Abstract
Objective
Variations in the noncoding single-nucleotide polymorphisms (SNPs) at positions 560 and 832 in the 5′ promoter region of the apolipoprotein E gene define genotypes that distinguish between high and low concentrations of plasma total and high-density lipoprotein cholesterol and triglycerides. We addressed whether these genotypes improve the prediction of ischemic heart disease (IHD) in subsamples of individuals defined by traditional risk factors and the genotypes defined by the ε2, ε3, and ε4 alleles in exon 4 of the apolipoprotein E gene.
Methods and Results
In a sample of 3686 female and 2772 male participants of the Copenhagen City Heart Study who were free of IHD events, 576 individuals (257 women, 7.0% and 319 men, 11.5%) were diagnosed as having developed IHD in 6.5 years of follow-up. Using a stepwise Patient Rule-Induction Method modeling strategy that acknowledges the complex pathobiology of IHD, we identified a subsample of 764 elderly women (≥65 years) with hypertriglyceridemia who had a history of smoking, a history of hypertension, or a history of both in which the A560T832/A560T832 and A560T832/A560G832 5′ 2-SNP genotypes had a higher cumulative incidence of IHD (172/1000) compared to the incidence of 70/1000 in the total sample of women.
Conclusions
Our study validates that 5′ apolipoprotein E genotypes improve the prediction of IHD and documents that the improvement is greatest in a subset defined by a particular combination of traditional risk factors in Copenhagen City Heart Study female participants. We discuss the use of these genotypes in medical risk assessment of IHD in the population represented by the Copenhagen City Heart Study.
Keywords: atherosclerosis, genetics, risk factors
Cholesterol accumulation in arterial walls is an important contributing factor in the development of ischemic heart disease (IHD).1 A plethora of variations in genes and their expressed products involved in lipid metabolism have been characterized.2–7 Fast and inexpensive gene measurement technologies can be used to identify genetic variations that predict interindividual variation in measures of lipid metabolism and risk of IHD.8–11 The promise is that the identified variants then would be used as additional information in risk assessment to guide the selection of nonpharmacological and pharmacological interventions to prevent initiation, progression, and severity of IHD.11
Variations in the gene coding the apolipoprotein E (apoE) protein have been implicated in predicting variation in plasma lipids and risk of IHD. APOE is a constituent of many atherogenic lipoprotein particles, such as triglyceride (TG)-rich chylomicrons and high-density lipoproteins (HDLs). The APOE molecule has 3 common isoforms (E2, E3, and E4) encoded by variation in 3 common alleles (ε2, ε3, and ε4) defined by 2 single-nucleotide polymorphisms (SNPs) in exon 4 of the APOE gene. Studies assessing the role of the APOE gene in lipid metabolism have repeatedly demonstrated that variations in plasma total cholesterol (T-C) and TG levels are associated with variation among genotypes defined by the ε2, ε3, and ε4 alleles.12,13 Both animals and humans carrying the ε4 allele also are at a higher risk for developing atherosclerosis.14–17
In a previous study, we15 demonstrated that common variations in the noncoding SNPs located at positions 560 and 832 in the 5′ promoter region of the APOE gene define 3 genotypes (A560T832/A560T832, A560T832/A560G832, and A560T832/T560T832) that distinguish between high and low concentrations of plasma T-C, HDL-cholesterol (HDL-C), and TG in 4 independent samples ascertained to represent the human population at large. This observation led us to hypothesize that interindividual differences in the risk of developing IHD may be associated with variation in these 5′ genotypes. A test of this hypothesis using a large population based sample ascertained by the Copenhagen Heart Study (CCHS) established an increased hazard of developing IHD in women carrying the A560T832/T560T832 genotype.18 Although quantitatively not very large, the estimated hazard remained statistically significant after the effects of dyslipidemia; other established risk factors and the genotypes defined by the ε2, ε3, and ε4 alleles were considered in the prediction model.
Assumptions that are implicit in the application of the Cox proportional hazards model that we used to evaluate IHD risk in the previous study may limit the medical use of the risk information obtained. Most important, use of a single model assumes that the expected relationship between disease status and variation in risk factor traits is the same for all individuals in the sample under study. The complex multifactorial nature of the pathobiology of IHD end points renders this assumption untenable. Development of IHD is an emergent property of interactions of many susceptibility genes and many environmental factors. There is no evidence that any of these factors act as independent agents whose phenotypic effects are additive, exchangeable with one another, and the same for each individual in the population at large. Furthermore, because the combined number of interacting genes and environments is large, every incident IHD case cannot have experienced effects of the same combination of genetic variants and exposures to high-risk environments. These considerations suggest that the goal of a model building strategy is not to determine which combination of risk factors in a single linear model predicts risk for every individual at risk, but how many models are necessary to best predict risk of disease. In this article, we explore whether variation in 5′ genotypes of the APOE gene that contribute to the prediction of IHD when a single prediction model is used makes a larger contribution to the prediction when multiple models are used to estimate risk of disease in subsamples of individuals defined by particular combinations of risk factor values.
The augmented Patient Rule-Induction Method (PRIM) developed by Dyson et al19,20 (G Dyson, CF Sing, submitted for publication) is a novel model building strategy for evaluating risk that acknowledges the etiologic heterogeneity of the disease. It is designed to identify combinations of risk factor values that characterize mutually exclusive subgroups of individuals that differ in average risk as measured by the cumulative incidence of the disease of interest. This analytical strategy addresses which combination of values of which subset of risk factors best predicts the disease of interest in which subset of individuals of the population from which the sample under study was drawn.
In this article, we use a stepwise application of the PRIM to test the hypothesis that information about 5′ APOE genotypes significantly improves the prediction of IHD in particular subsamples of individuals characterized by selected subsets of values of the traditional risk factors. We identified a subsample of 764 elderly women with hypertriglyceridemia who had a history of smoking, a history of hypertension, or a history of both in which the A560T832/A560T832 and A560T832/A560G832 5′ APOE genotypes had a significantly higher cumulative incidence of IHD (172/1000) compared to the cumulative incidence of 70/1000 in the total sample of 3686 women. The implications for the added value of this genetic information in the practice of medicine are discussed.
Methods
Study Participants
The CCHS is a prospective study of the Danish population at large age 20 years or older on entry into the study.21–23 The initial survey was carried out between 1976 and 1978. A follow-up survey was performed between 1991 and 1994. This follow-up survey serves as the baseline survey for the study reported here. Altogether, 16 563 individuals were invited to take this survey, 10 135 participated (response rate, 61%), and 9259 gave blood for DNA extraction. A subsample of middle-aged and elderly individuals who were at least 45 years old and free of IHD when they were seen for the follow-up survey was selected for our study. Clinical data and genotype information on the 4 APOE SNPs considered in this study were available on 3686 women and 2772 men who satisfied the selection criteria. Informed consent was obtained from all participants. More than 99% were Europeans of Danish descent. The study was approved by the Danish Ethics Committee for the City of Copenhagen and Frederiksberg (No. 100.2039/91).
Variable Definitions
All 6458 participants of the CCHS selected for this study were free of IHD at baseline. IHD was evaluated during the period from baseline to December 31, 1999. Information to establish the diagnoses of IHD (World Health Organization International Classification of Diseases, 8th edition, codes 410 to 414; 10th edition, codes I20 to I25) was gathered from the Danish National Hospital Discharge Register, the Danish National Register of Causes of Death, and medical records of general practitioners and hospitals. During the follow-up, 576 participants (257 of 3686 women, 7.0%; 319 of 2772 men, 11.5%) had developed IHD. The observed average follow-up time until an IHD event or the censure date of December 31, 1999, was 6.5 years (range, 0.01 to 8.2). The total exposure to risk of developing IHD was 39 648 person-years.
Baseline plasma HDL-C, TG, and T-C concentrations were measured by standard enzymatic assays (Boehringer Mannheim, GmbH Diagnostics, Mannheim, Germany) at the Department of Clinical Biochemistry, Rigshospitalet, Copenhagen University Hospital (Copenhagen, Denmark).21–22 Recommendations of the National Cholesterol Education Program Expert Panel, National Institutes of Health (Bethesda, Md) were used to define dyslipidemic subgroups (National Cholesterol Education Program, National Heart, Lung, and Blood Institute 2002).23 Dyslipidemia was diagnosed when an individual’s plasma T-C concentration was ≥200 mg/dL (5.18 mmol/L), TG was ≥150 mg/dL (1.60 mmol/L), or HDL-C was <40 mg/dL (1.04 mmol/L).
The definitions of smoking habit, glucose metabolism, and blood pressure used in our study are described in a previous report of the CCHS.24 Briefly, each factor was dichotomized to define a high-risk group; a history of smoking (current-smoker at any examination), a history of diabetes (self-reported disease, use of insulin, use of oral hypoglycemic drugs, nonfasting plasma glucose ≥11.1 mmol/L at any examination), or a history of hypertension (systolic blood pressure ≥140 mm Hg, diastolic blood pressure ≥90 mm Hg, use of antihypertensive drugs, or any combination of these at any examination).
The 4 SNPs in APOE were genotyped by polymerase chain reaction and restriction enzyme digestion as previously described in other studies.21,22,25
Statistical Analyses
The χ2 statistic was used to test homogeneity of the relative frequencies of dichotomous risk factor traits between genders.
The PRIM Algorithm
The PRIM algorithm was first introduced by Friedman and Fisher26 and augmented by Dyson et al19,20 (G Dyson CF Sing submitted for publication) for application to genetic studies. The PRIM enables one to partition the total sample of individuals into multiple subsamples, each defined by a subset of predictor variables. The augmented the PRIM algorithm selects the subset of statistically significant terms, defined by values of 1 or more predictor variables, that maximize the mean outcome of a response variable of interest in a selected subsample of individuals. The selected subsample satisfies the minimum size criterion denoted by the support parameter, β. β defines the minimum proportion of the sample of individuals not previously assigned to a subsample as a consequence of applying the PRIM algorithm who must be included in establishing a new subsample. The optimum value of β for a particular application of the PRIM is chosen according to the algorithm described by Dyson et al.19 We applied the PRIM algorithm using terms for the peeling and pasting processes defined by combinations of values of 2 predictor variables.20 The hypergeometric distribution was used to derive the theoretical null distribution (G Dyson, CF Sing, unpublished data) used to test whether the cumulative incidence of IHD associated with a particular peeling or pasting term was statistically significant. The multiple hypothesis tests conducted during the execution of the PRIM each uses a nominal significance threshold of 0.023, which corresponds to an experiment-wise significance level of 0.05.20 Multiple mutually exclusive subsamples of the total sample may be produced. Each subsample of the original sample includes individuals with the same values for a subset of predictor variables. The individuals who are not included in any of the subsamples produced by the peeling and pasting processes are assigned to a remainder subsample.
Stepwise Application of the PRIM Algorithm
The PRIM algorithm was first used to identify subsamples of individuals who are each characterized by different subsets of values for age, the 3 plasma measures of dyslipidemia (T-C, HDL-C, and TG), the 3 high-risk groups defined by 3 established risk factors (diabetes, smoking, and hypertension), and the combined {ε22, ε32}, {ε33}, and {ε42, ε43, and ε44} APOE genotype groups. We carried out a second application of the PRIM algorithm in each of the sub-samples to test whether variation in the 5′ APOE genotypes improved the prediction of the cumulative incidence of IHD in any of the subsamples. The improvement in prediction of the cumulative incidence of IHD using the 5′ APOE genotypes in one of the first step subsamples and not in another is interpreted as evidence for nonadditive relationships between the effects measured by the established risk factors that characterize the sub-samples and the genetic effects marked by the 5′ APOE genotypes. The values of the subset of risk factors that the PRIM selects to define a subsample are expected to vary from subsample to subsample because of the heterogeneity of the relationships between the risk of IHD and the etiologic causes among individuals in a representative sample of the population at large.
Results
A Descriptive Summary of the Sample
Summary statistics that describe the female and male samples are given in Table 1. Using a 5% test criterion, none of the SNP genotypes significantly deviated from the Hardy-Weinberg expectations in either of the genders, and the relative allele frequencies were not significantly different between genders. Relative frequencies of the four 5′ APOE genotypes and the 3 groups of traditional APOE genotypes defined by the ε2, ε3, and ε4 alleles are given in Table 2. Additionally, there was no statistically significant evidence for heterogeneity of relative frequencies of these 2 SNP genotypes between genders.
Table 1.
A Description of the CCHS Samples
| Trait | Women (n=3686) | Men (n=2772) | P-value (Using χ2 Test) |
|---|---|---|---|
| Cumulative incidence of IHD/1000 individuals at risk | 70 | 115 | <0.001 |
| Average follow-up time, y | 6.3* | 5.9* | |
| Predictors of IHD | |||
| Age older than 65 y, % | 51 | 43 | <0.001 |
| Blood measures of dyslipidemia, % | |||
| HDL-C <40 mg/dL | 6 | 20 | <0.001 |
| TG ≥150 mg/dL | 41 | 52 | <0.001 |
| T-C ≥200 mg/dL | 89 | 80 | <0.001 |
| Other established risk factor characters, % | |||
| Smoking | 57 | 70 | <0.001 |
| Diabetes | 4 | 8 | <0.001 |
| Hypertension | 62 | 69 | <0.001 |
The range of follow-up time was 0.01 to 8.2 years in both genders.
Table 2.
Relative Genotype Frequencies in the CCHS Samples
| Relative Frequency (N) |
|||
|---|---|---|---|
| Genotype Groups | Women | Men | P-value (Using χ2 test) |
| 5′ promoter region | |||
| A560T832/A560G832 | 0.35 (1290) | 0.36 (1010) | 0.438 |
| A560T832/A560T832 | 0.16 (583) | 0.15 (416) | |
| A560T832/T560T83 | 0.06 (213) | 0.06 (174) | |
| Others | 0.43 (1600) | 0.43 (1172) | |
| Exon 4 region | |||
| {ε33} | 0.56 (2066) | 0.58 (1596) | 0.335 |
| {ε22, ε32} | 0.13 (475) | 0.13 (362) | |
| {ε42, ε43, ε44} | 0.31 (1145) | 0.29 (814) | |
The widely acknowledged evidence that the natural history of IHD is gender specific, combined with the statistically significant differences in the gender-specific cumulative incidence of IHD and the statistically significant differences in gender-specific frequency distributions of the outcomes of the proposed predictor variables (Table 1), justifies carrying out model building and hypothesis testing strategies separately in the female and male samples.
Step 1 PRIM Analysis
For women, the optimum value of the support parameter was β = 0.20. The selected risk factors and their values that characterized each of 2 statistically significant, mutually exclusive subsamples (FS 1 and FS 2) and a remainder subsample (FS 3) based on the information obtained from the application of the PRIM are given in Table 3. The estimated cumulative incidences of IHD in these subsamples ranged from 33 to 139 cases/1000 women at risk. The highest incidence of IHD in the subsample (FS 1) of elderly women is almost twice as large as the estimate in the total sample of women (139 versus 70 cases/1000, respectively). Everyone in subsample FS 1 (n=764, 20.7% of the total sample) was aged 65 years or older at baseline; had hypertriglyceridemia (TG ≥150 mg/dL); and had a history of smoking, a history of having had hypertension, or a history of both. The second subsample (FS 2, n=839, 22.8% of the total sample), with the second highest estimate of cumulative incidence of IHD (99 cases/1000), consisted of 3 subgroups. The first subgroup consisted of 799 elderly women with low TG (<150 mg/dL) and a history of hypertension. The second subgroup consisted of 6 elderly women with low TG who had a history of smoking and a history of diabetes but no history of hypertension. The third subgroup consisted of 34 middle-aged women (45 to 64 years) who had a history of smoking and a history of diabetes. The estimated cumulative incidence of IHD in the remainder subsample (FS 3) of 2083 individuals (56.5%) who were not assigned to either of the 2 high-risk subsamples was 50% smaller than the estimate in the total female sample (33 versus 70 cases/1000, respectively).
Table 3.
The Subsamples of Women (n=3686, I=70) Defined by the Step 1 PRIM Analysis
| Subsample, n (%) | Cumulative Incidence (95% CI) | Description | Subgroup, n (%) |
|---|---|---|---|
| FS 1, 764 (20.7) | I=139 (119, 161) | Age ≥65 y and TG ≥150 mg/dL (and SMK and/or HYT) | 764 (20.7) |
| FS 2, 839 (22.8) | I=99 (82, 128) | Age ≥65 y and TG <150 mg/dL and HYT | 799 (21.7) |
| Or | |||
| Age ≥65 y and TG <150 mg/dL and SMK and no HYT and DIAB | 6 (0.2) | ||
| Or | |||
| Age 45 to 64 y and SMK and DIAB | 34 (0.9) | ||
| FS 3, 2083 (56.5) | I=33 (25, 41) | Age ≥65 y and no SMK and no HYT | 177 (4.8) |
| Or | |||
| Age ≥65 y and TG <150 mg/dL and SMK and no HYT and no DIAB | 157 (4.3) | ||
| Or | |||
| Age 45 to 64 y and (no SMK and/or no DIAB) | 1749 (47.4) |
I indicates number of cases per 1000 individuals at risk; HYT, history of hypertension; DIAB, history of diabetes; SMK, history of smoking.
For men, the optimum value of the support parameter was β=0.05. The selected risk factors and their values that characterized each of the 3 statistically significant, mutually exclusive subsamples (MS 1, MS 2, and MS 3) and a remainder subsample (MS 4, n=1698, 61.3% of the total sample) based on the information obtained from the application of the PRIM are given in Table 4. The estimated cumulative incidence of IHD in these 4 subsamples ranged from 65 to 209 cases/1000. On average, the estimates in the first 3 subsamples are 3 times larger (mean incidence=194 cases/1000) than the estimated cumulative incidence of IHD in the remainder subsample (65 cases/1000).
Table 4.
The Subsamples of Men (n=2772, I=115) Defined by the Step 1 PRIM Analysis
| Subsample, n (%) | Cumulative Incidence (95% CI) | Description | Subgroup, n (%) |
|---|---|---|---|
| MS 1, 234 (8.4) | I=209 (185, 273) | Age ≥65 y and HDL-C <40 mg/dL | 234 (8.4) |
| MS 2, 741 (26.7) | I=189 (171, 248) | Age ≥65 y and HDL-C ≥40 mg/dL and HYT | 741 (26.7) |
| MS 3, 99 (3.6) | I=192 (123, 296) | Age ≥ 65 y and HDL-C ≥ 40 mg/dL and DIAB and (ε33 or ε42 or ε43 or ε44) and no HYT | 10 (0.4) |
| Or | |||
| Age 45 to 64 y and DIAB and (ε33 or ε42 or ε43 or ε 44) | 89 (3.2) | ||
| MS 4, 1698 (61.3) | I=65 (54, 77) | Age ≥65 y and HDL-C >40 mg/dL and no HYT and [no DIAB and/or (ε22 or ε32)] | 213 (7.7) |
| Or | |||
| Age 45 to 64 y and no DIAB and/or (ε22 or ε32) | 1485 (53.6) |
I indicates number of cases/1000 individuals at risk; HYT, history of hypertension; DIAB, history of diabetes; SMK, history of smoking.
Most men assigned to the 3 high-risk subsamples were older than 65 years (985 of 1074 men). Subsample MS 1 consisted of 234 elderly men (8.4% of the total sample) with low plasma HDL-C concentration. Subsample MS 2 consisted of 741 elderly men (26.7% of the total sample) with HDL-C ≥40 mg/dL and a history of hypertension. Subsample MS 3 (n=99, 3.6% of the total sample) included 2 subgroups. The first subgroup consisted of 10 elderly men with HDL-C ≥40 mg/dL who had a history of diabetes, had 1 of the 4 genotypes (ε33, ε42, ε43, or ε44), and did not have a history of hypertension. The second subgroup included 89 middle-aged men (45 to 64 years) who had a history of diabetes and 1 of the 4 genotypes (ε33, ε42, ε43, or ε44).
Step 2 PRIM Analysis
We next applied the PRIM to select combinations of 5′ APOE genotypes that identify statistically significant genetic subgroups of each of the 2 subsamples and the remainder subsample of women and each of the 3 subsamples and the remainder subsample of men identified in the application of the PRIM to the predictor variables considered in step 1. A statistically significant high-risk genetic subgroup of the first female subsample (FS 1, Table 5) was identified. It included elderly women with hypertriglyceridemia who had a history of smoking, a history of having had hypertension, or a history of both and were carriers of one of the two 5′ genotypes (A560T832/T560T832 or A560T832/A560G832). The cumulative incidence of IHD in this genetic subgroup of 308 women is 172/1000 compared to 116/1000 in the subgroup with the A560T832/A560T832 or a genotype in the “others” group of genotypes. There was no statistically significant evidence that 5′ genotypes improved the prediction of IHD in the FS 2 and FS 3 subsamples (Table 5).
Table 5.
The Genetic Subgroups Defined by the Step 2 PRIM Analysis in Women
| Genetic Subgroup | Overall | Subsample FS 1 | FS 2* | FS 3* |
|---|---|---|---|---|
| A560T832/T560T832 or A560T832/A560G832 | N=1503, I=77 | N=308, I=172 (134, 217) (FS 1–1) | N=348, I=98 | N=847, I=34 |
| A560T832/A560T832 or others† | N=2183, I=65 | N=456, I=116 (82, 143) (FS 1–2) | N=491, I=100 | N=1236, I=32 |
The subsample in italics indicates the significant genetic subgroup from subsample S 1. I indicates number of cases/1000 individuals at risk (CI).
A CI cannot be calculated because the PRIM did not define this contrast in these subsamples. (See Dyson et al19 for more details.)
“Others” is the group of 5′ genotypes that does not include A560T832/T560T832, A560T832/A560G832, or A560T832/A560T832.
We did not detect a statistically significant genetic subgroup of any of the 3 subsamples or the remainder subsample identified in the first step the PRIM analysis of the male sample.
Discussion
A Prevailing Prevention Paradigm in Cardiology
A major goal of medical research is to identify subpopulations of individuals at increased risk of disease to efficiently allocate limited resources in a way that will maximize the reduction of individual suffering. Cardiovascular research has a long history of establishing the information collected on individuals that is useful in medical practice to identify those who are at increased risk of developing clinical symptoms of disease8,27–29 and those who would benefit from various pharmacological interventions to prevent, or even reverse, the progression of atherosclerosis.30,31 However, for most common chronic diseases having a complex multifactorial etiology (including IHD), only a fraction of individuals who develop disease are identified by the established risk factors.29,32–34 It is widely accepted that the ability to accurately predict those at risk may be significantly improved by considering genomic information.9,11
The immensity of the amount of genomic variation that may be considered and the complexity of how such variation may influence the initiation, progression, and/or severity of IHD8 makes identifying relevant variations a daunting task. Etiologic heterogeneity among those with disease and the primary role that interactions between genetic elements and environmental exposures play in determining risk are biological realities that make the search for genetic variations with possible predictive value a challenge that has not been adequately addressed. The applications of traditional multivariable linear regression to case-control data and Cox proportional hazards modeling of longitudinal data that identify predictors to evaluate the contribution of interactions of those predictors to risk of disease assume etiologic homogeneity among all individuals at risk and no correlations between predictor variables. These assumptions are untenable when analyzing data from observational studies designed to represent the population at large. Applications of such single-model approaches have led to the identification of genetic variations that make very small improvements to the prediction of common complex disease end points such as IHD.8a In the study reported here, we used a complementary analytic strategy designed to take etiologic heterogeneity and nonadditivity of predictor variable effects into consideration when evaluating the use of genetic variation in the identification of individuals at increased risk for IHD.
Biological Plausibility of the Risk Information Provided by PRIM Analyses
Using the traditional risk factors for atherosclerosis and 3 groups of traditional APOE genotypes defined by the ε2, ε3, and ε4 alleles, we found 2 statistically significant, mutually exclusive high-risk subsamples in women and 3 subsamples in men defined by different combinations of predictors (Tables 3 and 4). These subsamples are consistent with common knowledge that all the high-risk values for all the risk factors, which are involved in determining risk of IHD in the population at large, are not expected to be present in all individuals at risk.8 Our finding that different combinations of risk factors and their values are associated with different high-risk subsamples of women and men is consistent with well-established knowledge that genders differ in the natural history of the development of IHD.35–37
Variations in the values of a number of established risk factors, which may be internal (eg, plasma concentrations of T-C, TG, and HDL-C) or external (eg, exposure to tobacco smoking) to an individual, are hypothesized to combine with variations in the products of hundreds of genes to determine interindividual differences in initiation, progression, and severity of atherosclerosis, and these relationships are dynamic over the life cycle.8 The APOE gene is one of the few extensively studied candidate genes that has been repeatedly implicated in contributing to the determination of risk of IHD.7,17 In most studies, the contribution of the traditional genotypes defined by the ε2, ε3, and ε4 alleles add a small improvement in prediction of IHD risk beyond the traditional risk factors over a wide range of environmental and genetic backgrounds. We found in our previous study18 of the CCHS participants that the traditional genotypes defined by the ε2, ε3, and ε4 alleles do not statistically significantly improve the prediction of IHD in either gender. However, in the study reported here using the same CCHS sample, the traditional APOE genotypes improved the prediction of IHD in men in 2 of the 4 subsamples identified by the PRIM (Table 4).
In our earlier studies15,18 of the CCHS sample considered here, we established that particular 2 SNP 5′ genotypes influenced variability in plasma measures of lipid metabolism and variation in risk of IHD beyond that contributed by the traditional risk factors and the traditional APOE genotypes, particularly in women. This inference is supported by studies that report that estrogen response elements that modulate the response of the gene to estrogen are marked by the 5′ SNPs.38 Our current study further suggests that the small contribution of selected 5′ genotypes to improve risk prediction in the total sample of women is attributable to only 1 of the 3 subsamples obtained from the application of the PRIM (Table 6), as evidenced by a considerably larger statistically significant hazard ratio. That this validated effect is greatest in a sample of individuals with high TG suggests that the 5′ genotypes mark genetic elements that have pleiotropic effects on unmeasured or unknown intermediate traits that influence risk of IHD.
Table 6.
Hazard Ratio (P) for Genetic Subgroups Defined by the Step 2 PRIM Analysis in Women
| Subsample |
||||
|---|---|---|---|---|
| Genetic Subgroup | Overall | FS 1 | FS 2 | FS 3 |
| Hazard ratio (ignoring established risk factors) | ||||
| A560T832/T560T832 or A560T832/A560G832 | 1.242 (0.085) | 1.524 (0.031) (FS 1-1) | 0.998 (0.994) | 1.164 (0.538) |
| A560T832/A560T832 or others* | 1.0 | 1.0 (FS 1-2) | 1.0 | 1.0 |
| Hazard ratio (conditional on fitting established risk factors) | ||||
| A560T832/T560T832 or A560T832/A560G832 | 1.310 (0.038) | 1.517 (0.037) (FS 1-1) | 1.326 (0.243) | 1.066 (0.801) |
| A560T832/A560T832 or others* | 1.0 | 1.0 (FS 1-2) | 1.0 | 1.0 |
“Others” is the group of 5′ genotypes that does not include A560T832/T560T832, A560T832/A560G832, or A560T832/A560T832.
Consistent with the conceptual or theoretical biological modeling of the role of genetic variation in determining the risk of having a complex multifactorial disease,8,39–42 we find that the added value of genetic information for prediction depends on the genetic background and environmental contexts, which are characterized by the subsamples identified by the PRIM. This result supports the argument that traditional statistical approaches to evaluate genetic variation that estimate only marginal, context-independent effects are inconsistent with the ubiquity of biological interactions that determine the pathobiology of IHD.8,11,19,20,43–46 We next turn to a discussion of the implications of using genetic information for evaluating risk that come from our study of IHD.
Use of 5′ Genotypes of the APOE in Medical Risk Stratification
Any medical act requires the establishment of relevant information and a rationale for using it in making a decision. A traditional act in everyday medical practice is one in which the clinician, after logically considering all information available (ie, signs, symptoms, and laboratory results) assigns an individual under investigation to either a high-risk group or a low-risk group.47,48 The stepwise the PRIM algorithm used in this study is consistent with, and complementary to, such a clinical decision-making strategy. In the samples under study, women can first be stratified into 3 and men into 4 sub-samples that differ in their cumulative incidence of IHD. Because of the historical precedence established by clinical practice, these strata would be considered first in the evaluation of risk followed by consideration of the inclusion of genetic information. In our particular study, we found that 1 high-risk subsample of women (FS 1) can be further stratified into 2 subgroups based on their 5′ APOE genotype. This kind of context-dependent genetic information is expected to improve the traditional act of assignment of risk for IHD.
Several kinds of information may be used in making a decision about whether the statistically significant stratifications based on the 5′ APOE genotypes should be incorporated into the medical act of making a decision about whether a woman is at high risk for IHD: (1) age-specific propensity, (2) sensitivity and specificity of the stratification into high-and low-risk groups, and (3) the positive predictive value (+PV) of the decision based on the stratification strategy.
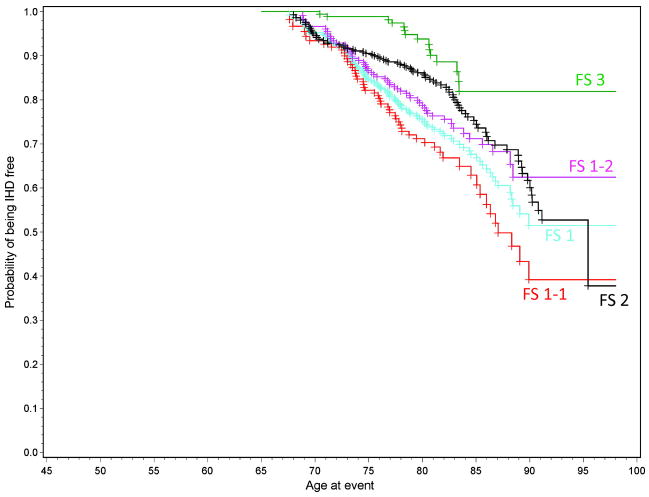
The age-specific propensities for developing IHD for individuals in a particular stratum can be derived from the probability of surviving free of IHD to a particular age. In the Figure, we present survival curves for elderly women who are included in subsamples FS 1, FS 2, and FS 3. Those in FS 1 who have high TG (≥150 mg/dL) and who have a history of smoking, a history of having had hypertension, or a history of both have approximately a 0.50 probability of developing IHD by 90 years of age compared to 0.20 among the elderly women in the low-risk subpopulation (FS 3). If the women in the FS 1 high-risk subpopulation carry either the A560T832/T560T832 or the A560T832/A560G832 2-SNP 5′ APOE genotype (genetic subgroup FS 1-1), their propensity for developing IHD by 90 years of age is ≈0.65 compared to <0.40 in the women who do not carry either of the proposed high-risk genotypes (genetic subgroup FS 1-2). The estimated age-dependent propensity of IHD in elderly women in the FS 2 subsample falls between the propensities observed for those elderly women in the FS 1 high-risk and those in the FS 3 low-risk subsamples. The observed variations in the gender, age, and genotype-dependent propensities of IHD among these subsamples serve as a compelling rationale for embracing a risk stratification algorithm that includes genotype information in making clinical decisions about the prevention of IHD.
Figure.
Age-dependent propensity of being IHD free in elderly women shown separately for the 3 subsamples and for the 2 genetic subgroups of subsample FS 1.
Sensitivity and specificity are properties of a risk stratification strategy that need to be taken into account when selecting a risk stratification algorithm in medical practice.49,50 From a clinical decision-making point of view, it is also important to know the probability that individuals in the high-risk stratum actually are at increased risk. The sensitivity and specificity do not give this information. Instead, the interpretation of the risk analysis needs to consider the +PV. The +PV is related to the sensitivity and specificity of a risk stratification algorithm and the prevalence of the disease of interest in the population from which the individuals are coming from through a mathematical formula derived from an application of the Bayes’ theorem of conditional probabilities.49
The sensitivity and specificity estimates for an algorithm that assigns women in the FS 1 and FS 2 to a high-risk stratum and those in the FS 3 subsample to a low-risk stratum are modest (0.735 and 0.588, respectively, Table 7). An estimate of +PV for the high-risk stratum is 0.118, which is ≈ 1.5 times higher than cumulative incidence of IHD of 0.070 (70 cases/1000 individuals at risk) in the overall sample of women. If the high-risk stratum includes the FS 1 subsample of women only, sensitivity of the stratification algorithm decreases, but its specificity increases to 0.808, and the estimate of the +PV (0.139) is slightly larger than the estimate for the stratum that includes both the FS 1 and the FS 2 subsamples.
Table 7.
Epidemiological Summaries of the Clinical Predictive Value of the PRIM-Defined Subsamples
| Subsample |
||||
|---|---|---|---|---|
| High Risk | Low Risk | Sensitivity | Specificity | +PV* |
| FS 2 and FS 1 | FS 3 | 0.735 | 0.588 | 0.118 |
| FS 1 | FS 3 and FS 2 | 0.412 | 0.808 | 0.139 |
| FS 1-1 | FS 3 and FS 2 and FS 1-2 | 0.206 | 0.926 | 0.172 |
| Marginal Estimates |
||||
|---|---|---|---|---|
| High Risk | Low Risk | Sensitivity | Specificity | +PV* |
| ATTT/ATAG | ATAT/others | 0.451 | 0.596 | 0.077 |
| Smoke† | No smoke | 0.642 | 0.435 | 0.079 |
| Hypertension‡ | No hypertension | 0.763 | 0.386 | 0.085 |
| TG ≥ 150 mg/dL | TG <150 mg/dL | 0.529 | 0.600 | 0.090 |
| >65 y | 45 to 65 y | 0.770 | 0.503 | 0.104 |
+PV reflects an expected proportion of IHD cases assigned to the related high-risk group. The estimate of +PV is related to sensitivity and specificity of a risk stratification algorithm that uses subsamples as a tool to identify individuals who are at increased risk and the prevalence of the proposed high-risk subsample in the population of interest through a mathematical formula that is derived from the application of the Bayes’ theorem of conditional probabilities (Fletcher and Fletcher49).
History of smoking.
History of hypertension.
If the high-risk stratum includes only the FS 1-1 genetic subgroup of elderly women with hypertriglyceridemia who have a history of smoking, a history of hypertension, or a history of both and who carry either the A560T832/T560T832 or the A560T832/A560G832 5′ genotype in the APOE promoter region, the sensitivity is low (0.206), but the specificity is highest (0.926). The estimated +PV for the FS 1-1 high-risk stratum (0.172) is higher than the estimate for the FS 1 high-risk stratum (0.139), ignoring genotype information, and ≈2.5 times higher than the estimate of 0.070 in overall female sample when assignments of individuals to sub-samples are ignored (Table 7).
Conclusions
Clinicians and patients need to know which trait(s) and which interventions work best in particular subsamples of individuals.8,51 The evaluation of the added value of a genetic variation using the traditional “one model describes all” approach provides information that has limited use in clinical practice.8,40,42 The stepwise PRIM algorithm that we used to evaluate the improvement in IHD prediction attributable to information about 5′ APOE genotypes acknowledges the biological reality of etiologic heterogeneity and nonadditive, context-dependent genetic effects that are consistent with what we know about the etiology of IHD. We propose that in Denmark this genetic information has added value when predicting an individual’s propensity to develop IHD only in a subpopulation of elderly women who have hypertriglyceridemia and a history of smoking, a history of hypertension, or a history of both. Intervention studies in Denmark are now needed to determine whether this context-dependent genetic information improves our ability to deliver the right treatment to the right patients at the right time, which is the primary goal of a modern healthcare system.
CLINICAL PERSPECTIVE.
Genomics research promises to provide DNA information that can be used in medical practice to guide selection of interventions to prevent or treat common diseases such as ischemic heart disease. A first step toward achieving this goal is to identify DNA sequence variants that are predictors of disease, disease risk factors, or both. To this end, we have demonstrated that 2 single-nucleotide polymorphisms in the 5′ regulatory region of the apolipoprotein E gene combine to define genotypes that predict dyslipidemia in samples from multiple populations. These genotypes also predicted risk of ischemic heart disease in Danish women. The positive predictive value of combinations of the 2 single-nucleotide polymorphism genotypes is, however, only marginally larger than the cumulative incidence (70/1000), suggesting that they would be a poor choice as a screening test. Using a Patient Rule-Induction Method modeling strategy, we identified a subsample of elderly Danish women with hypertriglyceridemia who had a history of smoking, a history of hypertension, or a history of both and who had particular 2 single-nucleotide polymorphism 5′ apolipoprotein E genotypes that had a statistically significant association with higher cumulative incidence of ischemic heart disease (172/1000). This statistical observation is consistent with the established biological reality that effects of genetic variation on diseases having a complex multifactorial pathobiology are context dependent. Our investigation also underscores the possibility that the most relevant question for genetic studies of complex diseases may extend beyond whether the phenotypic effect of a single-site DNA variation replicates across multiple populations to which multisite genotype is the best predictor of complex disease in which subgroup of the population at large.
Acknowledgments
We thank Ken Weiss for his devotion to carry out the statistical analyses and Christine Lusk, Tom Rea, and Kim Zerba for their critical review and suggestions for improving earlier drafts of this manuscript.
Sources of Funding
This work was supported by National Institutes of Health grants GM065509 and HL072905.
Footnotes
Disclosures
None.
References
- 1.Steinberg D. An interpretive history of the cholesterol controversy: part 1. J Lipid Res. 2004;45:1583–1593. doi: 10.1194/jlr.R400003-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet. 2005;6:287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 3.Department of Genome Sciences, University of Washington, Nickerson Group. Welcome to MDECODE. Available at: http://droog.gs.washington.edu/mdecode.
- 4.Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengård JH, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF. Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet. 2000;67:881–900. doi: 10.1086/303070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nickerson DA, Taylor SL, Fullerton SM, Weiss KM, Clark AG, Stengård JH, Salomaa V, Boerwinkle E, Sing CF. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res. 2000;10:1532–1545. doi: 10.1101/gr.146900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fullerton SM, Buchanan AV, Sonpar VA, Taylor SL, Smith JD, Carlson CS, Salomaa V, Stengård JH, Boerwinkle E, Clark AG, Nickerson DA, Weiss KM. The effects of scale: variation in the APOA1/C3/A4/A5 gene cluster. Hum Genet. 2004;115:36–56. doi: 10.1007/s00439-004-1106-x. [DOI] [PubMed] [Google Scholar]
- 7.Hegele RA, Dichgans M. Update on the genetics of stroke and cerebrovascular disease 2007. Stroke. 2008;39:252–254. doi: 10.1161/STROKEAHA.107.510503. [DOI] [PubMed] [Google Scholar]
- 8.Sing CF, Stengård JH, Kardia SL. Genes, environment, and cardiovascular disease. Arterioscler Thromb Vasc Biol. 2003;23:1190–1196. doi: 10.1161/01.ATV.0000075081.51227.86. [DOI] [PubMed] [Google Scholar]
- 9.Guttmacher AE, Collins FS. Realizing the promise of genomics in biomedical research. J Am Med Assoc. 2005;294:1399–1402. doi: 10.1001/jama.294.11.1399. [DOI] [PubMed] [Google Scholar]
- 10.van Asselt KM, Kok HS, van der Schouw YT, Peeters PH, Pearson PL, Grobbee DE. Role of genetic analyses in cardiology: part II: heritability estimation for gene searching in multifactorial diseases. Circulation. 2006;113:1136–1139. doi: 10.1161/CIRCULATIONAHA.105.563197. [DOI] [PubMed] [Google Scholar]
- 11.Humphries SE, Yiannakouris N, Talmud PJ. Cardiovascular disease risk prediction using genetic information (gene scores): is it really informative? Curr Opin Lipidol. 2008;19:128–132. doi: 10.1097/MOL.0b013e3282f5283e. [DOI] [PubMed] [Google Scholar]
- 12.Sing CF, Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet. 1985;37:268–285. [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson PWF, Myers RH, Larson MG, Ordovas JM, Wolf PA, Schaefer EJ. Apolipoprotein E alleles, dyslipidemia, and coronary heart disease: the Framingham Offspring Study. J Am Med Assoc. 1994;272:1666–1671. [PubMed] [Google Scholar]
- 14.Davignon J, Cohn JS, Mabile L, Bernier L. Apolipoprotein E and atherosclerosis: insight from animal and human studies. Clin Chim Acta. 1999;286:115–143. doi: 10.1016/s0009-8981(99)00097-2. [DOI] [PubMed] [Google Scholar]
- 15.Stengård JH, Kardia SL, Hamon SC, Frikke-Schmidt R, Tybjærg-Hansen A, Salomaa V, Boerwinkle E, Sing CF. Contribution of regulatory and structural variations in APOE to predicting dyslipidemia. J Lipid Res. 2006;47:318–328. doi: 10.1194/jlr.M500491-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 17.Wilson PWF, Schaefer EJ, Larson MG, Ordovas JM. Apolipoprotein E alleles and risk of coronary disease: a meta-analysis. Arterioscler Thromb Vasc Biol. 1996;16:1250–1255. doi: 10.1161/01.atv.16.10.1250. [DOI] [PubMed] [Google Scholar]
- 18.Stengård JH, Frikke-Schmidt R, Tybjærg-Hansen A, Nordestgaard BG, Sing CF. Variation in 5′ promoter region of the APOE gene contributes to predicting ischemic heart disease (IHD) in the population at large: the Copenhagen City Heart Study. Ann Hum Genet. 2007;71:762–771. doi: 10.1111/j.1469-1809.2007.00370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyson G, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A, Sing CF. An application of the Patient Rule-Induction Method for evaluating the contribution of the apolipoprotein E and lipoprotein lipase genes to predicting ischemic heart disease. Genet Epidemiol. 2007;31:515–527. doi: 10.1002/gepi.20225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson G, Frikke-Schmidt R, Nordestgaard BG, Tybjærg-Hansen A, Sing CF. Modifications to the Patient Rule-Induction Method that utilize non-additive combinations of genetic and environmental effects to define partitions that predict ischemic heart disease. Genet Epidemiol. 2009;33:317–324. doi: 10.1002/gepi.20383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frikke-Schmidt R, Nordestgaard BG, Agerlholm-Larsen B, Schnohr P, Tybjærg-Hansen A. Context-dependent and invariant associations between lipids, lipoproteins, and apolipoproteins and apolipoprotein E genotype. J Lipid Res. 2000;41:1812–1822. [PubMed] [Google Scholar]
- 22.Frikke-Schmidt R, Sing CF, Nordestgaard BG, Tybjærg-Hansen A. Gender- and age-specific contributions of additional DNA sequence variation in the 5′ regulatory region of the APOE gene to prediction of measures of lipid metabolism. Hum Genet. 2004;115:331–345. doi: 10.1007/s00439-004-1165-z. [DOI] [PubMed] [Google Scholar]
- 23.Schnohr P, Jensen G, Lange P, Scharling H, Appleyard M. The Copenhagen City Heart Study. Eur Heart J Suppl. 2001;3:H1–H83. [Google Scholar]
- 24.Frikke-Schmidt R, Sing CF, Nordestgaard BG, Steffensen R, Tybjærg-Hansen A. Subsets of SNPs define rare genotype classes that predict ischemic heart disease. Hum Genet. 2007;120:865–877. doi: 10.1007/s00439-006-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with Hhal. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 26.Friedman JH, Fisher NI. Bump hunting in high-dimensional data. Stat Comput. 1999;9:123–143. [Google Scholar]
- 27.Gordon T, Kannel WB. Multiple risk functions for predicting coronary heart disease: the concept, accuracy, and application. Am Heart J. 1982;103:1031–1039. doi: 10.1016/0002-8703(82)90567-1. [DOI] [PubMed] [Google Scholar]
- 28.Wilson PWF, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 29.Arnett DK, Baird AE, Barkley RA, Basson CT, Boerwinkle E, Ganesh SK, Herrington DM, Hong YL, Jaquish C, McDermott DA, O’Donnell CJ. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease: a scientific statement from the American Heart Association Council on Epidemiology and Prevention, the Stroke Council, and the Functional Genomics and Translational Biology Interdisciplinary Working Group. Circulation. 2007;115:2878–2901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- 30.Kannel WB, D’Agostino RB, Sullivan L, Wilson PW. Concept and usefulness of cardiovascular risk profiles. Am Heart J. 2004;148:16–26. doi: 10.1016/j.ahj.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Steinberg D. The statins in preventive cardiology. N Engl J Med. 2008;359:1426–1427. doi: 10.1056/NEJMp0806479. [DOI] [PubMed] [Google Scholar]
- 32.McGill HC., Jr Atherosclerosis: problems in endpoints for genetic analysis. Prog Clin Biol Res. 1979;32:27–49. [PubMed] [Google Scholar]
- 33.Zethelius B, Berglund L, Sundström J, Ingelsson E, Basu S, Larsson A, Venge P, Arnlöv J. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358:2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 34.Johnson KM, Dowe DA, Brink JA. Traditional clinical risk assessment tools do not accurately predict coronary atherosclerotic plaque burden: a CT angiography study. Am J Roentgenol. 2009;192:235–243. doi: 10.2214/AJR.08.1056. [DOI] [PubMed] [Google Scholar]
- 35.Barrett-Connor E, Giardina EG, Gitt AK, Gudat U, Steinberg HO, Tschoepe D. Women and heart disease: the role of diabetes and hyperglycemia. Arch Intern Med. 2004;164:934–942. doi: 10.1001/archinte.164.9.934. [DOI] [PubMed] [Google Scholar]
- 36.Barrett-Connor E. Women and cardiovascular disease. Can Med Assoc J. 2007;176:791–793. doi: 10.1503/cmaj.061677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim ESH, Menon V. Status of women in cardiovascular clinical trials. Arteroscler Thromb Vasc Biol. 2009;29:279–283. doi: 10.1161/ATVBAHA.108.179796. [DOI] [PubMed] [Google Scholar]
- 38.Lambert JC, Coyle N, Lendon C. The allelic modulation of apolipoprotein E expression by oestrogen: potential relevance for Alzheimer’s disease. J Med Genet. 2004;41:104–112. doi: 10.1136/jmg.2003.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss KM. Tilting at quixotic trait loci (QTL): an evolutionary perspective on genetic causation. Genetics. 2008;179:1741–1756. doi: 10.1534/genetics.108.094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noble D. The Music of Life: Biology Beyond the Genome. New York, NY: Oxford University Press; 2006. [Google Scholar]
- 41.Lewontin RC. The analysis of variance and the analysis of causes. Int J Epidemiol. 2006;35:520–525. doi: 10.1093/ije/dyl062. [DOI] [PubMed] [Google Scholar]
- 42.Lewontin RC. The Triple Helix: Gene, Organism, and Environment. Cambridge, MA: Harvard University Press; 2002. [Google Scholar]
- 43.Kaprio J, Ferrell RE, Kottke BA, Kamboh MI, Sing CF. Effects of polymorphisms in apolipoproteins E, A-IV, and H on quantitative traits related to risk for cardiovascular disease. Arterioscler Thromb. 1991;11:1330–1348. doi: 10.1161/01.atv.11.5.1330. [DOI] [PubMed] [Google Scholar]
- 44.Nelson LM, Bloch DA, Longstreth WT, Jr, Shi H. Recursive partitioning for the identification of disease risk subgroups: a case-control study of subarachnoid hemorrhage. J Clin Epidemiol. 1998;51:199–209. doi: 10.1016/s0895-4356(97)00268-0. [DOI] [PubMed] [Google Scholar]
- 45.Stengård JH, Kardia SL, Tervahauta M, Ehnholm C, Nissinen A, Sing CF. Utility of the predictors of coronary heart disease mortality in a longitudinal study of elderly Finnish men aged 65 to 84 years is dependent on context defined by ApoE genotype and area of residence. Clin Genet. 1999;56:367–377. doi: 10.1034/j.1399-0004.1999.560505.x. [DOI] [PubMed] [Google Scholar]
- 46.Stengård JH, Salomaa V, Rasi V, Vahtera E, Ehnholm C, Krusius T, Perola M, Vartiainen E. Utility of the Arg/Gin polymorphism of the factor VII (FVII) gene, serum lipid levels and body mass index in the prediction of the FVII:C and FVII:Ag in North Karelia: a cross-sectional and prospective study. Blood Coagul Fibrinolysis. 2001;12:445–452. doi: 10.1097/00001721-200109000-00004. [DOI] [PubMed] [Google Scholar]
- 47.Koss N, Feinstein AR. Computer-aided prognosis II. Development of a prognostic algorithm. Arch Intern Med. 1971;127:448–459. doi: 10.1001/archinte.127.3.448. [DOI] [PubMed] [Google Scholar]
- 48.Marshall RJ. The use of classification and regression trees in clinical epidemiology. J Clin Epidemiol. 2001;54:603–609. doi: 10.1016/s0895-4356(00)00344-9. [DOI] [PubMed] [Google Scholar]
- 49.Fletcher RW, Fletcher SW. Clinical Epidemiology: The Essentials. 4. Baltimore, MD: Lippincott Williams & Wilkins; 2005. [Google Scholar]
- 50.Kraft P, Wacholder S, Cornelis MC, Hu FB, Hayes RB, Thomas G, Hoover R, Hunter DJ, Chanock S. Beyond odds ratios—communicating disease risk based on genetic profiles. Nat Rev Genet. 2009;10:264–269. doi: 10.1038/nrg2516. [DOI] [PubMed] [Google Scholar]
- 51.Conway PH, Clancy C. Comparative-effectiveness research—implications of the Federal Coordinating Council’s Report. N Engl J Med. 2009;361:328–330. doi: 10.1056/NEJMp0905631. [DOI] [PubMed] [Google Scholar]