Abstract
BACKGROUND
Multiple reaction monitoring mass spectrometry (MRM-MS) of peptides with stable isotope–labeled internal standards (SISs) is increasingly being used to develop quantitative assays for proteins in complex biological matrices. These assays can be highly precise and quantitative, but the frequent occurrence of interferences requires that MRM-MS data be manually reviewed, a time-intensive process subject to human error. We developed an algorithm that identifies inaccurate transition data based on the presence of interfering signal or inconsistent recovery among replicate samples.
METHODS
The algorithm objectively evaluates MRM-MS data with 2 orthogonal approaches. First, it compares the relative product ion intensities of the analyte peptide to those of the SIS peptide and uses a t-test to determine if they are significantly different. A CV is then calculated from the ratio of the analyte peak area to the SIS peak area from the sample replicates.
RESULTS
The algorithm identified problematic transitions and achieved accuracies of 94%–100%, with a sensitivity and specificity of 83%–100% for correct identification of errant transitions. The algorithm was robust when challenged with multiple types of interferences and problematic transitions.
CONCLUSIONS
This algorithm for automated detection of inaccurate and imprecise transitions (AuDIT) in MRM-MS data reduces the time required for manual and subjective inspection of data, improves the overall accuracy of data analysis, and is easily implemented into the standard data-analysis work flow. AuDIT currently works with results exported from MRM-MS data-processing software packages and may be implemented as an analysis tool within such software.
The technology of choice for detecting and quantifying analytes in complex samples is multiple reaction monitoring mass spectrometry (MRM-MS).2 In MRM-MS, precursor ions for the analytes of interest are individually mass-selected and then fragmented by collisional or resonance excitation. One or more of the fragment ions (product ions) produced from the mass-selected precursor are monitored for purposes of identification and quantification. The most commonly used instrument for this type of analysis is the triple-quadrupole mass analyzer, in which quadrupole 1 (Q1) is used for mass selection, Q2 is used for fragmenting the analyte via collisional excitation with an inert gas such as nitrogen, and Q3 is used to monitor specific product ions (1). The primary motivation for use of MRM-MS and monitoring just a few fragment ions from the analytes of interest rather than acquiring full-scan tandem mass spectrometry (MS/MS) spectra is to improve the limit of quantification and increase analytical speed.
The vast majority of experience in the development and use of MRM-MS over the past 3 decades has come from scientists engaged in the analysis of small molecules, chiefly for preclinical and clinical monitoring of drug metabolism and pharmacokinetics (2, 3), for measuring concentrations of toxic or carcinogenic small molecules in the environment (2–4), and for assaying concentrations of hormones (5, 6) as well as small biological molecules in the context of inborn errors of metabolism (7, 8). These fields have long recognized that identifying analytes by monitoring and detecting only a few fragment ions is highly susceptible to false-positive identification and inaccurate quantification, a fact requiring additional measures to be taken to ensure correct identification and quantification (3, 6, 9, 10). The key factors contributing to these problems are interference and ion suppression by the components of the biological sample matrix (e.g., plasma, tissue, cell lines, urine) (11, 12). The components giving rise to these deleterious effects are often referred to as chemical and biological “noise,” but in fact they are often real sample constituents, just not the analytes of interest. False-positive identifications in MRM-MS occur when other coeluting sample constituents also produce the product ions monitored for the analyte of interest. Suppression manifests as a change (most often a decrease) in the ion current response in the mass spectrometer for the same amount of analyte analyzed from different samples. Suppression effects increase with the complexity of the biological matrix and with the presence and concentration of ionizable sample constituents (e.g., phospholipids) that are invariably found in biologically derived samples. Even in cases in which considerable efforts have been taken to remove nonvolatile salts and to prepare samples as identically as possible, small unseen variations in sample constitution or chromatographic elution can produce interference or suppression in a sample that is not observed in a seemingly identical sample (11–14). Although an exact and universally accepted physio-chemical explanation for suppression in electrospray ionization is still lacking, the phenomenon, its deleterious effect on quantitative analyses, and methods to compensate for the effect are well understood by the small-molecule community (11–14). The best way to deal with interference and suppression phenomena has been to use an internal standard, preferably an isotopically labeled version of the analyte itself. Such compounds behave identically to the analyte molecule with respect to chromatographic retention (with the exception of heavily deuterated analogs) and fragmentation behavior in the mass spectrometer but are distinguishable on the basis of their precursor mass and the masses of fragment ions containing the heavy isotopes. This methodology, referred to as “stable isotope dilution” (SID), has been used by the small-molecule community for decades (15–18) and is being increasingly adopted by new fields, including proteomics (19–22).
The past decade has seen a surge in research related to the use of MS for protein quantification. In this approach, 1 or more peptides derived by enzymatic digestion of a target protein are measured by MS. Detection of the peptide(s) and their abundance in the sample are surrogates for the presence and the amount of the target protein in the sample. The technology of choice for detecting and quantifying peptides in complex biological samples and biofluids is MRM-MS (23–28). Unfortunately, the lessons learned by the small-molecule MS community [namely that the use of liquid chromatography–MS/MS (LC-MS/MS) methods is by itself no guarantee of either specificity or quantitative accuracy] and the methods this community has developed to cope with these analytical problems are only slowly being appreciated by proteomics scientists and integrated into practice. The potential for false-positive identification and inaccurate quantification of peptides in highly complex samples such as plasma, tissue lysate, or cell lines is likely to be substantially greater than for small-molecule analysis of these same biological matrices. In small-molecule MRM-MS analyses of biofluids, tissues, and cell lines, proteins are removed by precipitation before analysis, thereby eliminating a huge potential source of interference. In proteomics, not only do we have to contend with the already highly complex and wide concentration range of the protein complement of the sample, but we must also be able to assay for peptides derived from these proteins after enzymatic digestion, which increases the complexity of the sample by another 50- to 100-fold (29).
Inaccurate quantification in peptide MRM-MS has many sources, including false-positive identification, suppression caused by matrix components, interference in 1 or more of the monitored product ion transitions, poor chromatography, MS instrument–related signal attenuation and saturation, and errors caused by the software used for peak detection and integration. We describe common sources of false-positive identifications and inaccurate quantification in peptide MRM-MS and present methods and protocols to detect and prevent such problems. Although many algorithms and methods have been devised to accelerate and simplify the design and implementation of MRM-based experiments in proteomics (30, 31), interferences in the MRM signals are not currently addressed and can drastically undermine the results obtained from quantitative experiments. Interferences are usually detected by painstaking and subjective manual examination of the raw data (32). Protein quantification for candidate biomarker verification in clinical proteomics (21, 25, 26, 33) and the application of MRM-MS methods for basic biological studies in model systems such as yeast (27) increasingly require the ability to assay many tens to hundreds of proteins. Clearly, manual inspection of such data is no longer possible or desired. We have developed a new algorithm for automated detection of inaccurate and imprecise transitions (AuDIT) in the application of MRM-MS for quantitative analyses of peptides. This algorithm can be used both in methods development and in routine testing of patient samples.
Materials and Methods
RESPONSE CURVE WITH 3 AND 5 MRM TRANSITIONS PER PEPTIDE
A response curve consisting of peptides spiked into plasma was prepared as described previously (34). In brief, for the 3-MRM data set, a 9-point response curve was generated by spiking 11 synthetic peptides (representing prostate-specific antigen, horseradish peroxidase, leptin, myelin basic protein, myoglobin, aprotinin, and C-reactive protein) into digested plasma (1 μg/μL) so as to span a concentration range of 1–500 fmol/μL, with corresponding isotopically labeled (13C/15N) peptide standards added at a fixed concentration of 50 fmol/μL to all samples [referred to as study I in (34)]. A 1-μL volume of each sample was analyzed in quadruplicate by LC-MRM-MS. Three data sets were acquired with a 4000 Q TRAP mass spectrometer (Applied Biosystems) at unit/unit resolution with a dwell time of 10 ms and an interscan delay time of 5 ms for each transition. One data set was acquired on a TSQ Quantum Ultra (Thermo Fisher Scientific) triple-quadrupole mass spectrometer with a 10-ms dwell time for each transition at unit/unit resolution. In all cases, only data from 10 peptides were analyzed, owing to insufficient detection of the 11th peptide.
For the 5-MRM data set, an equimolar mixture of the 7 proteins was digested, added to digested plasma, and then diluted with digested plasma to generate a 9-point response curve spanning the same concentration range (1–500 fmol/μL) in a digested-plasma background (1 μg/μL). Corresponding stable isotope–labeled internal standard (SIS) peptides were added to each sample at 50 fmol/μL [referred to as study II in (34)]. We monitored 5 MRM transitions for each analyte and SIS peptide on a 5500 Q TRAP mass spectrometer (Applied Biosystems) by scheduled MRM with a target cycle time of 0.5 s, a retention-time window of 90 s, and an interscan delay of 3 ms. The 2 additional transitions were selected from previous optimization experiments conducted with the synthetic peptides. The pooled and filtered human plasma used for all experiments was from Bioreclamation.
INDIVIDUAL SAMPLE DATA—CLINICAL SAMPLE SET
Samples were prepared and analyzed as described previously (26). The protocol for obtaining blood from patients was approved by the Massachusetts General Hospital Institutional Review Board, and all individuals gave written informed consent. A subset of the data (2 peptides from C-reactive protein) monitored from 1 patient (3 time points) was selected for evaluating algorithm performance. The data set consisted of 2 process replicates of each time point analyzed in 3 technical replicates on a 4000 Q TRAP mass spectrometer.
Three transitions were monitored for each peptide and its corresponding SIS peptide.
GENERATION OF EXTRACTED ION CHROMATOGRAMS (XICs) AND PEAK INTEGRATION
Data were imported into MultiQuant (version 1.1; Applied Biosystems/MDS Sciex) or Skyline (version 0.5; http://proteome.gs.washington.edu/software/skyline) (30), where XICs were integrated to generate an exportable table containing the following necessary sample information: sample name, concentration, replicate number, peptide and transition (precursor/product ion) information, peak area of analyte, and peak area of SIS peptide transitions.
PRE- AND POSTALGORITHM DATA NOTATION
After automatic integration of the data sets, we manually inspected the data (prealgorithm) to record any global aspects of the XICs or integration that might lead to incorrect quantification and added this information as a separate field or “note” in MultiQuant or Skyline, respectively. Observations included some of the following descriptions: inconsistent baseline integration for the analyte and SIS peptide transition; peak closely eluting to XIC that might obscure integration; interference; detector saturation; variable peak area ratio (PAR) in replicate samples; and the wrong peak integrated. After AuDIT was used to process the data, the same individual again manually inspected the data in a focused, AuDIT-directed manner to determine if the algorithm outcome agreed with the prealgorithm manual inspection and if not, which judgment (prealgorithm expert or algorithm) was correct. The comments were then amended postalgorithm to reflect the validity of the data for quantitative calculations.
DATA EXPORT AND PREPROCESSING
MRM-MS XICs that have been integrated with Multi-Quant or Skyline are exported into comma-separated value (csv) files. These files are then imported by custom programs written in R [(35), http://www.r-project.org/], a statistical programming language. The csv files are reformatted to:
Extract required columns (including sample name, replicate number, peptide identification, transition information, area for analyte, and SIS area) in addition to other optional fields (such as retention time, signal-to-noise ratio, and so on). The pre- and postalgorithm annotation is also captured.
Combine pre- and postalgorithm annotation for replicates so that any comments made for any of the replicates of a sample are consistently captured and maintained.
Remove any samples (e.g., QC runs, blank sample runs, and so forth) that are not to be included in the analysis.
The preprocessed data were written out in a consistent format that AuDIT could then use to assess the quality of the transitions.
ALGORITHM IMPLEMENTATION AND EXECUTION
AuDIT was designed to extensively use the concept of “relative ratio,” the ratio of the peak areas for any 2 transitions of the same precursor. All analyte (or all SIS) transition peak areas are used in pairs to calculate the ratio. The relative ratio is unlike the PAR, which is calculated as the ratio of analyte to SIS peak areas for a given transition of a specified precursor. The program reads the preprocessed data and executes the following steps:
Use all transitions of a peptide (peak area from XICs) to calculate relative ratios by either the minimal-pairs or all-pairs method. The minimal-pairs method calculates the relative ratio of a given transition by dividing its peak area by the peak area of 1 other transition from the same precursor. The all-pairs method calculates ratios for all possible transition pairs generated from 1 precursor. This process is performed for each peptide analyte and its corresponding SIS so that the relative ratios of the analyte can be compared with the relative ratios of the SIS.
Apply the t-test to determine a P value for the hypothesis that the relative ratios for the analyte are different from the relative ratios of the SIS.
Use the Benjamini–Hochberg false-discovery rate method to correct the nominal t-test P values to account for multiple hypothesis testing (36).
Disaggregate the corrected P values for the relative ratios into combined P values for each transition. Each transition is used to calculate either 2 ratios for the minimal-pairs method or n – 1 ratios for the all-pairs method (where n is the total number of observed transitions for each peptide). Calculation of the P value for determining if a transition is problematic requires combining the P values for the respective relative ratios. Because the same peak areas from a given transition were used in calculating all its ratios, the resulting P values are not independent. These dependent P values are combined by means of a previously outlined methodology (37, 38).
Calculate the CV for the PAR (analyte/SIS) from the results for all replicates in a transition for a given sample.
A transition is marked as “bad” if either the corrected combined P value for the transition is less than the P value threshold of 10−5 or if the CV is greater than the CV threshold of 0.2 (20%). Transitions not satisfying either of these conditions are classified as “good.” Although the chosen thresholds work well for all the data sets evaluated in this study, they can be changed to fine-tune the algorithm as needed.
ALGORITHM VALIDATION
AuDIT’s “good” and “bad” calls are compared with the global (prealgorithm) and focused (postalgorithm) annotation created by the expert. These comparisons are used to create 2 contingency matrices for each data set, as indicated in Table 1 and Table 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol56/issue2. ROC curves and plots of sensitivity vs specificity are then created with the ROCR library in R (39). The AuDIT software is available at http://www.genepattern.org/modules/AuDIT.html.
Table 1.
Definitions for the contingency matrices and AuDIT performance for all analyzed data sets.a
| A. Contingency matrices. | ||||||||
|---|---|---|---|---|---|---|---|---|
| Expert’s annotation |
||||||||
| AuDIT decision | Bad | Good | ||||||
| Bad | TN | FN | ||||||
| Good | FP | TP | ||||||
| B. AuDIT performance. | ||||||||
| Data set | TN, n | TP, n | FN, n | FP, n | Overall accuracy, %b | Sensitivity, %b | Specificity, %b | |
| 10-Peptide calibration curve, 3 transitions, MultiQuant | Site 1 | 117 | 144 | 1 | 8 | 97 | 99 | 94 |
| Site 2 | 23 | 247 | 0 | 0 | 100 | 100 | 100 | |
| Site 3 | 50 | 218 | 2 | 0 | 99 | 99 | 100 | |
| Site 4 | 81 | 174 | 14 | 1 | 94 | 93 | 99 | |
| 10-Peptide calibration curve, 3 transitions, Skyline | Site 1 | 56 | 206 | 8 | 0 | 97 | 96 | 100 |
| Site 2 | 15 | 254 | 1 | 0 | 100 | 100 | 100 | |
| Site 5 | 37 | 232 | 0 | 1 | 100 | 100 | 97 | |
| 10-Peptide calibration curve, 5 transitions, MultiQuant | Site 6 | 168 | 294 | 0 | 6 | 99 | 100 | 97 |
| Clinical samples, 3 transitions, MultiQuant | Cardiovascular peptides | 9 | 40 | 0 | 2 | 96 | 100 | 82 |
TN, true negative; FN, false negative; FP, false positive; TP, true positive. A transition is bad if it has some form of interference (i.e., it is imprecise or inaccurate). If not, the transition is labeled as good.
Overall accuracy = (TP + TN)/(TP + TN + FN + FP); sensitivity = TP/(TP + FN); specificity = TN/(TN + FP).
Results
SOURCES OF INACCURATE QUANTIFICATION IN PEPTIDE MRM-MS
Many factors can contribute to inaccurate quantification in MRM-MS, but the enormous sample complexity at the peptide level, combined with potential for overlapping precursor and product ion masses, synergistically exacerbate the potential for poor quantification in proteomics. When tryptic digests of plasma, tissue interstitial fluids, and tissue or cell lysates are analyzed by LC-MRM-MS, tens to hundreds of different peptides elute from the LC column into the MS system nearly simultaneously. Dealing with this high degree of complexity is made more difficult by the need both to use a mass window of 0.5–1 Da on triple-quadrupole mass analyzers to select the precursor ions of peptides for fragmentation and to select the product ions formed by fragmentation for detection. These requirements mean that 13C isotope peaks of non-targeted peptides and their product ions can “leak” into the selection and detection windows. Because only a few product ions from mass-selected precursors are being monitored (rather than acquisition of full MS and MS/MS scans) the analyst is blind to when such leakage is occurring. Although narrower mass windows would clearly produce higher selectivity for both precursor and product ions (40–43), the mass width of the selection windows is limited by the ion-transmission properties of triple-quadrupole mass analyzers, which produce a steep decline in signal as these windows are narrowed to <0.5 Da.
The lengths of most tryptic peptides are 8–20 amino acid residues, and these peptides are composed largely of subsets of the same 20 amino acids that differ only in their linear arrangement. Therefore, it is not uncommon in highly complex biological samples to have peptides of the same or nearly the same precursor mass but with different sequences that elute very close in time and are fragmented simultaneously with the analyte of interest (i.e., chimera spectra) (44). Furthermore, it is common for such peptides to fragment and produce one or more fragment ions with masses identical or nearly identical to the fragment ions of the analyte of interest. If one or more of these common fragment ions happen to be among the small number being monitored by MRM-MS (i.e., within ±0.5–1.0 Da of the mass of a monitored fragment ion from the desired analyte), then an interference results. Importantly, these types of interferences can arise from any ions, peptide or nonpeptide (e.g., detergent), that have the same nominal m/z precursor and product ion combination.
INACCURATE QUANTIFICATION IN THE ABSENCE OF AN INTERNAL STANDARD
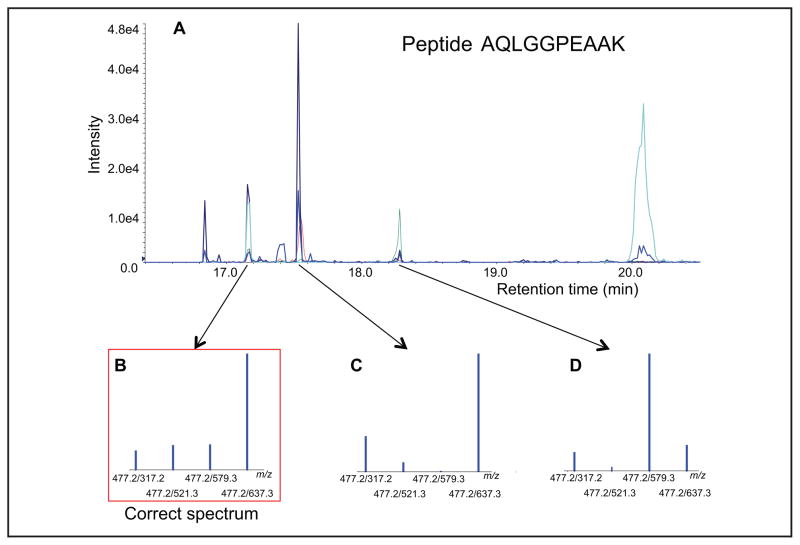
Fig. 1 illustrates how a peptide analyte can be falsely identified—and therefore be inaccurately quantified—in the absence of an SIS. Four product ion transitions from the doubly charged precursor of the peptide AQLG GPEAAK were monitored by MRM-MS in digested plasma. All 4 of these transitions coeluted in each of 3 distinct peaks in the chromatogram between 17 min and 18.5 min (Fig. 1, B–D). Identifying which of these peaks corresponds to the authentic analyte peptide in principle could be accomplished by comparing the product ion intensities of an external reference standard peptide run under identical MRM-MS conditions. Such identification, however, presumes the absence of interference in any of the analyte transitions and that the retention time for the external standard run in buffer is identical to that of the analyte in the matrix. Analyte quantification is also compromised because suppression of analyte signal (precursor and product ions) by matrix constituents cannot be accounted for by use of an external standard. Use of an SIS of the analyte can compensate for all of these effects.
Fig. 1. Inaccurate quantification in the absence of an internal standard.
(A), XIC for 4 product ion transitions from the doubly charged precursor of the peptide AQLGGPEAAK in digested plasma (monitored by MRM-MS m/z 477.2 to m/z 317.2, 521.3, 579.3, and 637.2). (B–D), All 4 transitions coelute in each of 3 distinct peaks in the chromatogram between 17 min and 18.5 min.
INACCURATE QUANTIFICATION FROM INTERFERENCES WITH USE OF SISs
Use of SID-MRM-MS methods for accurate peptide quantification relies on the physiochemical similarities between the analyte peptide and its stable isotope–labeled analog. Under identical conditions of chromatography, ionization, and collision-induced dissociation, each peptide and its corresponding fragment ions are sequentially detected on a millisecond time scale in the mass spectrometer. Quantification is accomplished by comparing the intensity of the analyte signal measured by the mass spectrometer with that of the SIS, which is added to the sample in a known quantity. Under the same conditions, the analyte and the SIS peptide dissociate to generate the same pattern of fragment ions, which differ only by m/z [reflecting addition of the stable isotope–labeled amino acid(s)] and absolute intensity. Importantly, the relative intensities of the complement of product ions formed by each precursor are nearly identical, as long as the amount of peptide detected is within the linear range of detection by MS (Fig. 2).
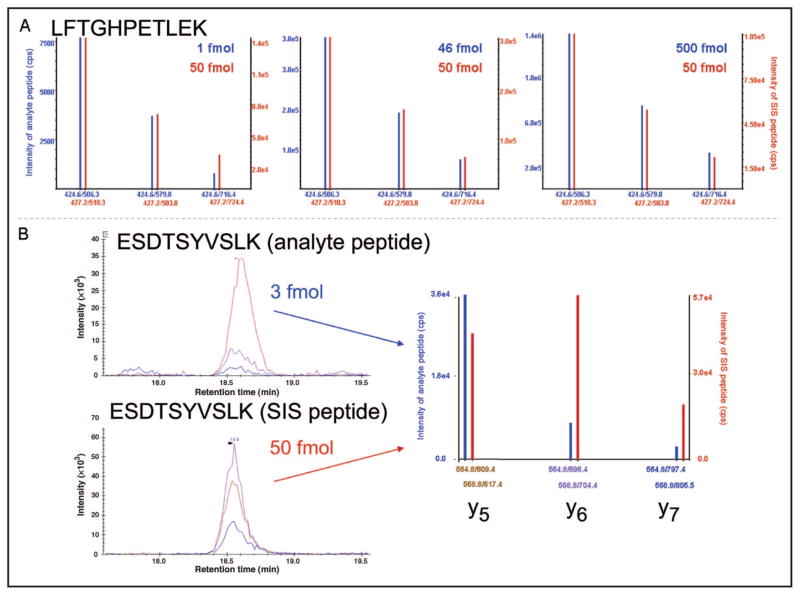
Fig. 2. Examples of MRM-MS spectra of analyte and SIS peptides in the absence (A) and presence (B) of a coeluting interference.
(A), MRM mass spectra of analyte peptide (LFTGHPETLEK, blue spectra) and the corresponding heavy SIS peptide (red spectra) in digested plasma matrix. Three separate LC-MRM-MS runs are shown with a fixed amount of SIS peptide (50 fmol) and varying analyte peptide (1, 46, and 500 fmol) spiked into 1 μg digested plasma. Although the absolute intensities of the fragments (y axes) vary with concentration and potentially as a function of sample introduction into the mass spectrometer, the relative intensities of the product ions at m/z 506.3, 579.8, and 716.4 maintain a constant relationship with one another. Furthermore, the relative intensities of the analyte fragment ions agree precisely with the intensities of the fragment ions from the heavy peptide. (B), Example of an interference in peptide ESDTSYVSLK (transition y5, m/z 564.8/609.4) in digested plasma, which gives rise to an altered MRM-MS profile. The XICs are shown for the 3 transitions monitored for analyte peptide (top) and SIS peptide (bottom).
In the context of MRM-based studies, in which far fewer product ions are detected and used to infer the identification of the target peptide (along with retention time), the relative intensities of the product ions for a given peptide can be used as an additional measure of the technique’s selectivity, an approach similar to that described for small-molecule MRM-MS (6). Fig. 2A illustrates the ideal situation in which interferences are absent. Spectra from LC-MRM-MS analyses of 3 digested plasma samples are shown. Analyte peptide is present at 3 different concentrations (1, 46, and 500 fmol/μL), and its corresponding SIS peptide is present at a constant concentration of 50 fmol/μL. The pattern of product ion intensities for the analyte peptide is identical to that of the SIS peptide (±5% variance in the relative ratios for each fragment), regardless of the concentration of peptide present, even in the presence of digested plasma matrix (provided the concentration is greater than the lower limit of quantification and less than that of saturation of the detector).
Unfortunately, in the context of digested plasma, interferences are more often the rule than the exception. Even in the presence of an SIS, coeluting interferences can confound identification and quantification. Fig. 2B illustrates the effect of a coeluting species with 1 similar Q1-to-Q3 transition. When multiple transitions are monitored, it is common to sum the signals from the monitored transitions, which hides the contribution of the interfering signal. The contribution from the y5 transition in the analyte (m/z 564.8/609.4) is much larger than the contributions from the y6 and y7 transitions, and their relative ratios do not agree with those of the SIS peptide. When the 3 transitions are evaluated individually and compared with the SIS peptide transitions, it becomes clear that 1 of the analyte transitions is receiving a contributing signal from another ion species and that this contributing signal will cause a particularly inaccurate quantification of the target peptide.
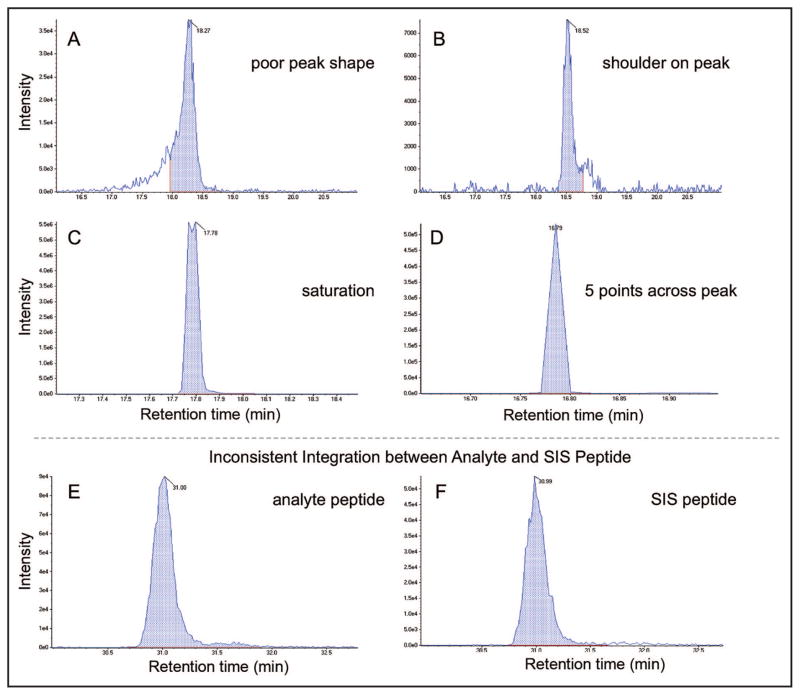
Inaccurate quantification can also arise from other sources of variation, including unstable electrospray, poor chromatography, chromatographic peaks that are too narrow (<6 points across), and inconsistency in peak integration between the analyte and SIS transitions. Examples of each of these factors are illustrated in Fig. 3. Each of these sources contributes substantial inconsistency to the quantification of the peptide analyte, producing poor reproducibility and high CVs that reduce overall confidence in the quantitative value of the assay. Early identification of sample-related issues (unstable electrospray, poor chromatography, and so forth) can often be rectified by an additional injection of the sample, whereas peak-integration issues can be resolved by the data-processing software. In either case, such issues are often not identified until the data set has been completely acquired, at which time the analyst becomes responsible for manually inspecting >2000 XICs at one time.
Fig. 3. Examples of sources of error in quantitative measurements by MRM-MS.
(A–E), XICs demonstrating common causes for incorrect quantification in SID-MRM-MS. (A), Example of poor chromatographic peak shape causing inconsistent automatic peak integration. (B), Presence of a peak with a closely eluting interference causing inconsistent integration. (C), Detector saturation at high analyte concentration. (D), Peak with <6 data points across, causing poor determination of peak area. (E, F), Comparison of the analyte and SIS peptides from the same sample, in which automatic peak integration did not use the same baseline start and stop times to determine peak area, causing inaccurate quantification.
ALGORITHM FOR AuDIT IN MRM-MS
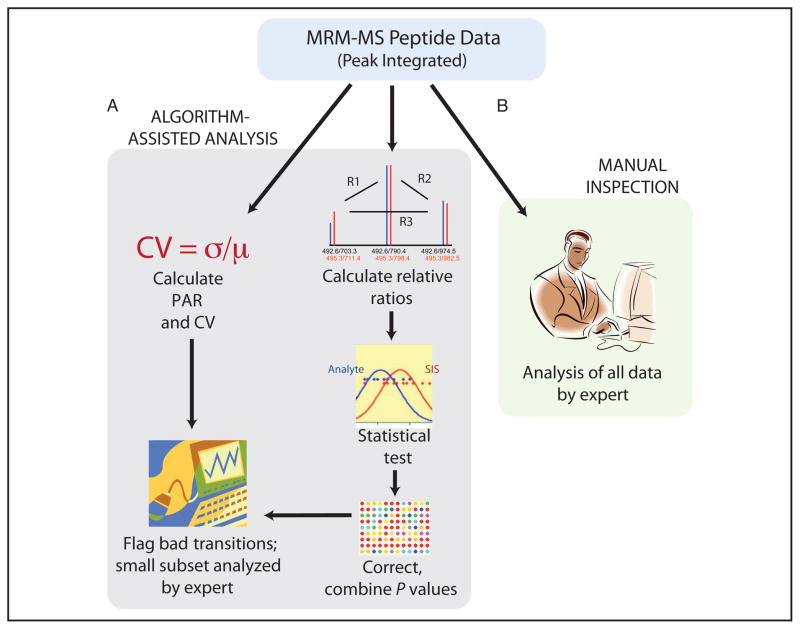
As described above, a considerable amount of tedious manual data inspection is required to minimize quantification errors from interferences (32). To speed up the analyses of large multianalyte MRM-MS data sets and to reduce the variation and error introduced by the differing skills of persons carrying out the manual and subjective data evaluation (45), we developed the AuDIT algorithm, which permits the automated filtering of data sets from SID-MRM-MS assays to identify problematic data to the researcher. AuDIT is specifically designed to identify inaccurate or imprecise transitions in the MRM-MS data that will produce inaccurate or highly variable quantitative measurements so that analysts can focus their attention on the peaks that require expert manual inspection. AuDIT is based on the fact that for a given precursor peptide, the relative ratios of observed product ion intensities are constant and independent of the analyte concentration over the linear operating range of the mass spectrometer (Fig. 2A). Fig. 4 presents an outline of algorithm operation in the context of the MRM-MS work flow.
Fig. 4. Analysis work flow for isotope dilution MRM-MS data with and without the use of AuDIT.
After LC-MRM-MS analysis of samples, transition peaks are identified and integrated with software from either the mass spectrometer vendor or another supplier. (A), Flow of data with use of the automated algorithm. The statistical test identifies problem transitions from the variation in the relative ratios for the analyte and the SIS. The CV of the PARs is used as a filter to flag transitions with unacceptably large variation. (B), The current standard practice of careful manual inspection of all transitions by an expert.
Relative ratios of observed transition intensities are calculated by 1 of 2 methods at the opposite ends of the complexity spectrum. The minimal-pairs method calculates the relative ratio of a given transition by dividing its intensity by the intensity of one other transition from the same precursor. In the case of 3 monitored transitions, 3 relative ratios are calculated from the 3 pairs of transitions. A natural way to implement this method is to impose an arbitrary order on the transitions and to use adjacent transitions to calculate relative ratios, with the last transition wrapping around to the first transition in the list. The minimal-pairs method calculates as many relative ratios as there are transitions and can identify individual errant transitions, as long as they are fewer than half the total number of recorded transitions.
The alternative method, which is at the other extreme in terms of the number of ratios calculated, is the all-pairs method. In this scheme, all possible pairs of transitions from a single precursor are chosen, and their observed intensities (peak areas from the XIC) are used to calculate as many relative ratios. In this method, each transition participates in 1 less ratio than the total number of monitored transitions. Thus, any number of problematic transitions can be identified.
The relative ratios are calculated separately for the analyte and the corresponding SIS peptide. AuDIT compares the relative ratios from the analyte peptide transitions with those of the SIS peptide to determine if they are similar. It then checks for aberrant transitions by applying a statistical test to these ratios. Replicates of a given ratio (derived from the respective sample replicates) are used in a t-test to check if the relative ratio for the analyte deviates from the corresponding ratio for the SIS. For an experiment that monitors several peptides with multiple transitions per peptide, a correction for multiple hypothesis testing is applied (36). Because a given transition is used in the calculation of multiple ratios, the corrected P values for these corresponding ratios are combined to estimate the P value for deciding whether a given transition is problematic. P values are combined by means of a method meant for multiple dependent P values (because the same intensities are used in multiple ratios, from which the P values to be combined are calculated) (37, 38). Transitions with corrected and combined P values less than a specified threshold (e.g., 10−5) are marked as “bad.”
Although the t-test evaluates whether the analyte and SIS ratios are similar, the test cannot estimate for either analyte or SIS peptides whether the ratios of product ions have unacceptably large variation. This orthogonal aspect is assessed with the CV of the PAR for the transition, which is based on measurements of replicate samples (either technical replicate injections of the same sample or process replicates). Any transitions with a PAR CV greater than a chosen limit (e.g., 20%) are deemed problematic, because such a large variation is unexpected in the linear-response region in MRM-MS assays. Such transitions are labeled as “bad.”
Finally, combining the orthogonal modalities allows a transition with either a significant P value or an unacceptable CV to be marked as “bad” as a whole and thus be deemed unacceptable for quantification.
ALGORITHM EVALUATION AND VALIDATION
There are currently no automated methods for identifying transitions with interferences (or other factors as discussed above) that can render them unsuitable for quantification. Consequently, the final decision on the true quality of a transition is subjective and until now has relied entirely on expert review of the data (6, 32). To evaluate our algorithm for detecting inaccurate and imprecise transitions, we compared the AuDIT results with those of an expert using a 2-step process. In the preliminary phase, the expert looks at all of the integrated XICs and creates an unbiased prealgorithm annotation that documents any potential problems, such as poor chromatography, inaccurate peak integration, and so forth. The expert records these annotations at the level of the MRM transition. The data are then run through AuDIT, and the algorithm’s good or bad classification is compared with the expert’s annotation (“global”; see Table 1 in the online Data Supplement). In cases in which AuDIT’s decision and the expert’s annotation disagree, the expert reevaluates the transitions to determine whether AuDIT’s assessment is justifiable—i.e., that the actual observations of questionable data quality or interferences are such that the relative ratio is not affected (and hence the transition may be used for quantification) or vice versa. This final phase of expert review creates a postalgorithm annotation that is performed with the same criteria and rigor as the first review but is primed for issues that might have been overlooked or wrongly assessed. This focused annotation is compared with AuDIT decisions to evaluate the algorithm’s efficacy in identifying inaccurate transitions. Table 1 and Table 1 in the online Data Supplement summarize the overall performance.
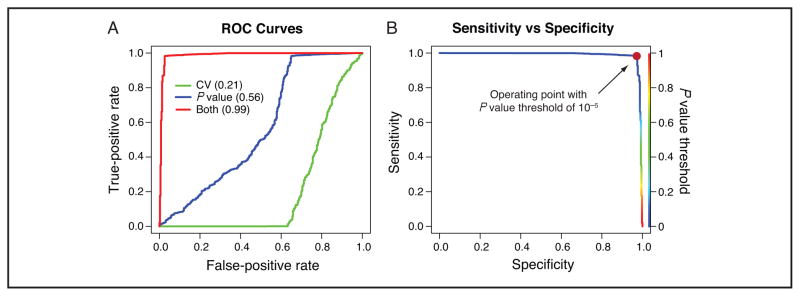
A 2 × 2 contingency matrix (Table 1A) is created to evaluate the performance of AuDIT on each data set, with a positive defined as a good transition call. Table 1A shows the various elements of the contingency matrix. Algorithm performance is estimated according to (a) overall accuracy, (b) sensitivity, and (c) specificity (Table 1). An ROC curve is used to evaluate the combined effect of incorporating P value and CV in the algorithm calculations for flagging inaccurate and imprecise peptides (Fig. 5A). The area under the ROC curve (AUC) is an indication of the quality of the classifier (46, 47). The ROC curve and the AUC also show that the t-test and the CV jointly achieve significantly better performance than either measure alone. The AUC for predicting transition quality with only the CV is <0.5, indicating that this modality is worse than a random predictor. The CV is affected only under specific circumstances, most of which are orthogonal to situations in which a significant t-test score will be obtained. It is therefore imperative that both the t-test and the CV be used to derive an accurate predictor of imprecise or inaccurate transitions. The P value for these experiments was set to 10−5, and the CV value was set to 0.2. Both the P value and CV thresholds are adjustable. Whereas the CV was set to an arbitrary value of 0.2, a sensitivity–specificity curve (Fig. 5B) was used to assess the effect of changing the P value threshold for comparing relative ratios of the fragment ions of the analyte and SIS peptides. As the P value threshold increases above 10−5, a concomitant decrease in sensitivity is observed. At P values <10−5, the specificity of the algorithm decreases. Thus, we selected a P value of 10−5 as the algorithm’s optimum threshold for sensitivity and specificity for identifying inaccurate or imprecise transitions.
Fig. 5. ROC curve and sensitivity–specificity plots summarizing performance of AuDIT in identifying inaccurate and imprecise transitions, as evaluated by an expert.
AuDIT uses the t-test P value and the CV of the PAR (ratio of analyte peak area to SIS peak area) to detect problem transitions. (A), Both the P value and the CV are required to achieve acceptable performance (i.e., as indicated by AUC values in parentheses). (B), Specificity and sensitivity values achieved as the P value threshold is varied from 0 to 1 (with a fixed CV threshold of 20%). The chosen P value threshold of 10−5 used for all of the analyzed data is indicated by the red circle (sensitivity, 98%; specificity, 97%). The rainbow color bar (right y axis) keys the location of the P value threshold on the sensitivity–specificity curve.
ALGORITHM REQUIREMENTS
AuDIT is based on comparing the relative-intensity ratios of the analyte’s transitions with those of the SIS peptide for all peptides being monitored. Therefore, an SIS with the same fragment ions as the analyte is a requirement for applying the algorithm. Furthermore, applying the t-test to ratios and then determining which transition is “inaccurate” requires that each transition be paired with 2 different transitions in calculating ratios. If a transition is used in only a single ratio and that ratio is problematic, it will not be possible to decipher which of the 2 transitions constituting the ratio is the cause. Thus, at least 3 transitions must be monitored for each peptide. Finally, although it is possible to apply the t-test to samples with only 2 replicates, additional replicates will render the result increasingly selective, robust, and reliable. We recommend a minimum of 3 to 4 replicate measurements.
ALGORITHM PERFORMANCE FOR CALIBRATION CURVE DATA
AuDIT has been validated primarily with data from a large-scale interlaboratory study of precision and reproducibility that was conducted under the auspices of the National Cancer Institute’s Clinical Proteomic Technology Assessment for Cancer program and used MRM-based measurements of proteins in plasma (34). AuDIT was used to assess calibration curves for 10 peptides from 4 sites that monitored 3 transitions per peptide (and each peptide’s corresponding SIS peptide) and 4 replicates per sample point. The results, summarized in Table 1, show that AuDIT can achieve overall accuracies of 94%–100% with sensitivities and specificities of 93%–100% and 94%–100%, respectively. Although specificities based on global annotation are low for some of the data sets (see Table 1 in the online Data Supplement), these results are due primarily to conservative calls by the expert and are significantly improved with the focused annotation. These data were generated with an Applied Biosystems 4000 Q TRAP triple-quadrupole mass spectrometer and processed with MultiQuant software.
Data from 2 of these sites (sites 1 and 2) and data from a new site (site 5) that used a Thermo TSQ Quantum triple-quadrupole mass spectrometer were also analyzed with Skyline software. Skyline has the advantage of being able to process data from mass spectrometers from different vendors, which enables the use of consistent settings for peak-integration parameters (e.g., identical start and stop times for integration for the analyte and SIS peptides and all transitions). Applying AuDIT to these data produces the performance results shown in Table 1. As is evident from this table, the software used for peak integration (MultiQuant or Skyline) does not have an effect on algorithm performance; however, the different mechanisms of peak integration for the 2 software packages may yield different numbers of good and bad assignments from the expert and AuDIT.
Fig. 5 presents ROC and sensitivity–specitivity plots for all MultiQuant (4 sites) and Skyline (3 sites) data from the 10-peptide response curve monitoring 3 transitions per peptide. Fig. 5A plots the false-positive rate against true-positive rate as the CV and P value thresholds are varied. The figure also compares the curves for the t-test alone and the CV alone with the current method that uses both the t-test results and the CV filter. Use of the CV or the P value alone to decide on transition quality yields a very marginal performance, with AUCs of 0.21 and 0.56, respectively. Combining the CV (<20%) and the P value (varies between 0 and 1) produces a classifier that performs significantly better (AUC, 0.99). This result indicates that P value and CV provide complementary information: The P value detects changes in the relative ratio caused by interferences, and the CV filters out transitions that have unacceptably large variation in replicate analyses. As is evident from Fig. 5B, sensitivities and specificities of about 97%–98% are achievable with appropriate values for the P value threshold.
Data for samples from study II of the National Cancer Institute’s Clinical Proteomic Technology Assessment for Cancer interlaboratory study (34) were also acquired at the Broad Institute on a Q TRAP 5500 instrument, and 5 transitions were monitored for each of the 10 peptides and their corresponding SIS peptides. For this set of samples, both the minimal-pairs and all-pairs methods were used to calculate relative ratios for the algorithm. These methods perform identically in terms of detecting problematic transitions. The all-pairs method evaluated each transition with more ratios than the minimal-pairs method. Although this difference did not matter for this data set, the all-pairs method has the potential to detect and pinpoint more erroneous transitions than the minimal-pairs method when the number of transitions exceeds 3; however, this advantage comes at the cost of calculating substantially more ratios. As more transitions are monitored per peptide, the number of ratios calculated with the all-pairs method increases markedly. The minimal-pairs method, on the other hand, always uses as many ratios as there are transitions and works most efficiently with odd numbers of ratios.
APPLICATION OF AuDIT TO CLINICAL SAMPLES
We also used AuDIT to analyze a data set of clinically relevant samples to identify any problematic transitions. The samples (26) consisted of patient plasma collected at 3 time points after an induced myocardial infarction. After abundant proteins were depleted, the samples were reduced, alkylated, digested with trypsin, and fractionated at the peptide level with strong cation-exchange chromatography. Peptides from 6 cardiovascular disease–related proteins were monitored in the various chromatography fractions, and a subset of 2 peptides was evaluated with AuDIT. For these data, AuDIT had a sensitivity of 100% (87% for global annotation) and a specificity of 82% (31% for global annotation), results that were comparable to those of the other data sets (Table 1).
Discussion
With increasing focus on biomarker verification as a bridge between biomarker discovery and clinical validation, greater attention must be paid to the analytical requirements for these quantitative assays. Fortunately, quantitative detection of small molecules with MRM-based assays has formed a good foundation for identifying the criteria that should be focused on and achieved (3). For example, analytical guidance criteria developed for small molecules have recommended that at least 3 fragment ions be detected for analyte identification with MS and that the relative abundances of structurally specific fragment ions be within ±20% of the reference standard (9, 10, 48).
Use of MRM-MS as the tool for precise, relative quantification of candidate biomarkers requires that 2 criteria be met: (a) sufficient information to correctly identify the target peptide in the sample matrix; and (b) interference-free signals from some or all of the fragment ions used to identify and quantify the peptide. As we have demonstrated here and in our prior publications (21, 26, 34), the only reliable method for achieving these criteria in complex proteomic samples is through the use of an SIS peptide. When an SIS peptide and monitoring for at least 3 transitions of both the analyte and the SIS forms of that peptide are used, we have observed that the relative ratios of the transitions are very constant throughout the linear range of the peptide (Fig. 2A) and that these ratios vary much less than 20% (often only 2%–5%) in replicate injections and across a wide range of concentrations. Incorporation of an SIS peptide adds an additional level of confidence in peptide identification by providing a reference to which the fragment ion ratios can be compared, as well as providing a precise retention time standard for the analyte of interest that is independent of the chromatographic conditions used. A greater deviation in the relative ratios of specific transitions for the analyte peptide compared with those of the SIS peptide (e.g., Fig. 2B) indicate to the analyst that a coeluting interference is likely present and that corrective measures must be taken to obtain useful quantitative information.
Use of external standards for quantification in peptide MRM-MS has been proposed (29). The advantages are that only the less expensive light (i.e., nonisotopic) form of the peptide need be synthesized and evaluated to define the MRM-MS fragmentation and chromatographic behavior. The variable suppression effects of the matrix are not accounted for in this method, however, and this deficiency could adversely affect the accuracy of the quantification, as the small-molecule community has previously demonstrated (11–13). In addition, false-positive identifications can lead to concomitant inaccurate quantification if the relative ratios of the fragment ions of the signature peptides are not also used for positive identification and quantification. Interfering components that are frequently present in blood give rise to all of the fragment ions being monitored, and some of these components elute very close in time to the authentic analyte. The observed reversed-phase HPLC retention time of an external standard peptide measured in buffer can shift by more than several minutes in the presence of a complex biological matrix. In addition, if an interference is present in one or more of the analyte transitions, there will not be a 1:1 match between the spectra of the external standard and the authentic analyte. Making both of these problems detectable through the use of the SIS enables corrective actions to be taken.
A common practice during SID-MRM-MS data processing is to observe the summed signals from all transitions of a single precursor and to compare them with those of the corresponding SIS peptide. This approach is valid and accurate only if each of the transitions monitored is known to be interference free. If the potential for interference is not assessed, the result may be gross miscalculation of analyte quantity. Therefore, we recommend that this method not be used unless the data have been shown to be interference free through the use of an SIS peptide.
In developing the first automated method to filter large MRM-MS data sets in this study, we flagged peptides with transitions of low quality while passing through for quantification peptide data that are interference free and of high quality. Such automated methods are clearly needed to aid processing of peptide MRM-MS data generated from analyses of complex biological samples. Even modest MRM-MS experiments produce very large amounts of data that presently require manual inspection. For example, an experiment consisting of an assay of 10 peptide targets (3 transitions for a light and heavy peptide) and 40 sample injections would produce >2000 XICs to inspect. This inspection process currently consumes enormous amounts of time of a highly trained analyst and is prone to human error. The development of this algorithm was driven and informed by our experience in analyzing many hundreds of large MRM-MS data sets, as well as the experiences of the small-molecule community that we have noted.
In our approach, the data not passing through the filter are marked for manual inspection on the basis on 2 criteria: (a) The relative ratios of the transitions of the analyte peptide do not agree with the relative ratios of the SIS peptide; or (b) the CV in the quantitative measurements of 3 to 4 sample replicates is too large (>20%). In the case of the first criterion, if the fragmentation pattern and the relative intensity of the analyte peptide do not agree with those of the SIS peptide, the quantitative measurement is inaccurate. In the case of the second criterion, the high variation of the data causes the quantitative measurement to be imprecise. In either case, if the cause for flagging the data is something that can be fixed, such as integration of the wrong peak or inconsistent baseline integration of the analyte and SIS peptides, then the analyst may annotate and resolve the integration issue. If instead the cause is bad chromatography or a bad injection, the sample could be queued to be rerun before much time is lost.
With the P value and the CV as 2 orthogonal dimensions used by AuDIT for identifying errant transitions, the method is geared toward flagging transitions that have the following: (a) an interference, saturation, or other confounding factor that changes the relative ratios for the analyte and SIS without substantially affecting the CV of the measurements; and (b) unacceptably large variation in replicate samples. The latter situation can arise from poor chromatography or an extremely lowsignal-to-noiseratio,whichcanincreasetheCVofthe measurement. Such problems can escape detection by the t-test because the increased variance of the ratios will reduce the statistical significance of any differences in the relative ratios of the analyte and the SIS.
From our comparative analysis of several large MRM-MS data sets, we found that use of AuDIT greatly increased the speed of quantitative analysis, from 2-fold to >10-fold. Analysis of the 8 large MRM-MS data sets (Table 1) with the traditional method of expert manual inspection of all transitions, flagged 9%–43% of the transitions as suspect, with a median of 23% across the 8 data sets. In the data sets containing 3 MRM transitions for 10 analyte and SIS peptides and a 9-point calibration curve with a total of 2160 XICs, having to inspect only 240 XICs (or 120 analyte and 120 transition pairs) is a 10-fold reduction in the time required for analyzing the data. This reduction translates into a time savings of approximately 3 to 4 h for a highly trained analyst. Of course, the overall quality of the data and how consistently it is integrated will have an effect on how many XICs are flagged for manual inspection.
The sensitivity, specificity, and overall accuracy of the data analyses also increased substantially with AuDIT compared with expert manual analysis (Table 1). In the most extreme case, increases of 75% in accuracy and >80% in specificity were achieved with AuDIT. These improvements were due in part to reducing the number of XICs requiring manual inspection to a smaller subset, thereby allowing the expert to evaluate the data more critically. In addition, AuDIT readily identifies issues with the data that are not immediately obvious to the expert, such as high imprecision (>20%) in the PAR of the analyte and SIS peptides in replicate samples, thus enabling the identification of transitions that do not deliver precise measurements.
We applied AuDIT to the analysis of real-world clinical samples for which the analyte concentrations were unknown. In this example, several XICs were flagged for manual inspection (18% of the total). AuDIT flagged 10% of the spectra with large variation in the technical replicates (i.e., CV >20%) that the expert had not identified as problematic data. AuDIT also identified 3 instances of interference or saturation in the data set (reflected in a small P value) that the expert had originally indicated. AuDIT did not identify 2 cases of interference in the SIS transition, however, because the CV criteria used were insufficiently stringent. In both cases, the calculated CVs for the transition in question were very close to 20%, just large enough to render the t-test ineffective. AuDIT has been designed so that the user can adjust the thresholds for P and CV values. Adjusting the CV criterion to a more stringent value of 15% correctly identified both false transitions without affecting any of the other performance criteria for the AuDIT analysis of this data set.
AuDIT can currently be applied to data exported from most analysis software. Embedding this algorithm in current peak-integration software programs would greatly reduce manual inspection and alert the researcher of potentially errant data at an early point in the data analysis, potentially allowing data for problematic samples with large CV values (due to column degradation and poor peak shape, for example) to be reacquired. In addition, the incorporation of AuDIT into these programs would streamline the current process, likely producing more efficient generation of accurate and precise quantitative data from SID-MRM-MS analyses.
In conclusion, with the increasing momentum observed in MRM-based methodologies for peptide and protein quantification, it is becoming clear that manual, subjective approaches to data analysis will shortly become the bottleneck in biomarker verification. To address this issue and to maintain this momentum, we developed and tested an algorithm that automatically detects inaccurate and imprecise transitions in MRM-MS data. Although peptide transitions may appear reliable and interference free in the method-development phase, in which use of a matrix of pooled samples is common, interferences can appear during routine analyses of a wide range of patient samples. Thus, this algorithm may be implemented as a necessary tool for data review at all stages of quantitative MRM-MS–based assays. The AuDIT software increases the speed of analysis of large MRM-MS data sets by minimizing the need for subjective, time-consuming inspection of the data. AuDIT also improves the overall accuracy of data analysis and works robustly when challenged with multiple types of interferences and problematic transitions. AuDIT currently works with results exported from MRM-MS data-processing software packages and may be readily implemented as an analysis tool in such software. We believe that AuDIT will accelerate adoption of MRM-MS assays by the proteomics and clinical chemistry communities by reducing the difficulty of analyzing MRM-MS peptide data.
Supplementary Material
Acknowledgments
Research Funding: This work was supported in part by grants to S.A. Carr from the National Cancer Institute (NCI; U24 CA126476) as part of the Clinical Proteomic Technologies for Cancer (CPTC) program, from the National Heart Lung and Blood Institute (grant R01 HL096738-01), and from The Women’s Cancer Research Fund of the Entertainment Industry Foundation, and by a grant to D.R. Mani from the National Cancer Institute Clinical Proteomic Technologies initiative (R01 CA126219).
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: MRM-MS, multiple reaction monitoring mass spectrometry; Q1, quadrupole 1; MS/MS, tandem MS; SID, stable isotope dilution; LC-MS/MS, liquid chromatography MS/MS; AuDIT, automated detection of inaccurate and imprecise transitions; SIS, stable isotope–labeled internal standard; XIC, extracted ion chromatogram; PAR, peak area ratio; csv, comma-separated value; AUC, area under the ROC curve.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.Yost RA, Enke CG. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal Chem. 1979;51:1251–64. doi: 10.1021/ac50048a002. [DOI] [PubMed] [Google Scholar]
- 2.Brumley WC, Sphon JA. Regulatory mass spectrometry. Biomed Mass Spectrom. 1981;8:390–6. doi: 10.1002/bms.1200080908. [DOI] [PubMed] [Google Scholar]
- 3.Sphon JA. Use of mass spectrometry for confirmation of animal drug residues. J Assoc Off Anal Chem. 1978;61:1247–52. [PubMed] [Google Scholar]
- 4.Vargo JD. Determination of sulfonic acid degradates of chloroacetanilide and chloroacetamide herbicides in groundwater by LC/MS/MS. Anal Chem. 1998;70:2699–703. doi: 10.1021/ac971365d. [DOI] [PubMed] [Google Scholar]
- 5.Draisci R, Palleschi L, Ferretti E, Lucentini L, Cammarata P. Quantitation of anabolic hormones and their metabolites in bovine serum and urine by liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2000;870:511–22. doi: 10.1016/s0021-9673(99)01293-5. [DOI] [PubMed] [Google Scholar]
- 6.Kushnir MM, Rockwood AL, Nelson GJ, Yue B, Urry FM. Assessing analytical specificity in quantitative analysis using tandem mass spectrometry. Clin Biochem. 2005;38:319–27. doi: 10.1016/j.clinbiochem.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Kuhara T. Noninvasive human metabolome analysis for differential diagnosis of inborn errors of metabolism. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855:42–50. doi: 10.1016/j.jchromb.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 8.Pitt JJ, Eggington M, Kahler SG. Comprehensive screening of urine samples from inborn errors of metabolism by electrospray tandem mass spectrometry. Clin Chem. 2002;48:1970–80. [PubMed] [Google Scholar]
- 9.Stein SE, Heller DN. On the risk of false positive identification using multiple ion monitoring in qualitative mass spectrometry: large-scale intercomparisons with a comprehensive mass spectral library. J Am Soc Mass Spectrom. 2006;17:823–35. doi: 10.1016/j.jasms.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Bethem R, Boison J, Gale J, Heller D, Lehotay S, Loo J, et al. Establishing the fitness for purpose of mass spectrometric methods. J Am Soc Mass Spectrom. 2003;14:528–41. doi: 10.1016/S1044-0305(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 11.Matuszewski BK, Constanzer ML, Chavez-Eng CM. Matrix effect in quantitative LC/MS/MS analyses of biological fluids: a method for determination of finasteride in human plasma at picogram per milliliter concentrations. Anal Chem. 1998;70:882–9. doi: 10.1021/ac971078+. [DOI] [PubMed] [Google Scholar]
- 12.King R, Bonfiglio R, Fernandez-Metzler C, Miller-Stein C, Olah T. Mechanistic investigation of ionization suppression in electrospray ionization. J Am Soc Mass Spectrom. 2000;11:942–50. doi: 10.1016/S1044-0305(00)00163-X. [DOI] [PubMed] [Google Scholar]
- 13.Tang L, Kebarle P. Dependence of ion intensity in electrospray mass spectrometry on the concentration of the analytes in the electrosprayed solution. Anal Chem. 1993;65:3654–68. [Google Scholar]
- 14.Tiller PR, Romanyshyn LA. Implications of matrix effects in ultra-fast gradient or fast isocratic liquid chromatography with mass spectrometry in drug discovery. Rapid Commun Mass Spectrom. 2002;16:92–8. doi: 10.1002/rcm.544. [DOI] [PubMed] [Google Scholar]
- 15.Browne TR. Stable isotopes in pharmacology studies: present and future. J Clin Pharmacol. 1986;26:485–9. doi: 10.1002/j.1552-4604.1986.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 16.Parsons HG. Stable isotopes in the management and diagnosis of inborn errors of metabolism. Can J Physiol Pharmacol. 1990;68:950–4. doi: 10.1139/y90-144. [DOI] [PubMed] [Google Scholar]
- 17.Moore LJ, Machlan LA. High accuracy determination of calcium in blood serum by isotope dilution mass spectrometry. Anal Chem. 1972;44:2291–6. doi: 10.1021/ac60322a014. [DOI] [PubMed] [Google Scholar]
- 18.Cohen A, Hertz HS, Mandel J, Paule RC, Schaffer R, Sniegoski LT, et al. Total serum cholesterol by isotope dilution/mass spectrometry: a candidate definitive method. Clin Chem. 1980;26:854–60. [PubMed] [Google Scholar]
- 19.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, et al. Isotope dilution–mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42:1676–82. [PubMed] [Google Scholar]
- 20.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–9. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bondar OP, Barnidge DR, Klee EW, Davis BJ, Klee GG. LC-MS/MS quantification of Zn-α2 glycoprotein: a potential serum biomarker for prostate cancer. Clin Chem. 2007;53:673–8. doi: 10.1373/clinchem.2006.079681. [DOI] [PubMed] [Google Scholar]
- 23.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat Biotechnol. 2006;24:971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 24.Whiteaker JR, Zhao L, Zhang HY, Feng LC, Piening BD, Anderson L, Paulovich AG. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal Biochem. 2007;362:44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, et al. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem. 2009;55:1108–17. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshishian H, Addona T, Burgess M, Mani DR, Shi X, Kuhn E, et al. Quantification of cardiovascular biomarkers in patient plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2009;8:2339–49. doi: 10.1074/mcp.M900140-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciccimaro E, Hanks SK, Yu KH, Blair IA. Absolute quantification of phosphorylation on the kinase activation loop of cellular focal adhesion kinase by stable isotope dilution liquid chromatography/mass spectrometry. Anal Chem. 2009;81:3304–13. doi: 10.1021/ac900204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prakash A, Tomazela DM, Frewen B, MacLean B, Merrihew G, Peterman S, MacCoss MJ. Expediting the development of targeted SRM assays: using data from shotgun proteomics to automate method development. J Proteome Res. 2009;8:2733–9. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin DB, Holzman T, May D, Peterson A, Eastham A, Eng J, McIntosh M. MRMer, an interactive open source and cross-platform system for data extraction and visualization of multiple reaction monitoring experiments. Mol Cell Proteomics. 2008;7:2270–8. doi: 10.1074/mcp.M700504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Z, Maher N, Torres R, Cotto C, Hastings B, Dasgupta M, et al. Isobaric metabolite interferences and the requirement for close examination of raw data in addition to stringent chromatographic separations in liquid chromatography/tandem mass spectrometric analysis of drug in biological matrix. Rapid Commun Mass Spectrom. 2008;22:2021–8. doi: 10.1002/rcm.3577. [DOI] [PubMed] [Google Scholar]
- 33.Abbatiello SE, Pan Y–X, Zhou M, Wayne AS, Veenstra TD, Hunger SP, et al. Mass spectrometric quantification of asparagine synthetase in circulating leukemia cells from acute lymphoblastic leukemia patients. J Proteomics. 2008;71:61–70. doi: 10.1016/j.jprot.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2008. [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 37.Brown MB. A method for combining non-independent one-sided tests of significance. Biometrics. 1975;31:987–92. [Google Scholar]
- 38.Kost JT, McDermott MP. Combining dependent p-values. Stat Probab Lett. 2002;60:183–90. [Google Scholar]
- 39.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 40.Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–82. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- 41.Spengler B. De novo sequencing, peptide composition analysis, and composition-based sequencing: a new strategy employing accurate mass determination by Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2004;15:703–14. doi: 10.1016/j.jasms.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7:1952–62. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman J, McKay MJ, Ashman K, Molloy MP. Unique ion signature mass spectrometry, a deterministic method to assign peptide identity. Mol Cell Proteomics. 2009;8:2051–62. doi: 10.1074/mcp.M800512-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun S, Meyer-Arendt K, Eichelberger B, Brown R, Yen CY, Old WM, et al. Improved validation of peptide MS/MS assignments using spectral intensity prediction. Mol Cell Proteomics. 2007;6:1–17. doi: 10.1074/mcp.M600320-MCP200. [DOI] [PubMed] [Google Scholar]
- 45.Holland JF, Sweeley CC, Thrush RE, Teets RE, Bieber MA. On-line computer controlled multiple ion detection in combined gas chromatography-mass spectrometry. Anal Chem. 1973;45:308–14. doi: 10.1021/ac60324a042. [DOI] [PubMed] [Google Scholar]
- 46.Fawcett T. ROC graphs: notes and practical considerations for researchers. Palo Alto (CA): HP Laboratories; 2004. [Google Scholar]
- 47.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–74. [Google Scholar]
- 48.Heller DN. U.S. Department of Health and Human Services, Food and Drug Administration (FDA), Center for Veterinary Medicine. Rockville (MD): FDA; 2003. Guidance for industry: mass spectrometry for confirmation of the identity of animal drug residues. Report nr: guidance document 118; p. 11. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.