with SigmaFAST™ 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium.
Flow cytometry
Cells were incubated with 20 μg/ml anti-HRP in PBS at 26 °C for 30 minutes, washed once with 5 ml of PBS, and then incubated with FITC-labelled anti-rabbit (Sigma) diluted 1:100 in PBS for another 30 min [24]. Cells were again washed with 5 ml of PBS and resuspended in PBS and subject to flow cytometry using a BD Biosciences FACS Calibur with a 488-nm argon Laser (15-milliwatt output power) in the standard configuration. FITC fluorescence was measured with a 530/30 BP filter in FL1.
Glycan preparation
N-glycans were prepared, from slime mould cultures or from secreted proteins, by enzymatic release of glycans from partially-purified glycopeptides using PNGase A as previously described for other organisms [25]. Generally, cells (ca. 1 g) were boiled in 5% formic acid and disrupted using a custom-made tight-fitting glass homogeniser followed by microscopic assessment of the degree of mechanical disruption. In one case, cells were lysed using a French press (American Instruments Co. Inc., Silver Spring, MD, USA) at 1200 p.s.i. in 5% formic acid. After treatment with pepsin (1 mg), glycopeptides were enriched by cation exchange and gel filtration chromatography prior to glycan release. As required, PNGase A was substituted by either PNGase F or endoglycosidase H. In one case, glycopeptides derived from a pronase digest of secreted proteins were prepared by gel filtration according to the method of Sharkey and Kornfeld [6] with omission of their ion-exchange columns and solvolysis steps, prior to release of N-glycans by PNGase A. Free N-glycans were pyridylaminated and purified by HPLC.
In a further experiment, following the procedure described by Davis [26], washed cells were suspended (4 × 106 cells/ml) in 10 mM Tris, pH 7.5, containing 1% sodium dodecyl sulphate and 10 mM dithiothreitol, incubated at 100 °C for 10 minutes and homogenised; the supernatant was subject to gel filtration (Sephadex G50, 1.5 × 50 cm) and the void fraction was precipitated with methanol. The resulting pellet was resuspended in 20 ml of 20 mM ammonium acetate, pH 8.8, containing 0.2% β-mercaptoethanol and 0.5% Igepal CA 530 and incubated with PNGase F (20 U) overnight at 37 °C. After heat treatment and centrifugation, the sample was again subject to gel filtration (G50 followed by G25 and finally G15) followed by a final purification on a small cation-exchange/reversed-phase column prior to further analysis.
For the small-scale preparation of glycans from fruiting bodies of the NC4 strain (obtained from a single 9 cm SM agar plate), the method of Rendić et al. [27] was employed; in brief, SDS-PAGE using a high percentage gel and a short run time was performed, the Coomassie-stained region excised and washed, the proteins subject to in gel tryptic digestion followed by PNGase A release of the N-glycans and purification using a mini-column of Dowex 50W×8 cation exchange and Lichroprep reverse phase resins prior to MALDI-TOF MS.
Glycan analysis
Complete N-glycomes were fractionated by either reversed-phase HPLC (Hypersil ODS; gradient of 1% methanol per minute using 0.1 M ammonium acetate, pH 4, as buffer; 1.5 ml/min) or normal-phase HPLC (Tosoh TSKgel Amide-80; gradient from 71.25% to 61.75% acetonitrile over 20 minutes followed by a gradient from 61.75% to 47.5% acetonitrile for a further 45 minutes using 10 mM ammonium formate, pH 7, as buffer; 1 ml/min). Glycans were respectively detected by fluorescence with excitation/emission wavelengths of 320/400 or 310/380 nm. Selected fractions were subject to 2D-HPLC with normal-phase HPLC in the first dimension (Takara Palpak Type N; gradient of 0.825% acetic acid/triethylamine, pH 7.3, in 74.5% acetonitrile up to 1.5% acetic acid /triethylamine, pH 7.3, in 50% acetonitrile from 5 to 40 minutes; 1 ml/min) and reversed-phase HPLC as above in the second. The pyridylaminated glycan standards MMF3 [28], Man5GlcNAc2 and Man9GlcNAc2 were derived from PNGase digestion of giant ragweed pollen extract, fungal α-amylase and soybean agglutinin, respectively.
Further analysis by HPLC or MS was performed after treatment overnight either with 1 mU endoglycosidase H (Roche), 5 mU of either jack bean α-mannosidase (Sigma), jack bean β-N-acetylhexosaminidase (Sigma) or bovine α-fucosidase (Sigma) in 0.1 M sodium citrate, pH 5, buffer at 37 °C, or with 2 μU α1,2-mannosidase from A. saitoi (Prozyme) using the supplied buffer. Monoisotopic MALDI-TOF MS was performed using a Bruker Ultraflex TOF-TOF instrument generally with 2,5-dihydroxybenzoic acid as matrix. For methylation analysis, the glycopeptides or glycans (approximately 1 nmol) were permethylated, hydrolysed, reduced and peracetylated prior to analysis of the obtained partially methylated alditol acetates by capillary GLC/MS using the instrumentation and microtechniques described elsewhere [29, 30].
LC-ESI-MS experiments with either a whole N-glycome or a purified fraction were carried out using a Q-TOF Ultima Global mass spectrometer (Micromass, Manchester, U.K.) and an upstream Micromass CapLC using a Thermo Aquastar 30 × 0.32 mm C18 guard column and a Thermo Hypercarb 100 × 0.32 mm porous graphitic carbon separation column and elution with acetonitrile containing 0.1% formic acid [31].
Cloning and expression of gmd and ger
Based upon the curated Genbank sequences XM_001134543 and XM_641512, the reading frames encoding GDP-Man dehydratase (GMD) and GDP-Fuc synthase (GER) were isolated by RT-PCR from both AX3 and HL250 Trizol-prepared RNA. The primers DdGMDfwd/BamHI (cgcggatccatgagtgaagaacgtaaagta), DdGMDrev/XhoI (ccgctcgagttaattatatttatcaccatt), DdGERfwd/BamHI (cgcggatccatgacagaaacaacaagcaaaa) and DdGERrev/XhoI (ccgctcgagttattttcttgcagtttcata) were used in combination with Expand polymerase (Roche) with additional MgCl2, dATP and dTTP as required. PCR fragments were cut with the relevant enzymes and ligated into pET30a; expression of the recombinant proteins in E. coli, Western blotting, assay and His-tag purification were performed as previously described for homologous enzymes [32].
Fucosyltransferase assay
Cells (1.25 × 106/ml) were vortexed and lysed by incubation on ice for 30 minutes in 50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% Triton X-100 and a 1:100 dilution of a protease inhibitor cocktail (Sigma), prior to centrifugation for 30 minutes. The core fucosyltransferase activity was assayed using a dansylated Man5GlcNAc2-Asn-Ser peptide derived by dansylation of a glycopeptide purified from a pronase digest of fungal α-amylase. The dansyl-glycopeptide (180 μM) was typically incubated at room temperature (23 °C in PCR tubes) for four hours in the presence or absence of GDP-Fuc (1 mM) at a final concentration of 40 mM HEPES, pH 8.0, 5 mM MgCl2 and 5 mM MnCl2 with 2 μl of Dictyostelium lysate (final volume 5 μl). The incubation mixture was then subject to RP-HPLC as previously described [24]; in the case of a larger-scale reaction, collected fractions were analysed by MALDI-TOF MS with 2,5-dihydroxybenzoic acid as matrix as well as by GLC/MS. For comparison, a purified form of Caenorhabditis core α1,3-fucosyltransferase FUT-1 [33] was employed.
Results
Axenic Dictyostelium cultures express novel N-glycans
As part of our strategy of analysing the glycome of non-mammalian model organisms, we examined the Asn-linked oligosaccharides of the slime mould Dictyostelium discoideum. The initial experiments on axenic cultures of pseudo-‘wild-type’ AX3 cells were directed at gaining an impression of the overall N-glycomic capacity of this organism as well as re-investigating the phenomenon that it may [13] express anti-HRP epitopes, as do other model organisms such as Arabidopsis, Drosophila and Caenorhabditis [34]. To assess whether these epitopes are based on cross-reactivity of the anti-HRP1 antiserum to core α1,3-fucose or β1,2-xylose (as part of the N-glycan structure), a Dictyostelium mutant (HL250; modC) with a defect in fucose metabolism [17] was also examined. This strain lacking fucosylated glycans is somewhat slower growing in axenic culture than the standard AX3 strain (Figure 1A); less efficient germination of older spores from this strain and slightly abnormal sheath properties have been previously noted [17, 19].
Figure 1. Anti-horseradish peroxidase epitopes of axenically-cultured Dicytostelium discoideum.
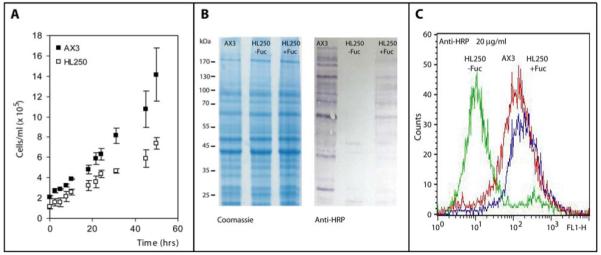
(A) The AX3 strain exhibits somewhat quicker growth during the pre-logarithmic phase of axenic culture in comparison to the HL250 (modC) strain; (B) Western blotting shows the presence of anti-HRP epitopes in axenically-grown AX3, but not in axenically-grown HL250, consistent with a defect in fucose metabolism in that strain; a partial recovery of anti-HRP epitopes was observed when HL250 cells were grown in the presence of exogenous fucose. The Coomassie-stained gel shows that protein loading was approximately equal. (C) Flow cytometry also shows the absence of anti-HRP epitopes in the HL250 strain grown in the absence of fucose (green) as compared to AX3 (red) or HL250 grown in the presence of exogenous fucose (blue).
As shown by both Western blotting and flow cytometry, axenic AX3 cells express anti-HRP epitopes on a number of proteins in whole extracts and on the cell surface, whereas such epitopes are basically absent in the HL250 strain (Figures 1B and 1C); however, when the medium was supplemented with fucose, anti-HRP staining was restored in the HL250 strain, a result suggestive of the presence of a fucose ‘salvage’ pathway [35] in this organism. These data are consistent with core α1,3-fucose (as present on plant, fly and nematode N-glycans), but not β1,2-xylose (as present on plant glycans), being the source of cross-reactivity of Dicytostelium proteins towards the antibody. In order to examine this in more detail, N-glycans from cell pellets of both these Dictyostelium strains were subject to pyridylamination and analysed by HPLC and mass spectrometric methods.
This approach brought a further indication of a lack of xylose on the N-glycans, due to the absence of any glycan species with an m/z consistent with the presence of xylose in the complete N-glycome mass spectrum. Indeed, upon release with PNGase A, the major N-glycan species in mid-log phase AX3 has the composition Hex8HexNAc4Fuc1, a glycan absent from the HL250 glycome (compare Figure 2A and 2B). Comparable data were obtained by analysis in the negative mode of pyridylaminated glycans (with the ions in the [M-2H+Na]− form; Supplementary Figure 1) and by ESI-MS of unlabelled N-glycans (Supplementary Figure 2). We also examined PNGase A-released glycans isolated from the proteins secreted by washed axenic cultures as well as from whole AX3 cells subject to homogenisation with a French press. The ‘secreted’ N-glycome contained the same two major species as the whole AX3 axenic cells, although the presence of a glycan with the composition Hex5HexNAc4Fuc1 and the absence of typical oligomannosidic glycans is also obvious, whereas the French press lysed cells appeared to have a similar glycosylation pattern as those which were disrupted with a glass homogeniser (Supplementary Figure 3).
Figure 2. The N-glycome of axenically-cultured Dicytostelium discoideum.
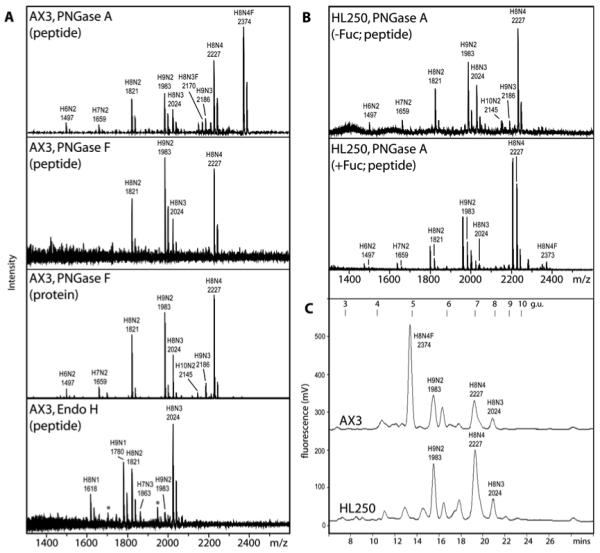
(A) MALDI-TOF MS of pyridylaminated N-glycans from axenically-grown mid-log phase AX3 either released from glycopeptides using PNGase A, PNGase F or EndoH or from intact proteins treated with PNGase F showing the absence of fucosylated glycans from the latter three glycan pools. The peaks marked with an asterisk in the spectrum of the EndoH-released glycans are non-pyridylaminated forms of the Hex9HexNAc1 (m/z 1703) and Hex8HexNAc3 (m/z 1947) species. The data with pyridylaminated PNGase A-released glycans is compatible to those of the unlabelled, free N-glycans (see ESI-MS data in Supplementary Figure 2). (B) MALDI-TOF MS of PNGase A-released pyridylaminated N-glycans from axenically-grown mid-log phase HL250 grown in the absence or presence of exogenously-added fucose, showing the appearance of a fucosylated H8N4F glycan (m/z 2373) in the latter. C) RP-HPLC pyridylaminated PNGase A-released N-glycans from axenically-grown mid-log phase AX3 and HL250 grown in the absence of exogenous fucose shows a difference in elution properties; peaks were collected and subject to MALDI-TOF MS (major species are annotated) and compared to an isomaltooligosaccharide standard (g.u., glucose units). The major [M+Na]+ species are annotated in the form HxNyF0-1, where H signifies hexose, N is N-acetylhexosamine and F is fucose.
Consistent with the lack of core fucose on HL250 glycans is the distinct nature of the reversed-phase HPLC profile of the N-glycans from this strain as compared to that of the AX3 N-glycans. The shift to the left of the major AX3 glycan, as compared to the major HL250 glycan (Figure 2C), is compatible with the presence of core α1,3-linked fucose on this structure; a shift to the right, on the other hand, would have been an indication of core α1,6-fucosylation [36]. Furthermore, this major Hex8HexNAc4Fuc1 structure is absent from AX3 glycans released using PNGase F, an enzyme incapable of removing core α1,3-fucosylated glycans [37]; indeed, PNGase F treatment either of glycopeptides or of intact, denatured proteins resulted in spectra showing the presence primarily of Hex8HexNAc2-4 and Hex9HexNAc2-3 (see Figure 2A). On the other hand, no information about modifications of the reducing-terminal GlcNAc residue can be inferred from the Endo H digest due to the mode of action of this enzyme, which cleaves between the two GlcNAc residues of the chitobiose core; however, other than the absence of fucose from the released glycans, the Endo H-derived data is compatible with the other digests in terms of the observed species (Figure 2A). The presence of Hex8HexNAc4Fuc1 is regained to a minor extent in HL250 cells grown in a fucose-containing medium (Figure 2B), consistent with a partial restoration of anti-HRP staining.
Collection of the individual RP-HPLC fractions and their analysis by mass spectrometry verifies that the major AX3 N-glycan has indeed the composition Hex8HexNAc4Fuc1 (for the [M+H]+ form, m/z 2351). Mild acid hydrolysis also resulted in a mass spectral ‘ladder’ of species compatible with this composition (data not shown). The modification of the core region was also verified by the results of MS/MS. In particular, the fragment of m/z 446 indicates the presence of core fucose on the reducing-terminal pyridylaminated N-acetylglucosamine residue, while this fragment was lacking from the non-fucosylated form Hex8HexNAc4 (Figure 3). The position of the four HexNAc residues in both glycans was also determined by this method: as usual for an N-glycan, there are two within the reducing-terminal core, whereas the other two are ‘branching’ modifications of mannose residues. One is in the so-called ‘bisecting’ position on the central β1,4-linked mannose residue, as indicated by the m/z 1014 fragment, whereas the other is putatively linked to the α1,6-linked mannose residue, compatible with the m/z 1379 fragment. In an older study on Dictyostelium glycans, a residue in the latter position was named an ‘intersecting’ GlcNAc [8]. As previously suggested, these intersecting/bisecting residues confer resistance to glycosidase digestion: combined α-mannosidase and β-hexosaminidase digestion results in the loss of only up to three hexose and one N-acetylhexosamine residues, as judged by MALDI-TOF MS (data not shown). The Hex8HexNAc4Fuc1 structure is, though, sensitive to hydrofluoric acid treatment, which resulted in a loss of m/z 146, and insensitive to both bovine kidney fucosidase and endoglycosidase H; together with the absence of this structure from the glycan pool released by PNGase F, its reversed phase HPLC behaviour, the occupation of 2-position of the reducing terminal GlcNAc by the N-acetyl moiety and substitution of the 4-hydroxyl of this residue by the second core GlcNAc, these data are indicative of the presence of core α1,3-fucose. Linkage analysis of partially-methylated alditol acetates verified the putative positions of the bisecting and intersecting residues and also showed that the composition of the major glycans with and without core fucose corresponds to Man8GlcNAc4Fuc0-1 (Table 1). Due to the pyridylamination of the reducing-terminal GlcNAc, the actual position of the fucose on this residue cannot be confirmed by GLC/MS; however, as described above, the α1,3-linkage can be inferred from the glycan’s other properties.
Figure 3. MS/MS analysis of the major N-glycans of axenically-grown mid-log cells.
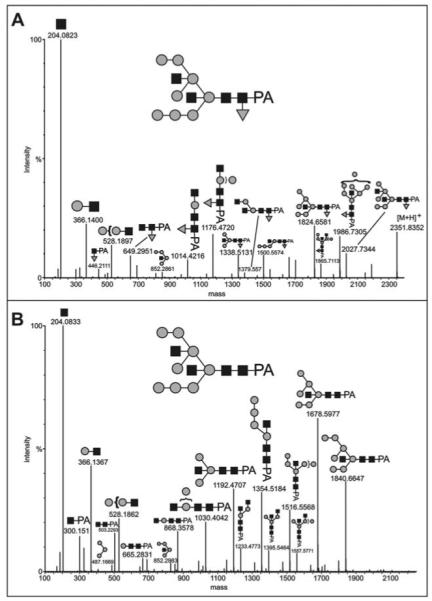
The major AX3 pyridylaminated N-glycans were purified by sequential NP-HPLC (Palpak) and RP-HPLC prior to analysis by LC-ESI-MS/MS; the MaxEnt3 transformed data for (A) the m/z 1176.93 [M+2H]2+ (Man8GlcNAc4Fuc1) and (B) the m/z 1103.41 [M+2H]2+ (Man8GlcNAc4) molecular ions are shown. Fragments of the postulated complete glycan structures are depicted using the nomenclature of the Consortium for Functional Glycomics (black square, N- acetylglucosamine; grey circle, mannose; grey triangle, fucose). The composition and linkages of the intact glycans were proven by GLC-MS (see Table 1); up to three mannose and one GlcNAc residues could be released by combined α-mannosidase and β-N-acetylhexosaminidase digestion (data not shown). Based on the expected prior processing by ER mannosidase, the Man8B isomer is taken as the basis for these structures.
Table 1. Linkage analysis of the Man8GlcNAc4 and Man8GlcNAc4Fuc1 N-glycans.
Both the purified pyridylaminated H8N4 and H8N4F1 glycans derived from axenically-grown AX3 were subject to permethylation. Partially methylated alditol acetates, obtained after hydrolysis, reduction and peracetylation, were separated on an RTX200 column and analysed by GLC/MS in the positive-ion CI-mode. The innermost reducing terminal GlcNAc is not detected because of PA-labelling. The data obtained reveal the presence of terminal bisecting and intersecting GlcNAc in both cases, which is corroborated by the exclusive detection of 3,4,6-trisubstituted Man and the lack of 3,6-disubstituted Man. The results obtained were as follows:
| Methylated sugar derivative |
H8N4-PA | H8N4F-PA | Linkage |
|---|---|---|---|
| 2,3,4-Fuc | − | (+)a | Fuc(1- |
| 2,3,4,6-Man | + | + | Man(1- |
| 3,4,6-Man | + | + | -2)Man(1- |
| 2,4-Man | − | − | -3,6)Man(1- |
| 3,4-Man | − | − | -2,6)Man(1- |
| 2,3-Man | − | − | -4,6)Man(1- |
| 2-Man | + | + | -3,4,6)Man(1- |
| 3,4,6-GlcNAc | + | + | GlcNAc(1- |
| 3,6GlcNAc | + | (+)a | -4)GlcNAc(1- |
Trace amounts, due to low amount of material.
A gene putatively defective in HL250 cells encodes a GDP-mannose dehydratase
As shown above, the HL250 strain lacks fucosylated N-glycans; previous studies have shown that this strain is, in general, fucose-deficient and lacks the ability to synthesise GDP-Fuc [17], the donor required for the enzymes which catalyse fucose transfer. In order to study the GDP-Fuc synthesis pathway in vitro, we cloned cDNAs encoding the two relevant Dictyostelium enzymes, GDP-mannose dehydratase (GMD; EC 4.2.1.47) and GDP-keto-6-deoxymannose 3,5-epimerase/4-reductase (GER or FX protein; EC 1.1.1.271) from both the AX3 and HL250 strains. Both cDNAs originating from AX3 cells directed expression of recombinant proteins in E. coli, which when co-incubated were enzymatically active in the conversion of GDP-Man to GDP-Fuc as judged by HPLC-based assays of either soluble E. coli extracts or the purified proteins; the optimum of the combined reaction was at 30 °C and at pH 7-8. Similarly, the ger cDNA derived from HL250 cells also encoded an active enzyme as judged by formation of the GDP-Fuc product when HL250 GER was co-incubated with AX3 GMD (Figure 4A); thus, the biochemical function of the GMD and GER orthologues in Dictyostelium is thereby proven. However, neither gmd clone derived from HL250 cDNA encoded an active enzyme, although the recombinant HL250 protein was expressed (Figure 4B); both these clones contained a mutation as compared to the AX3 clones in which a Gly codon (GGT) is changed to a Glu (GAT) (Figure 4C). At this position, an uncharged Ala or Gly residue is present in GMD sequences from other species.
Figure 4. Reconstitution of Dictyostelium GDP-Fucose biosynthesis in vitro.
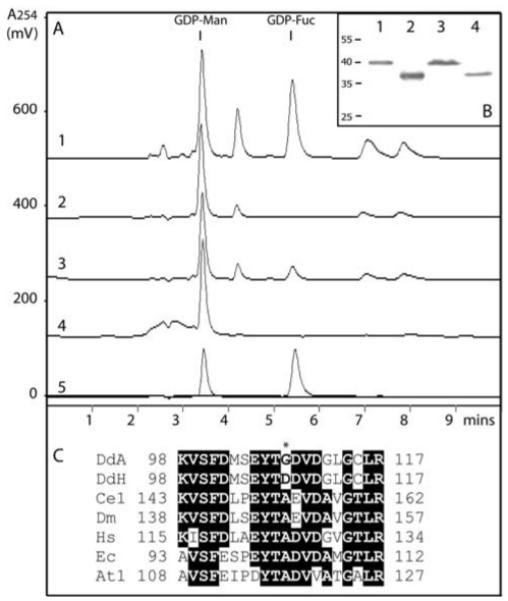
(A) Recombinant forms of the Dictyostelium GMD and GER encoded by cDNAs derived from either the AX3 or the HL250 strain were assayed in the following combinations: 1, AX3 GMD with AX3 GER; 2, HL250 GMD with AX3 GER; 3, AX3 GMD with HL250 GER; 4, HL250 GMD with HL250 GER; 5, overlaid chromatograms of GDP-Man and GDP-Fuc standards; other peaks in the RP-HPLC chromatograms derive from the bacterial lysates. (B) Expression of protein from all four clones was demonstrated by anti-His Western blotting: 1, AX3 GMD; 2, AX3 GER; 3, HL250 GMD; 4, HL250 GER. (C) Comparison of the protein sequences of various GDP-Man dehydratases in the region surrounding the site corresponding to the mutation in the HL250 GMD cDNA; DdA, Dicytostelium discoideum AX3 (residues 98-117); DdH, Dicytostelium discoideum HL250; Ce1, Caenorhabditis elegans GMD-1 (BRE-1); Dm, Drosophila melanogaster GMD; Hs, Homo sapiens GMD; Ec, Escherichia coli GMD; At1, Arabidopsis thaliana GMD1. Amino acids identical in a majority of the selected sequences are highlighted and the Gly → Asp change in the HL250 sequence is marked with an asterisk. On the other hand, the AX3 and HL250 ger cDNA sequences were found to be identical, although expression and activity levels differed in the experiments shown in panels A and B.
Dictyostelium extracts contain a core fucosyltransferase activity
A second prerequisite, other than the ability to synthesise GDP-Fuc, for the expression of fucosylated glycans is the presence of relevant fucosyltransferases. The structure of these fucosylated oligosaccharides in Dictyostelium is unusual, since these are neither of the ‘standard’ biantennary complex nor paucimannosidic types, such as those which can be found in mammals, plants or insects. Thus, the modification of an oligomannosidic type glycan with fucose (as well as ‘intersecting’ and ‘bisecting’ GlcNAc residues) indicates that the fucosyltransferase acceptor substrate in Dictyostelium is novel. Considering the glycopeptides available in our laboratory, we tested an oligomannosidic (Man5GlcNAc2; derived from fungal amylase) as well as biantennary (GlcNAc2Man3GlcNAc2; GnGn, derived from IgG) and paucimannosidic (Man3GlcNAc2; MM, also derived from IgG) substrates. With both AX3 and HL250 extracts as enzyme sources, we observed an alteration in retention time with only the first of these glycans (Figure 5A). Indeed, two new peaks appeared in incubations of a dansylated Man5GlcNAc2 glycopeptide. Mass spectral analysis (Figure 5B) of the collected fractions indicated that the first peak, which only appeared when GDP-Fuc was added to the incubation, was a fucosylated form of the substrate, the second peak was identical to the original substrate and the third peak eluting at 23 minutes is a degradation product (GlcNAc-glycopeptide), indicative of an endogenous endoglycosidase in the extracts, which is active especially around pH 6 and, thus, is probably equivalent to the Endo S previously described [38]. By cleavage of the fucosyltransferase substrate, the endoglycosidase reduces the yield of fucosylated product.
Figure 5. Demonstration of a novel core α1,3-fucosyltransferase activity in Dictyostelium.
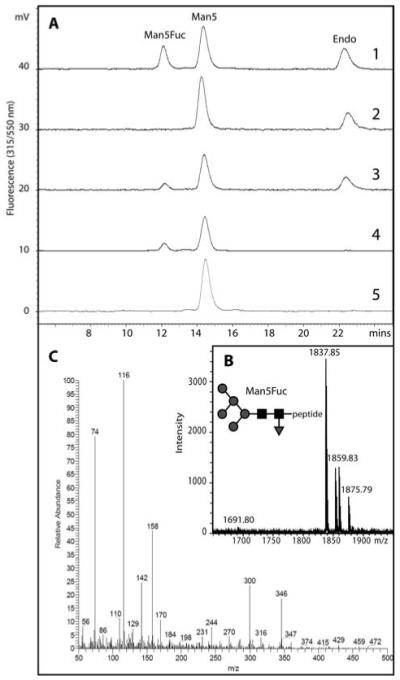
(A) A dansylated glycopeptide (dansyl-Asn-Ser-Man5GlcNAc2; Man5) was incubated in the presence of (1) a lysate of mid-log phase Dictyostelium AX3 cells and GDP-Fuc, (2) an AX3 lysate without GDP-Fuc, (3) an HL250 lysate and GDP-Fuc, (4) purified recombinant Caenorhabditis elegans FUT-1 and GDP-Fuc or (5) no enzyme. The shift to a lower retention time results in formation of a fucosylated glycopeptide (Man5Fuc). (B) The collected Man5Fuc fraction was subject to MALDI-TOF MS verifying the addition of fucose (m/z [M+Na]+ = 1837; as compared to the non-fucosylated from with m/z 1691). (C) The core 1,3-linkage of the transferred fucose was demonstrated by the presence of 3,4-disubstituted GlcNAc (2-deoxy-2-(N-methyl)acetamido-6-O-methyl-glucitol) in the obtained EI spectrum.
The earlier retention time of the first peak in comparison to the substrate was suggestive, based on previous studies on the properties of dansylated glycopeptides [39], that the fucose was α1,3-linked to the core; this supposition was supported by the co-elution of this peak with the Man5GlcNAc2Fuc1 product of FUT-1 (see also Figure 5A), an authentic recombinant Caenorhabditis core α1,3-fucosyltransferase [33]. The Dicytostelium fucosyltransferase product was also subject to linkage analysis by GLC-MS: assuming a standard pentasaccharide core structure in which the reducing-end GlcNAc is substituted at C-4 by the penultimate GlcNAc residue, the detection of a 1,3,4,5-tetra-O-acetyl-2-deoxy-2-(N-methyl)acetamido-6-O-methyl-glucitol derivative with its characteristic electron impact mass spectrum (Figure 5C) clearly confirms the presence of an N-acetylhexosamine residue which is substituted at both C-3 and C-4. This proved that the fucose was indeed attached to the core reducing-terminal GlcNAc in a 1,3-linkage; thus, we show that Dictyostelium indeed possesses a core α1,3-fucosyltransferase, which generates a linkage of the type recognised by anti-horseradish peroxidase. The presence of this activity in both AX3 and HL250 is compatible with a defect in the gmd gene, but not in a fucosyltransferase gene, being the molecular basis for the lack of anti-HRP epitopes in the HL250 strain.
Developmental alteration of glycan expression
In the older literature, changes in the glycosylation of Dictyostelium during the transition from amoebae, through the ‘tip’ stage, to fruiting body formation (‘culmination’) have been inferred from elution profiles of gel filtration or other chromatographic separations [4-6, 10]. However, no mass spectrometric analysis of the N-glycans of the different stages was ever performed. In order to propose exact structures for those glycans which are putatively differentially expressed during slime mould development, we placed concentrated AX3 axenic cultures on filter paper; development was observed and material with the expected morphology was collected after various time points (approximately 1 g of each stage). The material was mechanically disrupted and N-glycans were released using PNGase A and pyridylaminated as described for the axenic cultures. The subsequent analyses indeed show a distinct shift in the N-glycome of Dictyostelium during development as shown by both RP-HPLC and MALDI-TOF MS analyses. Particularly striking is the accumulation of smaller glycans (Figure 6), in comparison to the axenic culture, during development, as well as of glycans with reduced retention times on a normal-phase HPLC column and altered retention times on a reversed-phase HPLC column (Supplementary Figure 4); for instance, there is a reduction in the occurrence of the Hex8HexNAc4Fuc1 glycan eluting at 4.8 g.u. (m/z 2373) and an increase in Hex5HexNAc2Fuc1 eluting at 5.2 g.u. (m/z 1481).
Figure 6. MALDI-TOF MS analysis of Dictyostelium N-glycans expressed during development.
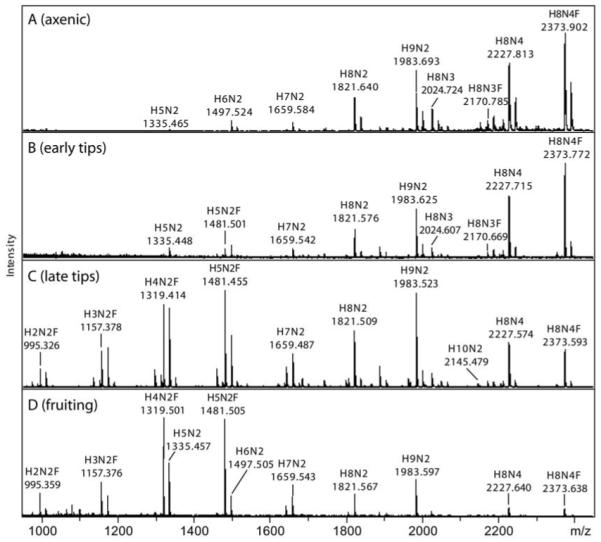
Pyridylaminated N-glycans derived from PNGase A digestion of axenic (A; 1.5 × 106 cells per ml), early tips (B; 10-12 hrs), late tips (C; 16 hrs) and fruiting bodies (D; 24 hrs) of the AX3 strain were analysed by MALDI-TOF MS. Glycans from each stage were prepared two or three times independently; the relative intensities of these typical spectra were calculated and are presented in Supplementary Table 1. The major [M+Na]+ species are annotated in the form HxNyF0-1
A closer examination of the data (see also Supplementary Table 1), shows that, in mid-log axenically-grown cells, glycans with the compositions Hex9HexNAc2, Hex8HexNAc4 and Hex8HexNAc4Fuc1 predominate, whereas the early tips also express some Hex5HexNAc2Fuc1. In late tips, the shift is more pronounced and Hex3-5HexNAc2Fuc0-1 species are present. The Hex3HexNAc2Fuc1 glycan from late tips has the same RP-HPLC elution time as a core α1,3-fucosylated glycan (so-called MMF3) previously examined from giant ragweed pollen (data not shown) and MS/MS analyses of the Hex3-5HexNAc2Fuc1 glycans demonstrated the presence of core fucosylation (see Figure 7 for an example). Finally, in AX3 fruiting bodies, Hex4-5HexNAc2Fuc0-1 are the major peaks, while the Hex8HexNAc4Fuc1 glycan is nearly absent; the Hex5HexNAc2 and Hex9HexNAc2 species have the same two-dimensional HPLC elution properties as standard Man5GlcNAc2 and Man9GlcNAc2. The smaller fucosylated structures from late tips and fruiting bodies were only apparent after release by PNGase A (Supplementary Figure 5); an indication that these are core α1,3-fucosylated is the data showing their completely absence from the PNGase F-released sample. The mannosidase sensitivity of Hex5HexNAc2, Hex5HexNAc2Fuc1 and Hex9HexNAc2 from fruiting bodies was tested by RP-HPLC and MALDI-TOF MS (data not shown). Of these three structures, only Hex9HexNAc2 was sensitive to Aspergillus saitoi α1,2-mannosidase (as judged by a change in RP-HPLC retention time and the reduction in m/z from 1961 to 1313), consistent with the presence of four α1,2-mannose residues on standard eukaryotic Man9GlcNAc2. Jack bean mannosidase, on the other hand, resulted in digestion of multiple mannose residues from all three glycans, resulting in products with m/z 827 for the putative Man5GlcNAc2 and Man9GlcNAc2 or m/z 973 for the putative Man5GlcNAc2Fuc1 structures.
Figure 7. Analysis of a putatively core α1,3-fucosylated N-glycan from Dictyostelium late tips.
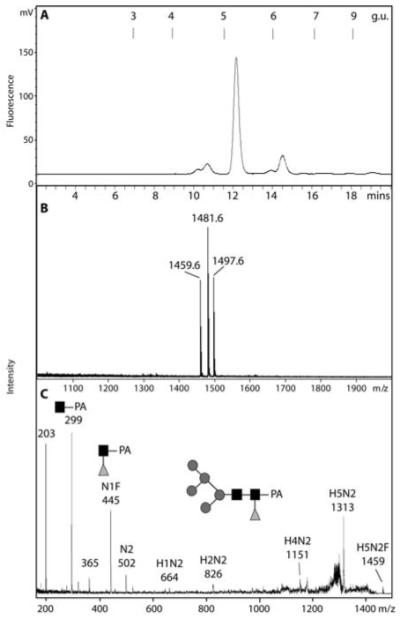
A) A normal-phase HPLC fraction (Palpak Type N; 8 g.u.) of late tip glycans released by PNGase A was analysed by RP-HPLC and shown to contain a major species with a retention time of 5.1 g.u., consistent with core α1,3-fucosylation. B) MALDI-TOF MS of the purified 5.1 g.u. fraction showed it to contain a species with m/z 1459.75 (Hex5HexNAc2Fuc1-PA; H+-adduct) as well as the corresponding Na+ and K+ adducts. C) The MS/MS fragmentation of this glycan shows the presence of an m/z 445 fragment consistent with the composition GlcNAc1Fuc1PA; key fragment ions are annotated. A comparable MS/MS spectrum was obtained from the corresponding glycan in a TSKgel Amide-80 (7.1 g.u.) fraction; the Hex5HexNAc2Fuc1 glycan was insensitive to both Aspergillus α1,2-mannosidase and bovine kidney α-fucosidase, but was sensitive to jack bean α-mannosidase (data not shown). The predicted structure of the glycan is shown according to the nomenclature of the Consortium for Functional Glycomics (black square, N-acetylglucosamine; grey circle, mannose; grey triangle, fucose).
N-glycomic comparison of axenic and non-axenic strains
Many Dictyostelium studies are based on using the AX3 strain; in the present study, using this axenic strain has the advantage of facilitating the generation of sufficient material for HPLC-based analyses; nevertheless, we sought to compare the N-glycomes of the fruiting bodies of axenic AX3 and the non-axenic NC4 strains. In the latter case, as the overall yield of fruiting bodies was limited, a recently-developed mini-method [27] was employed for the preparation of N-glycans. While some smaller putatively bisected or intersected glycans (Hex4-5HexNAc3Fuc1; m/z 1444 and 1606) were only present in the NC4 glycome, mass spectrometry indicated that the N-glycosylation repertoire of the axenic and non-axenic strains is rather similar as judged by the presence of Hex3-5HexNAc2Fuc1, Hex8-9HexNAc2 and Hex8HexNAc4Fuc0-1 in both samples (Figure 8).
Figure 8. Comparison of N-glycans from the fruiting bodies of axenic and non-axenic strains.
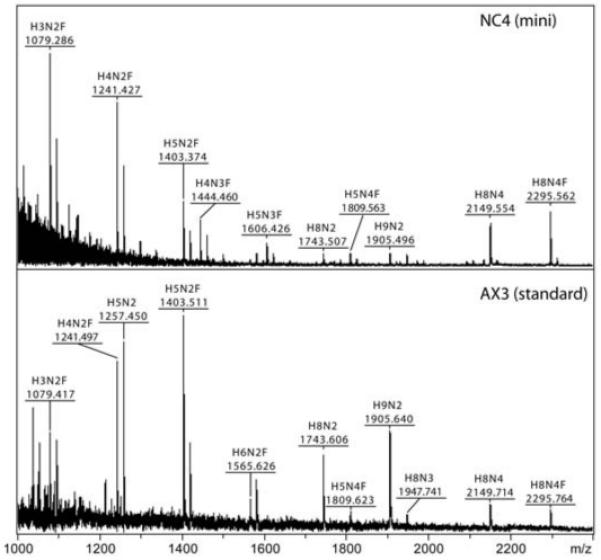
Underivatised PNGase A-released N-glycans derived from NC4 and AX3 fruiting bodies were subject to MALDI-TOF MS in the positive mode. The NC4 glycans were prepared using a small-scale method as described, whereas the AX3 glycans are from the standard large-scale preparation (for the results with the corresponding pyridylaminated glycans, see Figure 7 and Supplementary Figure 4). The major [M+Na]+ species are annotated in the form HxNyF0-1.
Discussion
Glycan analysis in Dictyostelium
The cellular slime mould Dictyostelium is a unique and interesting model for studying development, due to the transition from a unicellular state through to a fruiting body. Over twenty years ago, some structural glycobiologists also recognised the potential for examining the role of glycosylation in development using this system; in the past ten years, the most progress has been made in understanding O-glycosylation in the slime mould [40-43]. Considering the recent availability of the whole genome sequence, it is time, though, to reappraise the N-glycosylation of this organism. Particularly, the means of releasing the glycans from the parent glycopeptides and glycoproteins and the reliance on radiolabelling is one reason why the full potential of the N-glycome of Dictyostelium discoideum could not earlier be appraised. Current models of N-glycans in this species, as summarised in a review article [22] and which suggest difucosylation of the core and the presence of xylose, methylphosphate, sulphate or ‘intersecting’ GlcNAc residues, are based on a composite of various data from many sources and do not represent any single proven glycan structure.
In this study, we show for the first time that the major N-glycan of axenically-grown Dictyostelium indeed has a novel structure, containing core α1,3-fucose in addition to both intersecting and bisecting GlcNAc residues. During this study, we used release by either PNGase A (a plant enzyme capable of removing core α1,3-fucosylated glycans), PNGase F (a bacterial enzyme which cannot remove core α1,3-fucosylated glycans) or endoglycosidase H (a bacterial enzyme which can cleave vertebrate oligomannosidic or hybrid N-glycans). The fact that older studies based on use of endoglycosidase H did not specifically reveal the presence of core fucose or bisecting GlcNAc is a limitation of the enzyme itself. Since PNGase F cannot cleave core α1,3-fucosylated glycans [37], only the use of PNGase A could (and does) allow us to observe these structural elements for the first time.1 On the other hand, none of the structures we detected or characterised contain either xylose, methylphosphate or sulphate, regardless of the employed mode of either preparation or mass spectrometry; thereby, the apparent incorporation of [35S]sulphate or of [3H]methyl groups into slime mould glycopeptides or N-linked oligosaccharides [9, 16, 44] is not explained by our results.
Some of the older studies used the secreted lysosomal proteins from axenic cells incubated in buffer [9, 45]; we also prepared glycans from secreted proteins and the mass spectrum we obtained is rather similar to that with the whole axenic cells. Furthermore, we treated intact, denatured proteins with PNGase F, following a previously published protocol and detected the same glycans, lacking core fucose, as those released from pepsin-derived glycopeptides. Also lacking fucose are the glycans released by Endo H; however, endoglycosidase digestions of slime mould samples are known not to release all glycans, but a solvolysis procedure, based on one used to remove sulphate from proteoglycans, increased endoglycosidase sensitivity of Dictyostelium glycans [9]. One study used the phrase ‘sulphate and/or fucose’ to describe those residues released by this procedure [6]; thus, an uncertainty regarding the presence of core sulphate can be inferred from the older literature. It is, therefore, possible that the core modification released by solvolysis or by mild acid hydrolysis of solely [3H]mannose-labelled glycans is core α1,3-fucose and its removal would explain the ability to subsequently gain sensitivity to endoglycosidase H.
The anti-HRP epitope in Dictyostelium
In other species, core α1,3-fucose or β1,2-xylose linked to N-glycans are sources of cross-reactivity to anti-HRP, an antiserum raised against a plant glycoprotein containing both these modifications [34]; thus, in theory one or both of these modifications may be present in Dictyostelium. Previous studies have suggested that indeed fucose and/or xylose are components of the glycans of the slime mould. Both [3H]fucose labelled glycopeptides which are either sensitive or resistant to endoglycosidase H treatment have been described [4, 6]. In the case of the endoglycosidase-sensitive glycans produced late in development, the fucose label is not retained, suggesting that it is ‘lost’ with the GlcNAc-peptide portion; however, the nature of the linkage was not proven. In the structures we find in the axenic cells, a core α1,3-fucosylated bisected/intersected glycan is dominant and its nature is compatible with the endoglycosidase-resistant forms of fucosylated glycans being core α1,3-fucosylated. The presence of core α1,3-fucose also accounts for the anti-HRP reactivity of AX3 cells, while its absence in HL250 cells is compatible with the fucose-deficiency of this strain; the apparent lack of xylose from the N-glycans is also compatible with data indicating that the ‘anti-xylose’ fraction of anti-HRP does not cross-react with slime mould proteins [13]. The previous suggestion that xylose may be part of a slime mould N-glycan was based on the apparent identification of an unusual glycan with a single core GlcNAc residue [10], a finding which was not reproduced in our study.
It has been previously shown by others [13] that the intensity of anti-HRP reactivity (as judged by Western blotting) increases during tip formation and culmination. However, the overall occurrence of fucosylated N-glycans does not drastically change; certainly, it is noteworthy that the size of core fucosylated glycans tends to be smaller in late tips and fruiting bodies, with Hex3-5HexNAc2Fuc1 glycans apparent in the AX3 and NC4 strains at this stage. This may mean that the core α1,3-fucose is more accessible to the anti-HRP antibodies; certainly, previous data suggests that extension of the trimannosyl core region of an N-glycan has a negative effect on recognition by anti-HRP [46]. Thus, increased epitope recognition may reflect not increased core α1,3-fucosylation but an increased exposure of these epitopes in later development. Interestingly, another antibody (CAB4), raised against Dictyostelium, has been described to recognise core α1,6-fucosylation [13, 47] and reactivity to this antibody decreased during development [13]; considering the apparent lack of core α1,6-fucose on the glycans released, detected and characterised in our study, we have no explanation for this phenomenon. Furthermore, we observe no obvious ‘peripheral’ fucose, as was previously suggested to occur on slime mould N-glycans [11].
Glycogenomic potential of Dictyostelium
As judged by the N-glycans detected in this study, it is obvious that the glycosylation capacity of the slime mould is highly unique, at least in comparison to organisms whose glycans have been studied to date. N-glycans with the composition of Hex8HexNAc3-4Fuc0-1 carrying core fucose and intersecting/bisecting residues have not been found in other protozoa, yeast, plants, insects, nematodes and mammals. Thus, the biosynthesis and the underlying genetic basis of N-glycosylation in the slime mould are expected to show major differences as compared to other species. Most likely, the processing of the N-glycan precursor and transfer of Glc3Man9GlcNAc2 to protein will be indeed the same as in the majority of other eukaryotes, since dolichol-linked oligosaccharides with this composition have been detected [16] and the normal complement of alg genes is present in the slime mould genome [48]. Indeed, the different α1,2-mannosidase sensitivity of the structures of the Hex5HexNAc2 and Hex9HexNAc2 glycans is compatible with Glc3Man9GlcNAc2 being the major oligosaccharyltransferase substrate and indicates that Dictyostelium does not transfer Man5GlcNAc2 carrying two α1,2-linked mannose residues to proteins as is the case with Trichomonas vaginalis [48]; instead, the Man5GlcNAc2 present in tips and fruiting bodies is of the same structure as the Golgi intermediate in higher eukaryotes and probably the result of processing by class I mannosidases. Thereby, the initial trimming by ER mannosidase to yield the Man8B isomer of Man8GlcNAc2 is probably the same as in most eukaryotes, since no Hex9HexNAc4 glycans were detected in our study. Of the defined Dictyostelium N-glycosylation mutants, two (HL241 and HL243) display defects in dolichol-linked oligosaccharide biosynthesis [16] and another, modA, is an α-glucosidase II mutant, affecting the second trimming event in the endoplasmic reticulum [15].
The presence of bisecting GlcNAc in the slime mould has been previously only inferred from the properties of a GlcNAc transferase in cell extracts [49]; the apparent absence of bisected GlcNAc from previously-characterised slime mould glycans may be due to resistance to endoglycosidase H digestion, even though some other bisected N-glycans can be cleaved by this enzyme [50]. On the other hand, the capacity to generate structures containing intersecting GlcNAc has been shown by both glycan analysis and enzymatic assay [8, 49]. More recently, it has been proposed that β1,4-GlcNAc transferase homologues in the Dictyostelium genome are responsible for biosynthesis of such structures [22]. A reduction in the activity of the intersecting transferase during development has been previously postulated [51] and a reduction in peripheral HexNAc is confirmed by our glycan analyses.
Whether the additional GlcNAc residues are transferred before or after core fucosylation is not clear, since we show that a core α1,3-fucosyltransferase activity can be detected in slime mould extracts, which is capable of modifying Man5GlcNAc2. In this respect, the relevant enzyme is similar to the FUT-1 core α1,3-fucosyltransferase from Caenorhabditis elegans; however, the slime mould enzyme, unlike that from the worm, cannot accept a Man3GlcNAc2 (MM) structure. In contrast to plant and insect core α1,3-fucosyltransferases, both the worm and slime mould enzymes do not require the prior action of GlcNAc-TI [33]; in this respect, our data contradicts a previous study which suggested that Dictyostelium extracts can transfer fucose to a standard biantennary glycan (GnGn) containing non-reducing terminal GlcNAc [13]. Whereas the older study characterised the putative core α1,3-fucosyltransferase product by anti-HRP binding, we have verified its nature by structural analysis.
Potentially, the Dictyostelium fucosyltransferase(s) may accept a wide range of glycan substrates ranging from Man5GlcNAc2 through to the Man8GlcNAc4 structures. Which gene(s) encode(s) the core α1,3-fucosyltransferase activity is unclear; there are indeed ten slime mould members of GT10, the glycosyltransferase family whose members are all α1,3-fucosyltransferases. Further work is, therefore, necessary to determine more about these enzymes, their specificities and their effects on the glycome.
Certainly, though, all fucosylation reactions in the slime mould require the capacity to generate GDP-Fuc; in this study we have also verified the identity of the enzymes (GMD and GER) required for its de novo synthesis. One of these, GMD, is defective in the HL250 strain [17] and, based on sequencing, we postulate that a single amino acid substitution may be the basis of the mutation in the so-called modC gene. This mutation is presumed to account for the lack of anti-HRP epitopes in the HL250 strain (this study) and for the loss of reactivity to other antibodies (83.5 and MUD62) which recognise O-phosphoglycan epitopes [13, 19]. We do not postulate that the lack of anti-HRP epitopes and of core fucose necessarily has any biological effect, even though the HL250 strain is somewhat slow-growing and displays a spore germination defect; the presence of fucose within the cytoplasmic glycans modifying Skp1 [52], which has a role in mediating signals during Dictyostelium development, may account for the biological phenotypes of this strain. The ability to partially rescue the anti-HRP reactivity defect of HL250 by inclusion of exogenous fucose in the medium is consistent with the presence of potential fucokinase (FKP; XM_641090) and fucose-1-phosphate guanylyltransferase (FGPT; XM_633203) genes in Dictyostelium. The homologous enzymes in mammals are associated with a second (salvage) pathway of GDP-Fuc biosynthesis [53, 54].
One aspect of the glycomic changes during development is the overall trend towards increased occurrence of a population of smaller N-glycans [4]. Such glycans may be present in the whole tip or fruiting body or be restricted to a certain sub-population of cells in these stages; however, antibody or lectin reagents are not necessarily able to distinguish glycans sufficiently in order to show the exact location of these glycan types. Other data indicate there is no obvious difference in size and endoglycosidase sensitivity between the N-glycans of prestalk and prespore cells within slime mould slugs, but there may be a difference in the amount of fucose incorporated into these structures [55]. Regardless of which exact cells express these glycans, the switch to smaller glycans during development is quite possibly due to changes in the expression of processing α-mannosidases, although which of the seven class 38 and one class 47 homologues may be involved is yet to be resolved. However, a trend to increased α-mannosidase activity during development has been described in the M4 strain which lacks lysosomal α-mannosidase [51]. Certainly, knockouts will be required to ascertain the actual role of mannosidases and other enzymes in the developmental alteration of N-glycan structure.
Conclusion
In this study, we determined the structures of neutral N-glycans of the cellular slime mould Dictyostelium discoideum and show that these vary during the transition from axenic cells through to culmination. The use of modern methodologies allows us to identify, for the first time, a range of novel N-glycan structures in this organism; many aspects, particularly the type of fucosylation, of these glycans are also defined and enable us to explain the presence of anti-HRP epitopes in this organism. The foundation is, therefore, in place for a fresh look at the role of glycosylation in slime mould development and to define the molecular basis for the unusual modifications of its N-glycans.
Acknowledgments
The authors gratefully acknowledge the contribution of Stefan Karl, project student in the laboratory, to the characterisation of the Dictyostelium GMD and GER enzymes, Lukas Sobczak, MSc student, for the FUT-1 used as a control, Dr. Matthias Wieser for help with the flow cytometry, Dr. Friedrich Altmann for access to the Micromass Global ESI-Q-TOF MS. Dr. Katharina Paschinger for her critical reading of the manuscript and Prof. Wolfgang Nellen for his demonstration of Dictyostelium techniques. This work was supported by a grant to I.B.H.W. from the Austrian Fonds zur Förderung der wissenschaftlichen Forschung [P19615].
The abbreviations used are
- CI
chemical ionisation
- EI
electron impact ionisation
- ESI
electrospray ionisation
- FITC
fluorescein isothiocyanate
- GLC
gas liquid chromatography
- GER
GDP-keto-6-deoxymannose 3,5-epimerase/4-reductase (EC 1.1.1.271)
- GMD
GDP-mannose dehydratase (EC 4.2.1.47)
- g.u.
glucose units
- HRP
horseradish peroxidase
- MALDI-TOF
matrix-assisted laser desorption ionisation/time-of flight
- MMF3
Manα1-6(Manα1-3)Manβ1-4GlcNAcβ1-4(Fucα1-3)GlcNAcβ1-Asn
- MS
mass spectrometry
- PA
pyridylamino
- PBS
phosphate-buffered saline
- PNGase A
peptide:N-glycosidase from almonds
- RP-HPLC
reversed phase high pressure liquid chromatography
Footnotes
References
- 1.Devreotes P. Dictyostelium discoideum: a model system for cell-cell interactions in development. Science. 1989;245:1054–1058. doi: 10.1126/science.2672337. [DOI] [PubMed] [Google Scholar]
- 2.Ziska SE, Henderson EJ. Cell surface oligosaccharides participate in cohesion during aggregation of Dictyostelium discoideum. Proc Natl Acad Sci U S A. 1988;85:817–821. doi: 10.1073/pnas.85.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frazier WA, Rosen SD, Reitherman RW, Barondes SH. Purification and comparison of two developmentally regulated lectins from Dictyostelium discoideum. Discoidin I and II. J Biol Chem. 1975;250:7714–7721. [PubMed] [Google Scholar]
- 4.Ivatt RL, Das OP, Henderson EJ, Robbins PW. Glycoprotein biosynthesis in Dictyostelium discoideum: developmental regulation of the protein-linked glycans. Cell. 1984;38:561–567. doi: 10.1016/0092-8674(84)90510-5. [DOI] [PubMed] [Google Scholar]
- 5.Tschursin E, Riley GR, Henderson EJ. Differential regulation of glycoprotein sulfation and fucosylation during growth of Dictyostelium discoideum. Differentiation. 1989;40:1–9. doi: 10.1111/j.1432-0436.1989.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 6.Sharkey DJ, Kornfeld R. Developmental regulation of asparagine-linked oligosaccharide synthesis in Dictyostelium discoideum. J Biol Chem. 1991;266:18485–18497. [PubMed] [Google Scholar]
- 7.Gabel CA, Costello CE, Reinhold VN, Kurz L, Kornfeld S. Identification of methylphosphomannosyl residues as components of the high mannose oligosaccharides of Dictyostelium discoideum glycoproteins. J Biol Chem. 1984;259:13762–13769. [PubMed] [Google Scholar]
- 8.Couso R, van Halbeek H, Reinhold V, Kornfeld S. The high mannose oligosaccharides of Dictyostelium discoideum glycoproteins contain a novel intersecting N-acetylglucosamine residue. J Biol Chem. 1987;262:4521–4527. [PubMed] [Google Scholar]
- 9.Freeze HH, Yeh R, Miller AL, Kornfeld S. Structural analysis of the asparagine-linked oligosaccharides from three lysosomal enzymes of Dictyostelium discoideum. Evidence for an unusual acid-stable phosphodiester. J Biol Chem. 1983;258:14874–14879. [PubMed] [Google Scholar]
- 10.Amatayakul-Chantler S, Ferguson MAJ, Dwek RA, Rademacher TW, Parekh RB, Crandall IE, Newell PC. Cell surface oligosaccharides on Dictyostelium during development. J Cell Sci. 1991;99:485–495. [Google Scholar]
- 11.Ivatt RJ, Das OP, Henderson EJ, Robbins PW. Developmental regulation of glycoprotein biosynthesis in Dictyostelium. J Supramol Struct Cell Biochem. 1981;17:359–368. doi: 10.1002/jsscb.380170407. [DOI] [PubMed] [Google Scholar]
- 12.Crandall IE, Newell PC. Changes in cell surface glycoproteins during Dictyostelium development analysed using monoclonal antibodies. Development. 1989;107:87–94. doi: 10.1242/dev.107.1.87. [DOI] [PubMed] [Google Scholar]
- 13.Srikrishna G, Wang LY, Freeze HH. Fucoseβ-1-P-Ser is a new type of glycosylation: using antibodies to identify a novel structure in Dictyostelium discoideum and study multiple types of fucosylation during growth and development. Glycobiology. 1998;8:799–811. doi: 10.1093/glycob/8.8.799. [DOI] [PubMed] [Google Scholar]
- 14.Free SJ, Schimke RT, Freeze H, Loomis WF. Characterization and genetic mapping of modA. A mutation in the post-translational modification of the glycosidases of Dictyostelium discoideum. J Biol Chem. 1978;253:4102–4106. [PubMed] [Google Scholar]
- 15.Freeze HH, Yeh R, Miller AL, Kornfeld S. The mod A mutant of Dictyostelium discoideum is missing the α1,3-glucosidase involved in asparagine-linked oligosaccharide processing. J Biol Chem. 1983;258:14880–14884. [PubMed] [Google Scholar]
- 16.Freeze HH, Willies L, Hamilton S, Koza-Taylor P. Two mutants of Dictyostelium discoideum that lack a sulfated carbohydrate antigenic determinant synthesize a truncated lipid-linked precursor of N-linked oligosaccharides. J Biol Chem. 1989;264:5653–5659. [PubMed] [Google Scholar]
- 17.Gonzales-Yanes B, Mandell RB, Girard M, Henry S, Aparicio O, Gritzali M, Brown RD, Erdos GW, West CM. The spore coat of a fucosylation mutant in Dicytostelium discoideum. Dev. Biol. 1989;133:576–587. doi: 10.1016/0012-1606(89)90060-2. [DOI] [PubMed] [Google Scholar]
- 18.Freeze HH, Bush JM, Cardelli J. Biochemical and genetic analysis of an antigenic determinant found on N-linked oligosaccharides in Dictyostelium. Dev Genet. 1990;11:463–472. doi: 10.1002/dvg.1020110523. [DOI] [PubMed] [Google Scholar]
- 19.Champion A, Griffiths K, Gooley AA, Gonzalez BY, Gritzali M, West CM, Williams KL. Immunochemical, genetic and morphological comparison of fucosylation mutants of Dictyostelium discoideum. Microbiology. 1995;141:785–797. doi: 10.1099/13500872-141-4-785. [DOI] [PubMed] [Google Scholar]
- 20.Freeze HH, Lammertz M, Iranfar N, Fuller D, Panneerselvam K, Loomis WF. Consequences of disrupting the gene that encodes α-glucosidase II in the N-linked oligosaccharide biosynthesis pathway of Dictyostelium discoideum. Dev Genet. 1997;21:177–186. doi: 10.1002/(SICI)1520-6408(1997)21:3<177::AID-DVG1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Pang TL, Wu CJ, Chen PA, Weng YL, Chen MY. Dictyostelium gnt15 encodes a protein with similarity to LARGE and plays an essential role in development. Biochem Biophys Res Commun. 2007;360:83–89. doi: 10.1016/j.bbrc.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 22.West CM, van der Wel H, Coutinho PM, Henrissat B. Glycosyltransferase Genomics in Dictyostelium discoideum. In: Loomis WF, Kuspa A, editors. Dictyostelium Genomics. Horizon Scientific Press; Norwich: 2005. pp. 235–264. [Google Scholar]
- 23.Fey P, Kowal AS, Gaudet P, Pilcher KE, Chisholm RL. Protocols for growth and development of Dictyostelium discoideum. Nat Protoc. 2007;2:1307–1316. doi: 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- 24.Rendić D, Linder A, Paschinger K, Borth N, Wilson IBH, Fabini G. Modulation of neural carbohydrate epitope expression in Drosophila melanogaster cells. J. Biol. Chem. 2006;281:3343–3353. doi: 10.1074/jbc.M508334200. [DOI] [PubMed] [Google Scholar]
- 25.Altmann F, Fabini G, Ahorn H, Wilson IBH. Genetic model organisms in the study of N-glycans. Biochimie. 2001;83:703–712. doi: 10.1016/s0300-9084(01)01297-4. [DOI] [PubMed] [Google Scholar]
- 26.Davis SJ. Oligosaccharide heterogeneity of glycoproteins sulfated during the vegetative growth of Dictyostelium discoideum. J Cell Biochem. 1988;38:77–86. doi: 10.1002/jcb.240380202. [DOI] [PubMed] [Google Scholar]
- 27.Rendić D, Wilson IBH, Lubec G, Gutternigg M, Altmann F, Leonard R. Adaptation of the “in-gel release method” to N-glycome analysis of low-milligram amounts of material. Electrophoresis. 2007;28:4484–4492. doi: 10.1002/elps.200700098. [DOI] [PubMed] [Google Scholar]
- 28.Wilson IBH, Altmann F. Structural analysis of N-glycans from allergenic grass, ragweed and tree pollens: Core α1,3-linked fucose and xylose present in all pollens examined. Glycoconjugate Journal. 1998;15:1055–1070. doi: 10.1023/a:1006960401562. [DOI] [PubMed] [Google Scholar]
- 29.Paz-Parente J, Cardon P, Leroy Y, Montreuil J, Fournet B, Ricard G. A convenient method for methylation of glycoprotein glycans in small amounts by using lithium methyl-sulfinyl carbanion. Carbohydr. Res. 1985;141:41–47. doi: 10.1016/s0008-6215(00)90753-5. [DOI] [PubMed] [Google Scholar]
- 30.Geyer R, Geyer H. Saccharide linkage analysis using methylation and other techniques. Methods Enzymol. 1994;230:86–108. doi: 10.1016/0076-6879(94)30009-7. [DOI] [PubMed] [Google Scholar]
- 31.Pöltl G, Kerner D, Paschinger K, Wilson IBH. N-Glycans of the porcine nematode parasite Ascaris suum are modified with phosphorylcholine and core fucose residues. FEBS J. 2007;274:714–726. doi: 10.1111/j.1742-4658.2006.05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhomberg S, Fuchsluger C, Rendić D, Paschinger K, Jantsch V, Kosma P, Wilson IBH. Reconstitution in vitro of the GDP-fucose biosynthetic pathways of Caenorhabditis elegans and Drosophila melanogaster. FEBS J. 2006;273:2244–2256. doi: 10.1111/j.1742-4658.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- 33.Paschinger K, Rendić D, Lochnit G, Jantsch V, Wilson IBH. Molecular basis of anti-horseradish peroxidase staining in Caenorhabditis elegans. J. Biol. Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- 34.Paschinger K, Rendić D, Wilson IBH. Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj J. 2009;26:385–395. doi: 10.1007/s10719-008-9155-3. [DOI] [PubMed] [Google Scholar]
- 35.Ishihara H, Heath EC. The metabolism of L-fucose III. The biosynthesis of guanosine diphosphate L-fucose in porcine liver. J Biol Chem. 1968;243:1110–1115. [PubMed] [Google Scholar]
- 36.Kubelka V, Altmann F, Kornfeld G, März L. Structures of the N-linked oligosaccharides of the membrane glycoproteins from three lepidopteran cell lines (Sf-21, IZD-Mb-0503, Bm-N) Arch. Biochem. Biophys. 1994;308:148–157. doi: 10.1006/abbi.1994.1021. [DOI] [PubMed] [Google Scholar]
- 37.Tretter V, Altmann F, März L. Peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached α1→3 to the asparagine-linked N-acetylglucosamine residue. Eur. J. Biochem. 1991;199:647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- 38.Freeze HH, Etchison JR. Presence of a nonlysosomal endo-β-N-acetylglucosaminidase in the cellular slime mold Dictyostelium discoideum. Arch Biochem Biophys. 1984;232:414–421. doi: 10.1016/0003-9861(84)90557-5. [DOI] [PubMed] [Google Scholar]
- 39.Roitinger A, Leiter H, Staudacher E, Altmann F. HPLC method for the determination of Fuc to Asn-linked GlcNAc fucosyltransferases. Glycoconjugate J. 1998;15:89–91. doi: 10.1023/a:1006951802623. [DOI] [PubMed] [Google Scholar]
- 40.Mreyen M, Champion A, Srinivasan S, Karuso P, Williams KL, Packer NH. Multiple O-glycoforms on the spore coat protein SP96 in Dictyostelium discoideum. Fuc(α1-3)GlcNAc-α-1-P-Ser is the major modification. J Biol Chem. 2000;275:12164–12174. doi: 10.1074/jbc.275.16.12164. [DOI] [PubMed] [Google Scholar]
- 41.Van Der Wel H, Morris HR, Panico M, Paxton T, Dell A, Kaplan L, West CM. Molecular cloning and expression of a UDP-N-acetylglucosamine (GlcNAc):hydroxyproline polypeptide GlcNAc-transferase that modifies Skp1 in the cytoplasm of Dictyostelium. J Biol Chem. 2002;277:46328–46337. doi: 10.1074/jbc.M208024200. [DOI] [PubMed] [Google Scholar]
- 42.Wang F, Metcalf T, van der Wel H, West CM. Initiation of mucin-type O-glycosylation in Dictyostelium is homologous to the corresponding step in animals and is important for spore coat function. J Biol Chem. 2003;278:51395–51407. doi: 10.1074/jbc.M308756200. [DOI] [PubMed] [Google Scholar]
- 43.Ercan A, Panico M, Sutton-Smith M, Dell A, Morris HR, Matta KL, Gay DF, West CM. Molecular characterization of a novel UDP-galactose:fucoside α3-galactosyltransferase that modifies Skp1 in the cytoplasm of Dictyostelium. J Biol Chem. 2006;281:12713–12721. doi: 10.1074/jbc.M513664200. [DOI] [PubMed] [Google Scholar]
- 44.Freeze HH, Wolgast D. Biosynthesis of methylphosphomannosyl residues in the oligosaccharides of Dictyostelium discoideum glycoproteins. Evidence that the methyl group is derived from methionine. J Biol Chem. 1986;261:135–141. [PubMed] [Google Scholar]
- 45.Freeze HH, Wolgast D. Structural analysis of N-linked oligosaccharides from glycoproteins secreted by Dictyostelium discoideum. Identification of mannose 6-sulfate. J Biol Chem. 1986;261:127–134. [PubMed] [Google Scholar]
- 46.Bencúrová M, Hemmer W, Focke-Tejkl M, Wilson IBH, Altmann F. Specificity of IgG and IgE antibodies against plant and insect glycoprotein glycans determined with artificial glycoforms of human transferrin. Glycobiology. 2004;14:457–466. doi: 10.1093/glycob/cwh058. [DOI] [PubMed] [Google Scholar]
- 47.Srikrishna G, Varki NM, Newell PC, Varki A, Freeze HH. An IgG monoclonal antibody against Dictyostelium discoideum glycoproteins specifically recognizes Fucα1,6GlcNAcβ in the core of N-linked glycans - Localized expression of core-fucosylated glycoconjugates in human tissues. J Biol Chem. 1997;272:25743–25752. doi: 10.1074/jbc.272.41.25743. [DOI] [PubMed] [Google Scholar]
- 48.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. The diversity of dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc Natl Acad Sci U S A. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharkey DJ, Kornfeld R. Identification of an N-acetylglucosaminyltransferase in Dictyostelium discoideum that transfers an “intersecting” N-acetylglucosamine residue to high mannose oligosaccharides. J Biol Chem. 1989;264:10411–10419. [PubMed] [Google Scholar]
- 50.Tai T, Yamashita K, Ito S, Kobata A. Structures of the carbohydrate moiety of ovalbumin glycopeptide III and the difference in specificity of endo-β-N-acetylglucosaminidases CII and H. J Biol Chem. 1977;252:6687–6694. [PubMed] [Google Scholar]
- 51.Sharkey DJ, Kornfeld R. Developmental regulation of processing α-mannosidases and “intersecting” N-acetylglucosaminyltransferase in Dictyostelium discoideum. J Biol Chem. 1991;266:18477–18484. [PubMed] [Google Scholar]
- 52.West CM. Evolutionary and functional implications of the complex glycosylation of Skp1, a cytoplasmic/nuclear glycoprotein associated with ubiquitination. Cell Mol Life Sci. 2003;60:229–240. doi: 10.1007/s000180300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pastuszak I, Ketchum C, Hermanson G, Sjoberg EJ, Drake R, Elbein AD. GDP-L-fucose pyrophosphorylase - Purification, cDNA cloning, and properties of the enzyme. J Biol Chem. 1998;273:30165–30174. doi: 10.1074/jbc.273.46.30165. [DOI] [PubMed] [Google Scholar]
- 54.Hinderlich S, Berger M, Blume A, Chen H, Ghaderi D, Bauer C. Identification of human L-fucose kinase amino acid sequence. Biochem. Biophys. Res. Commun. 2002;294:650–654. doi: 10.1016/S0006-291X(02)00541-7. [DOI] [PubMed] [Google Scholar]
- 55.Riley GR, West CM, Henderson EJ. Cell differentiation in Dictyostelium discoideum controls assembly of protein-linked glycans. Glycobiology. 1993;3:165–177. doi: 10.1093/glycob/3.2.165. [DOI] [PubMed] [Google Scholar]
- 56.Feasley CL, West CM, Chia CP. N-Glycosylation of gp130 Implicated in Cell Interactions in Dictyostelium. Glycobiology. 2008;18:975–976. Abstract 145. [Google Scholar]


