Abstract
Human cytidine deaminases, including APOBEC3G (A3G) and A3F, are part of a cellular defense system against retroviruses and retroelements including non-LTR retrotransposons LINE-1 (L1) and Alu. Expression of cellular A3 proteins is sufficient for inhibition of L1 and Alu retrotransposition, but the effect of A3 proteins transferred in exosomes on retroelement mobilization is unknown. Here, we demonstrate for the first time that exosomes secreted by CD4+ H9 T cells and mature monocyte-derived dendritic cells encapsidate A3G and A3F and inhibit L1 and Alu retrotransposition. A3G is the major contributor to the inhibitory activity of exosomes, however, the contribution of A3F in H9 exosomes cannot be excluded. Additionally, we show that exosomes encapsidate mRNAs coding for A3 proteins. A3G mRNA, and less so A3F, was enriched in exosomes secreted by H9 cells. Exosomal A3G mRNA was functional in vitro. Whether exosomes inhibit retrotransposons in vivo requires further investigation.
Keywords: Exosomes, APOBEC3G, APOBEC3F, LINE-1, Alu, Retrotransposon
Introduction
Long interspersed elements (L1) are the only active autonomous non-LTR retrotransposons in the human genome. While L1s make up about 17% of the human genome, only 80 to 100 of the over 500,000 copies can retrotranspose (Kazazian, 2004). L1s are about 6 kb long, harbor an internal polymerase II promoter and encode two open reading frames (ORFs): ORF1p coding for a 40 kDa protein with RNA binding and RNA chaperone activity (Holmes, Singer, and Swergold, 1992; Kolosha and Martin, 2003; Martin, Li, and Weisz, 2000), and ORF2p coding for a 150 kDa protein with endonuclease (Feng et al., 1996), reverse transcriptase (Mathias et al., 1991), and C-terminal cysteine-rich domains (Fanning and Singer, 1987). Upon translation, L1 RNA assembles with ORF1p and ORF2p and the formed ribonucleoprotein complex moves to the nucleus, where an endonuclease makes a single-stranded nick. The reverse transcriptase uses the nicked DNA to prime reverse transcription of the L1 RNA, leading ultimately to integration of L1 cDNA (Babushok and Kazazian, 2007). In contrast to L1, Alu elements are non-autonomous and are mobilized by the L1 retrotransposition machinery (Dewannieux, Esnault, and Heidmann, 2003; Ostertag and Kazazian, 2001). As insertions of both Alu and L1 elements into the human genome may cause a number of human genetic disorders (Callinan and Batzer, 2006; Wallace, Belancio, and Deininger, 2008), under normal conditions retrotransposons are silenced. Mechanisms protecting cells against retrotransposition include repression of transcription by DNA methylation (Roman-Gomez et al., 2005), premature polyadenylation (Perepelitsa-Belancio and Deininger, 2003), aberrant splicing (Belancio, Hedges, and Deininger, 2006) or RNA interference (Yang and Kazazian, 2006). Another strategy is editing of nucleic acids by the APOBEC3 (A3) family of cellular cytidine deaminases.
The most prominent member of this family, APOBEC3G (A3G), first identified through its ability to inhibit replication of vif-deficient HIV-1 (Sheehy et al., 2002), also functions as a post-entry restriction factor against wild-type viruses (Chiu et al., 2005; Khatua et al., 2009; Pion et al., 2006; Vetter and D'Aquila, 2009; Wang et al., 2008a). Although deaminase-independent activities of A3G are postulated to contribute to its antiviral function (Bishop, Holmes, and Malim, 2006; Holmes et al., 2007; Newman et al., 2005), there is strong evidence that HIV-1 restriction requires A3G deaminase activity when the protein is not overexpressed (Browne, Allers, and Landau, 2009; Miyagi et al., 2007; Schumacher et al., 2008). In mammals, A3G also restricts replication of a variety of endogenous retroviruses (Esnault et al., 2005; Esnault et al., 2006; Esnault et al., 2008; Schumacher et al., 2008) and retroelements including Alu (Bogerd et al., 2006b; Chiu et al., 2006; Hulme et al., 2007) and L1 (Bogerd et al., 2006b; Chen et al., 2006; Kinomoto et al., 2007; Muckenfuss et al., 2006; Niewiadomska et al., 2007; Schumann, 2007; Stenglein and Harris, 2006; Tan et al., 2009; Turelli, Vianin, and Trono, 2004). The expansion of a single A3 gene in mice to the seven A3 genes found in primates correlates with the significant enhancement of restriction of endogenous retroelements seen in primates. This supports the suggestion that A3 proteins provide a cellular defense mechanism against endogenous and exogenous invaders (Esnault et al., 2005).
We recently reported that functional A3G associated with exosomes, is secreted by CD4+ H9 T cells (Khatua et al., 2009). This was corroborated by the observation that a fraction of cellular A3G associates with multivesicular bodies (MVB)/late endosomes (Alce and Popik, 2004), organelles that release intralumenal vesicles as exosomes after fusion with the plasma membrane (de Gassart et al., 2004; Stoorvogel et al., 2002; Thery, Zitvogel, and Amigorena, 2002). This observation suggests that A3G and possibly other A3 proteins are transmitted between cells, in exosomes, and contribute to an antiretroviral defense mechanism (Khatua et al., 2009).
The biological role of exosomes is not fully understood. Secretion of transcription factors (Ratajczak et al., 2006a; Ratajczak et al., 2006b), morphogenic proteins (Liegeois et al., 2006) as well as cellular mRNAs and miRNAs (Bruno et al., 2009; Deregibus et al., 2007; Valadi et al., 2007) in association with exosomes suggests that these vesicles may epigenetically reprogram the recipient cells and may play a role during embryogenesis. Although retrotransposition occurs sporadically in somatic cells, L1 and Alu transpositions are most active in germ cells and during early embryogenesis (Garcia-Perez et al., 2007; Prak et al., 2003; van den Hurk et al., 2007). We speculate that secretion of exosomes including A3G or other A3 proteins, in combination with functional mRNAs coding for these proteins, provides an additional line of defense against retroelements, especially in recipient cells that do not express or express only suboptimal levels of these proteins.
In the present study, we tested whether exosomes secreted by cells expressing A3G can restrict retrotransposition in recipient cells. We demonstrate that exosomes secreted by cells expressing A3G and A3F potently restrict L1 and Alu retrotransposition. This leads us to speculate that exosomes contribute to anti-retroelement defenses in vivo.
Materials and methods
Cells
SupT1, H9 and peripheral blood mononuclear cells (PBMCs) were propagated in RPMI supplemented with 10% FCS and gentamicin (50 µg/ml; complete RPMI). HeLa and 293T cells were cultivated in DMEM supplemented with 10% FCS and gentamicin (complete DMEM). PBMCs were obtained by Ficoll-Paque (Amersham) density centrifugation from healthy blood donors (New York Blood Center). Monocyte-derived dendritic cells (M-DC) were obtained from the plastic adherent fraction of PBMC. After 3h adherence to plastic dishes, adherent cells were washed, collected by scraping and plated in T75 flask at 2 × 106 cells/ml. The cells were cultivated for 6 days with 5 ng/ml interleukin-4 (IL-4; Humzyme) and 5 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF; Humzyme) in complete RPMI. Maturation of M-DC was induced by incubation of the immature M-DC with 10 ng/ml of lipopolysaccharide (LPS; E.coli strain O55:B5, Sigma-Aldrich) as described (Wang et al., 2007a). The M-DCs were more than 94% pure by HLA-DR and CD11c staining (FACS analysis).
Plasmids
Human A3-HA expression vectors were obtained from B. R. Cullen and A3H-V5 from Y. Zheng. L1RP-EGFP and EFO5J were obtained from E. T. L. Prak and H. H. Kazazian. Alu neoTet and p220.CMV L1-RP was provided by T. Heidmann.
L1 retrotransposition assay
293T cells were plated one day before transfection in 12-well plates at 2 × 105 cells/well in complete DMEM. The cells were transfected using the PolyFect reagent (Qiagen) and 1 µg of the L1 retrotransposition reporter plasmid without or with 0.5 µg of A3-HA or A3H-V5-His (Dang et al., 2008). The total amount of transfected DNA was adjusted to 1.5 µg with control pcDNA3.1 vector. After 3 h, the transfection medium was replaced with complete DMEM for another 3h. Exosomes (20 µg) were added where indicated and incubated with cells for 24 h. The following day, new aliquots of exosomes were added and incubated with cells for another 24 h. Three days post-transfection, the cells were trypsinized and transferred to 60-mm dishes. On day 5–7 post-transfection, the cells were analyzed for GFP expression by fluorescent microscopy and flow cytometry (FACSCalibur, BD). FACS analysis was performed on fixed cells and retrotransposition frequency was determined by gating GFP-positive cells.
L1-mediated Alu retrotransposition assay
HeLa cells (1.1 × 105 cells/well) were cotransfected using PolyFect (Qiagen) in 12-well plates with 1 µg of neoTet-marked Alu and 0.5 µg pCMV-L1 (p220 L1-RP). In control experiments, the cells were additionally cotransfected with A3G-V5 or GFP vectors. Negative controls were mock transfected or transfected with Alu, L1, or Alu and GFP. The transfected cells were incubated with different exosome preparations (20 µg), starting 6 h post-transfection with an additional dose of exosomes added next day. Three days post-transfection, the cells were trypsinized and transferred to 60-mm dishes. Starting at day 7 post-transfection, the cells were selected in the presence of G418 (1 mg/ml; Invitrogen) and the media were changed daily. One week later, the G418-resistant cell foci were fixed with 3% paraformaldehyde, stained with 1% crystal violet, washed, air-dried and counted using Qantity One software (ChemiDoc XRS, Bio-Rad).
Quantitative real-time RT-PCR analysis
Total RNA was extracted and purified using a total RNA purification kit (Norgen) and contaminating DNA was digested with Turbo DNA-free DNase (Ambion). Purified RNA (200 ng) was reverse transcribed to cDNA using an iScript cDNA synthesis kit (Bio-Rad) and subjected to real-time PCR using primers specific for human A3 genes (SABiosciences). Amplification of A3 genes was normalized to GAPDH as an endogenous control. Normalized expression of A3C in H9 cells or H9 exosomes was set as 1.0. The experiment was repeated twice with triplicate cDNA samples used for PCR.
Exosome isolation protocol
Exosome preparation was performed as previously described (Khatua et al., 2009). In short, cells were grown in culture media supplemented with 10% FCS depleted of endogenous exosomes. Culture supernatants were collected every 24 h and were first centrifuged at 300 × g for 10 min to pellet cells. Cell debris was removed from the resulting supernatant by centrifugation at 2000 × g for 15 min followed by 30 min centrifugation at 10,000 × g. The clarified supernatant was filtered through a 0.45 µm filter and ultracentrifuged at 100,000 × g using a SW32 rotor to pellet exosomes. Exosomes were washed by resuspension in PBS and pelleted for 1 h at 100,000 × g using a SW41 rotor. After suspension in a small volume (50–100 µl) of PBS, purified exosomes were aliquoted and stored at −80°C. Protein concentration of exosome preparations was determined using Micro BCA protein assay (Thermo Scientific).
PolyA+ mRNA isolation and in vitro translation
PolyA+ mRNA was isolated directly from H9 exosomes using Oligotex direct mRNA protocol according to the manufacturer’s specifications (Qiagen). One µg of polyA+ mRNA was used for in vitro translation using human in vitro protein expression kit according to manufacturer’s recommendation (Thermo Scientific). A sample without added RNA was used as a negative control. GFP mRNA was used as a positive control. Translation reactions (25 µl) were performed for 90 min at 30°C and total reaction volumes were subjected to Western blot analysis.
Western blot analysis
Proteins were separated on SDS-10% polyacrylamide gels and analyzed as described (Khatua et al., 2009). Antibodies for Western blotting were obtained from the following sources; rabbit anti-Alix from H. Stenmark (Oslo, Norway), rabbit anti-A3G from K. Strebel (NIH), rabbit anti-GFP, monoclonal anti-HA and anti-V5 antibodies from Invitrogen, rabbit anti-Rab7 from Santa Cruz Biotechnology. Rabbit anti-A3F antibodies were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Statistical analysis
All experiments were repeated at least three times with similar results. The variations in single experiments were expressed by calculating the standard deviation (SD) from triplicate samples and presented as the mean values ± SD.
Results
Exosomes secreted by H9 CD4+ T cells and mature monocyte-derived dendritic cells (M-DCs) inhibit L1 retrotransposition
Previous reports have shown that multiple human A3 proteins inhibit retrotransposition of L1 to different degrees, with A3A, A3B, A3C and A3F being the most effective and A3DE, A3G, and A3H having a smaller or negligible effect on L1 mobilization (Bogerd et al., 2006b; Chen et al., 2006; Kinomoto et al., 2007; Muckenfuss et al., 2006; Niewiadomska et al., 2007; Stenglein and Harris, 2006; Turelli, Vianin, and Trono, 2004). We previously showed that A3G is specifically packaged and secreted by cells in association with exosomes that confer resistance to HIV-1 after internalization by recipient cells (Khatua et al., 2009). To determine whether exosomes secreted by cells expressing A3G or other A3 proteins show inhibitory activity against retroelements, we analyzed the effect of isolated exosomes on L1 retrotransposition using an established 293T cell culture-based retrotransposition reporter assay (Brouha et al., 2003; Ostertag et al., 2000; Wei et al., 2000).
For this assay, we used a construct in which an active L1RP (L1) element (Kimberland et al., 1999; Ostertag et al., 2000) was tagged with a retrotransposition indicator EGFP cassette. The EGFP gene, under control of a CMV promoter, was interrupted by a γ-globin intron and was cloned in a transcriptional orientation opposite to L1. EGFP was expressed only after the full-length L1 was transcribed, the intron was removed by cellular splicing machinery, and the RNA was reverse transcribed and integrated into the host genome. 293T cells were transfected with the L1 reporter construct alone or in combination with plasmids expressing various human A3 proteins (Fig. 1). In addition, cells transfected with L1 were exposed to exosomes secreted by H9 CD4+ T cells, activated M-DC, and SupT1 CD4+ T cells, or exosomes isolated from the blood plasma of normal blood donors. 293T cells were also transfected with EF05J, a negative control derived from L1RP containing disabling mutations in ORF1 (Farkash et al., 2006).
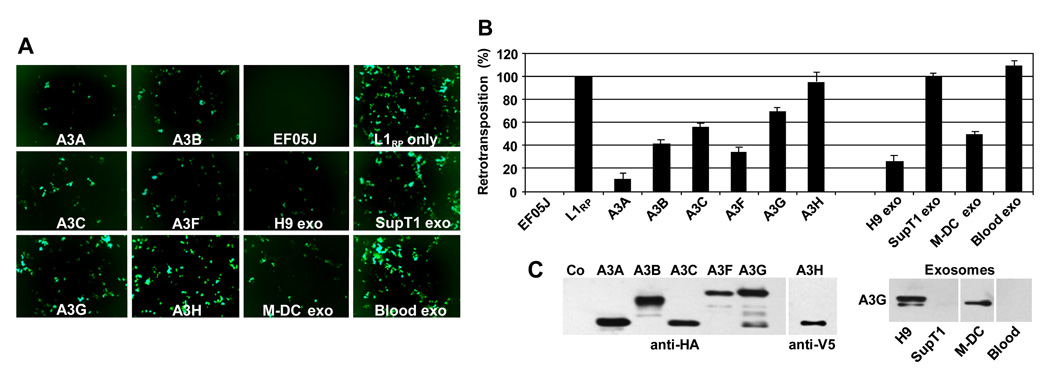
Fig. 1. Effect of A3 proteins and exosomes on L1 retrotransposition.
293T cells were transfected with a pL1RP tagged with an EGFP retrotransposition indicator cassette and HA- or V5-tagged A3 expression plasmids. Cells transfected with pL1RP-EGFP only were exposed to exosomes (20 µg/ml) secreted by H9, SupT1, M-DC, or isolated from peripheral blood plasma (Blood exo). (A) After 5 days, cells were observed by fluorescence microscopy and (B) trypsinized, fixed with 3% paraformaldehyde and subjected to flow cytometry analysis. The retrotransposition level in cells transfected with pL1RP-EGFP (in the absence of A3 proteins or exosome treatment) was set as 100%. EF05J = negative control with inactive pL1RP-EGFP. Error bars, mean values ± SDs of triplicate samples. Similar results were obtained in 4 independent experiments. (C) Expression of A3G proteins in transfected cells (left panel) or in exosomes (right panel) was analyzed by SDS-PAGE followed by immunoblotting with anti-HA or anti-V5. A3G in exosomes was detected with anti-A3G antibodies.
Retrotransposition events were monitored by the appearance of EGFP-positive cells 5–7 days after transfection (Fig. 1A) and quantitated by flow cytometry (Fig. 1B). Expression of A3 proteins, used as controls, showed an anticipated inhibition of L1 retrotransposition, with cells containing A3A, A3B, and A3F having the most potent inhibitory activity. Retrotransposition events were also inhibited by A3C (about 43%) and by A3G (about 30%). A3H showed no activity against L1 under the conditions of assay. Observed differences in L1 inhibition were most likely A3 dependent since all A3 proteins were expressed in 293T cells at roughly comparable levels (Fig. 1C). Similarly, exosomes isolated from various cellular sources had different anti-L1 activities. While the effect of transient expression of A3G on L1 was only moderate (30% decrease) (Fig. 1B), H9 and M-DC exosomes with A3G inhibited L1 retrotransposition (in contrast to A3G-negative exosomes secreted by SupT1 cells or derived from blood plasma). In particular, exosomes secreted by A3G-positive H9 cells inhibited L1 transposition by about 75%, while exosomes released from mature M-DCs inhibited L1 transposition by about 50%. Together, these results suggest that in addition to A3G, other A3 proteins present in exosomes contribute to L1 inhibition.
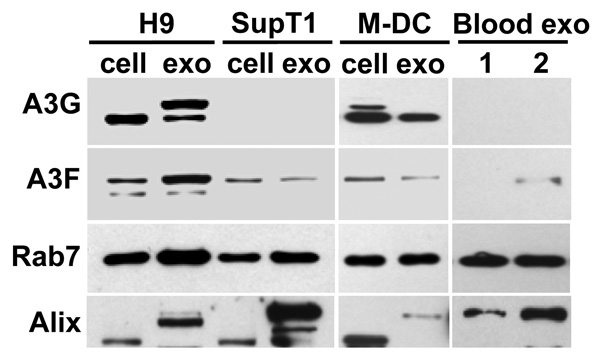
Since A3F expressed in 293T cells was more potent in inhibiting L1 than A3G (Fig. 1B), we determined if A3F was detectable in H9-, SupT1-, mature M-DC-, and plasma-derived exosomes, and thus might contribute to the capacity of these exosomes to inhibit L1 retrotransposition. As shown by Western blot analysis (Fig. 2), A3G was not only expressed by H9 T cells and mature M-DC, but also detected in exosomes secreted by these cells. Two forms of A3G were detected in H9 exosomes, while only faster migrating A3G was present in H9 cells (Khatua et al., 2009). In addition to H9 cells and M-DC, A3F was also detectable in both SupT1 cells and exosomes, as well as in blood exosomes derived from one of the two blood donors. However, unlike H9 exosomes that appeared to concentrate A3F about 3-fold in comparison to H9 cells, the expression of A3F in exosomes secreted by M-DC and SupT1 cells was lower than in their respective cell samples. Comparable levels of Rab7 expression in cells and exosomes suggest that observed differences in A3G and A3F were not caused by unequal protein loading on a gel. Different forms of Alix, a typical exosomal marker, were also detected in exosomes and in cells, with slower migrating forms of Alix detected predominantly in exosomes (Khatua et al., 2009). Together, H9 exosomes expressed about 10 times more A3F protein than SupT1 or M-DC exosomes. Thus, the presence of both A3G and A3F in exosomes may translate into the higher potency of H9 exosomes against L1 mobilization.
Fig. 2. Expression of A3G and A3F in exosomes.
Exosomes were purified from cell culture supernatants or from blood plasma of 2 different healthy blood donors. Cell and exosome lysates (20 and 10 µg/lane, respectively) were separated by SDS-PAGE and analyzed by Western blotting for the presence of A3G, A3F, and exosome markers Rab7 and Alix. In addition to the 70–75 kDa form of Alix detected predominantly in cells, higher molecular forms of Alix (95–105 kDa) were detected, particularly in exosomes. A slower migrating form of A3G was specifically detected in H9 exosomes. Note that A3F was detected in exosomes purified from blood plasma of donor 2 but not donor 1.
H9 cells secrete exosomes enriched in A3G and A3F mRNAs
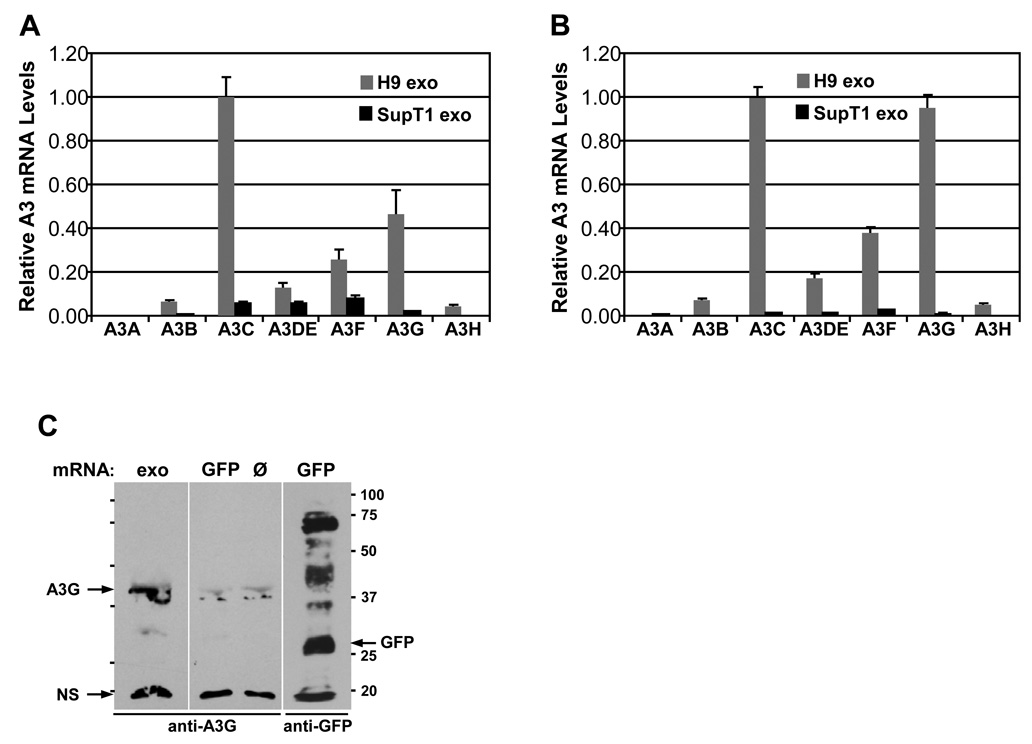
In addition to hundreds of proteins, exosomes encapsidate numerous cellular mRNAs that can be delivered and translated into corresponding proteins and in turn profoundly affect gene expression in target cells (Deregibus et al., 2007; Ratajczak et al., 2006a; Skog et al., 2008; Valadi et al., 2007). To investigate the possibility that H9 exosomes incorporate mRNAs coding for A3 proteins, total RNA was isolated from H9 and SupT1 cells and exosomes and analyzed for the presence of A3 specific mRNAs using real-time RT-PCR. Expression of A3 mRNAs was normalized against GAPDH mRNA. Since the expression of A3C mRNA was the highest among tested A3 mRNAs in H9 cells and exosomes, it was set as 1 (100%) and the levels of other A3 mRNAs were expressed relative to the level of A3C mRNA. H9 cells expressed A3G and A3F mRNAs at a level of about 48% and 27%, respectively, of A3C mRNA (Fig. 3A). In contrast, SupT1 cells expressed significantly lower levels of A3 mRNAs with the highest levels of A3C, A3DE and A3F mRNAs ranging between 7 and 10% of H9 A3C mRNA level. In SupT1 exosomes, A3 mRNAs were almost undetectable with the exception of A3F mRNA (Fig.3B). The very low levels of A3 mRNAs in SupT1 exosomes is in keeping with the low expression of these proteins in these cells. H9 exosomes, on the other hand, were enriched in A3G and A3F mRNAs in addition to A3C mRNA, with A3F mRNA expressed at 38% and A3G expressed at 97% of A3C mRNA level in H9 exosomes. To investigate whether exosomal A3G mRNA is functional, we performed an in vitro translation assay using polyA+ mRNA isolated directly from H9 exosomes (Fig.3C). Using rabbit anti-A3G antibodies, we detected a protein product around 43 kDa that corresponds to A3G. Low intensity bands at around 37 kDa and stronger bands around 20 kDa were detected in all in vitro reactions even in the absence of added mRNA, suggesting that they represent non-specific proteins. In conclusion, H9 exosomes encapsidate several A3 mRNAs and some of them can be translated in vitro into proteins. In contrast, exosomes secreted by SupT1 cells were generally depleted of A3 mRNAs.
Fig. 3. Expression of A3 mRNAs in CD4+ T cells and exosomes.
Total RNA was extracted from H9 and SupT1 cells and from exosomes secreted by these cells and subjected to real-time RT-PCR. Levels of A3 mRNAs in cells (A) and in exosomes (B) were normalized against GAPDH mRNA levels and were expressed relative to A3C mRNA levels that were set as 1.0 in both H9 cells and H9 exosomes. Error bars, mean values ± SD of triplicate samples. (C) A3G mRNA present in H9 exosomes is functional. PolyA+ mRNA (1 µg) isolated from H9 exosomes (exo) was subjected to an in vitro translation using a human in vitro protein expression assay. Samples without RNA (Ø, negative control) or containing GFP mRNA (positive control) were included. Translation reaction products were separated by SDS-PAGE and analyzed by Western blotting for the presence of A3G and GFP. Molecular weight markers (in kDa) are shown. NS, non-specific protein.
A3G is a major determinant of restriction of Alu retrotransposition by exosomes
Recent studies have shown that A3G strongly restricts Alu retrotransposition, possibly by sequestering Alu RNAs in cytoplasmic high molecular mass (HMM) ribonucleoprotein complexes thus preventing access of Alu RNAs to the nuclear L1 machinery (Bogerd et al., 2006b; Chiu et al., 2006; Gallois-Montbrun et al., 2008; Hulme et al., 2007). To examine the ability of exosomes to inhibit retrotransposition of Alu in human cells, we performed an L1-dependent Alu retrotransposition assay using the neoTet marked Alu reporter construct as described (Dewannieux, Esnault, and Heidmann, 2003). HeLa cells were cotransfected with an Alu reporter and L1 expression vector p220.CMV-L1 (Esnault et al., 2006) and treated for 48 h with exosomes of different cell origins. The appearance of neomycin-resistant colonies provided a direct measure of successful Alu retrotransposition. The colonies were stained with crystal violet and counted two weeks later. To ensure measurement of L1-dependent Alu transposition, we included negative controls by transfecting HeLa cells with Alu and a GFP expression vector or transfecting the cells only with either the neoTet marked Alu reporter or with L1 vector. No retrotransposition events were detected in negative controls (Fig. 4).
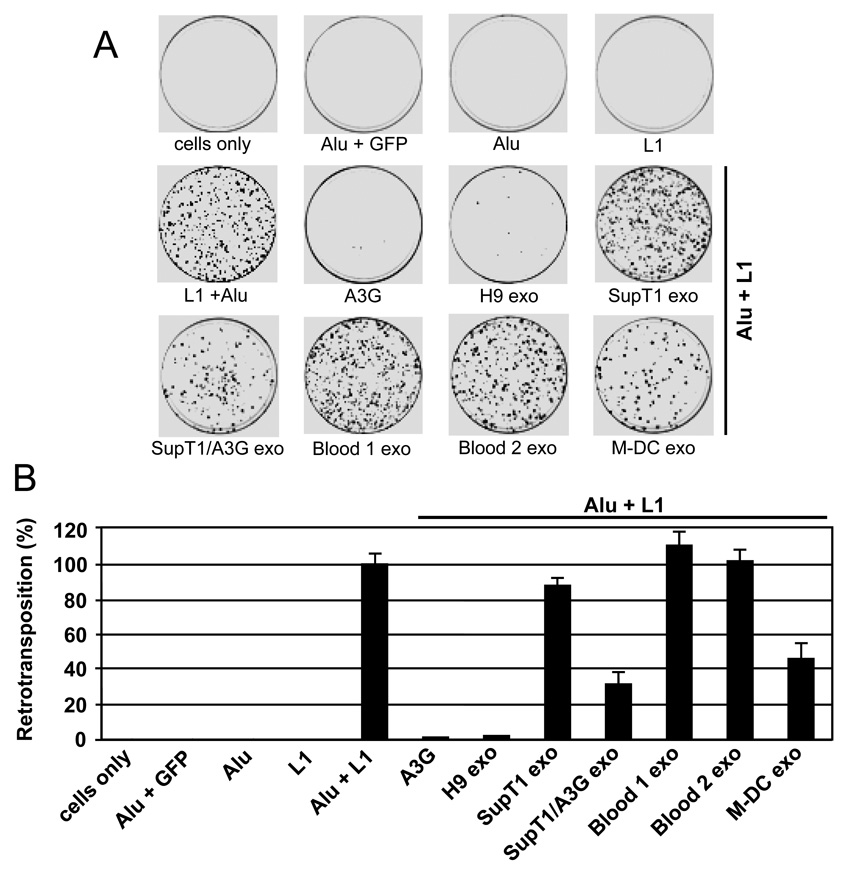
Fig. 4. Effect of exosomes on L1-mediated Alu retrotransposition.
HeLa cells were cotransfected with an Alu construct tagged with the neoTet retrotransposition indicator cassette and an L1 construct (Alu + L1), with or without an A3G expression vector. Cells transfected with Alu + L1 were exposed with different exosomes (20 µg/ml). Three days post-transfection, cells were subjected to selection in G418 for 14 days and resistant colonies were fixed, stained with crystal violet (A) and counted (B). As negative controls, cells were mock transfected (cells only) or transfected with L1 alone, Alu neoTet alone (Alu) or together with irrelevant GFP gene (Alu + GFP). Retrotransposition efficiency was expressed relative to the positive control (Alu + L1), which was set as 100%. Values represent the means ± SD of triplicate samples. Similar results were obtained in 4 independent experiments.
Consistent with previous observations (Bogerd et al., 2006b; Chiu et al., 2006; Hulme et al., 2007), expression of A3G from transfected vector inhibited over 98% of Alu retrotransposition events. H9 exosomes were almost equally potent and inhibited on average 92–96% of Alu retrotranspositions. To investigate the contribution of A3G to the observed Alu inhibition by H9 exosomes, we first tried down-regulating the expression of A3G in H9 cells by using short interfering RNA. However this approach did not substantially reduce A3G protein expression in either H9 cells or in exosomes (Khatua et al., 2009), and thus did not appreciably reduce the inhibitory effect previously seen on Alu transposition (not shown).
To overcome this problem, we transfected A3G-negative SupT1 cells with GFP-A3G or GFP expression vectors and collected exosomes to be used in retrotransposition assays. Exosomes secreted by SupT1 cells transfected with GFP-A3G (SupT1/A3G exo) inhibited Alu retrotransposition by 68%, while exosomes derived from GFP-transfected cells (not shown) or parental A3G-negative SupT1 cells inhibit Alu retrotransposition only marginally (Fig. 4). Similarly, exosomes isolated from peripheral blood plasma samples of 2 different healthy donors showed no inhibitory effect on Alu retrotransposition. In contrast, exosomes secreted by mature M-DC that expressed A3G (Fig. 1C) inhibited Alu transposition by 55%. Since exosomes isolated from blood plasma (Fig. 1C) and secreted by SupT1 (Fig. 2) did not express detectable levels of A3G, these results suggest that the inhibitory effect of exosomes against Alu retrotransposition are affected, at least in part, by the presence of encapsidated A3G.
Discussion
Retrotransposons constitute about 45% of the human genome. To protect genome stability, eukaryotic cells have developed several strategies to restrict proliferation of transposable elements. One of the strategies adopted by cells is silencing the mobilization of retroelements by cytidine deaminases belonging to the family of A3 proteins. Human A3 proteins show some preferences with regard to the inhibition of non-LTR transposons, L1 or Alu. For instance, A3A and A3B inhibit both L1 and Alu retrotransposition, A3C and A3F mostly inhibit L1 (Bogerd et al., 2006b; Chen et al., 2006; Muckenfuss et al., 2006; Stenglein and Harris, 2006), while A3G preferentially inhibits Alu mobilization, with a marginal effect on L1 (Bogerd et al., 2006b; Chiu et al., 2006; Hulme et al., 2007). However, a stronger A3G mediated inhibitory activity on L1 retrotransposition has also been reported (Kinomoto et al., 2007; Niewiadomska et al., 2007). Recently, a inhibitory activity against Alu retrotransposition has been attributed to A3H and A3DE (Tan et al., 2009) as well as A3F (Gallois-Montbrun et al., 2008). Since cells may express suboptimal levels or different forms of A3 proteins, it is likely that the supply of specific A3 proteins in exosomes may influence the extent and specificity of anti-retroelement activity in the recipient cell. Indeed, we have shown that A3G transmitted in exosomes and functionally expressed in HIV-1 susceptible cells strongly inhibits replication of HIV-1 in exosome-receiving cells (Khatua et al., 2009).
Exosomes are small membrane vesicles released by various types of cells, and carry molecular imprints of cells from which they were secreted. Exosomes originate from intralumenal vesicles of multivesicular bodies (MVBs) that form during the inward budding of limiting membranes of endosomes. Fusion of MVBs with the plasma membrane releases exosomes into intercellular space where they are believed to play a role in intercellular communication by transferring biologically active molecules to recipient cells. Detection of mRNA and microRNA in exosomes has led to the proposition that exosomes are also involved in exchange of genetic information between cells (Valadi et al., 2007).
In this study, we tested the hypothesis that exosomes secreted by cells expressing A3G and A3F restrict mobilization of L1 and Alu retroelements in target cells. Using an in vitro assay to quantify the frequency of L1 and Alu retrotransposition, we have confirmed that transiently overexpressed A3 proteins inhibit these non-LTR retroelements to different extents, with A3A, A3B and A3F restricting L1, and A3G strongly inhibiting Alu retrotransposition. Somewhat surprisingly, H9 exosomes that were originally found to encapsidate A3G and inhibited HIV-1 replication (Khatua et al., 2009) had a strong inhibitory activity against L1. Since A3G only weekly reduced L1 activity in our assays, this suggests that H9 exosomes encapsidate other A3 protein(s) that are able to inhibit L1. Indeed, we have identified A3F in H9 exosomes suggesting that this cytidine deaminase, alone or in combination with A3G or other restriction factors, is responsible for the observed inhibitory effect on L1 retrotransposition (Muckenfuss et al., 2006). Therefore, the moderate activity of exosomes secreted by mature M-DC may be explained by the lower levels of A3F detected in these exosomes. However, it is possible that other A3 proteins enhance the activity of H9 exosomes against L1 retrotransposition. One such candidate could be A3C which modestly inhibits L1 (Bogerd et al., 2006b; Kinomoto et al., 2007; Muckenfuss et al., 2006). The possibility that exosomal mRNAs are translated in recipient cells after exosome internalization has been well documented (Deregibus et al., 2007; Ratajczak et al., 2006a; Skog et al., 2008; Valadi et al., 2007). We have found that H9 exosomes encapsidate mRNAs coding for A3C, A3F and A3G. Further analysis showed that A3G mRNA isolated from exosomes is functional and supports protein synthesis in an in vitro translation system. Based on these observations, we speculate that transfer of functional A3 proteins and corresponding mRNAs modulate or enable cells to resist invading or endogenous retroelements.
Alu retrotransposition was profoundly inhibited by H9 exosomes and moderately inhibited by M-DC exosomes. Since H9 exosomes encapsidated not only A3G but also A3F, we investigated whether A3G in exosomes inhibits Alu mobilization. A short interfering RNA (siRNA) approach to downregulate A3G in H9 cells was unsuccessful. However, introduction of A3G gene into A3G-negative CD4+ SupT1 cells that expressed low levels of A3F showed that exosomes secreted by the cells reduced Alu retrotransposition by 68%. This result confirms that exosomal A3G inhibits Alu but does not rule out the possibly that additional factors (e.g. A3F), likely in combination with A3G, limit Alu activity. Blood-derived microvesicles were negative for A3G and one preparation (donor 2) contained small amounts of A3F. However, all tested blood-derived microvesicles (3 different donors) did not show any effect on L1 or Alu retrotransposition.
The mechanism of exosome entry and delivery of its cargo into the cell cytoplasm without degradation in the lysosomal compartment is unknown. However, the observation that some cellular receptors (Mack et al., 2000), transcription factors (Ratajczak et al., 2006a) or mRNAs (Deregibus et al., 2007; Ratajczak et al., 2006a; Valadi et al., 2007) transferred by microvesicles or exosomes remain functional in recipient cells, supports the notion that molecules delivered by exosomes escape degradation.
Another unanswered question is how A3 proteins are packaged into exosomes. One possibility is that cytoplasmic molecules may be engulfed during the inward budding of limiting membranes of MVBs. However, this non-specific mechanism does not explain specific enrichment of some molecules (e.g. A3F or A3G mRNA) in exosomes. The ability of A3 proteins to interact with certain mRNAs and small noncoding RNAs (Chiu et al., 2006; Gallois-Montbrun et al., 2008; Khan et al., 2007; Kozak et al., 2006; Wang et al., 2007b; Wang et al., 2008b) suggests that A3 proteins may localize into exosomes indirectly by interaction with cellular RNAs. Although packaging of A3G into HIV-1 particles is mediated by HIV-1 Gag and cellular RNAs (Alce and Popik, 2004; Cen et al., 2004; Douaisi et al., 2004; Luo et al., 2004; Navarro et al., 2005; Schafer, Bogerd, and Cullen, 2004; Svarovskaia et al., 2004; Zennou et al., 2004), packaging into exosomes obviously does not require HIV-1 Gag. Our findings that exosomes encapsidate large quantities of 7SL RNA (not shown) together with previous observations that A3G interacts with 7SL RNA (Bach et al., 2008; Wang et al., 2007b; Wang et al., 2008c), suggests that 7SL RNA may mediate A3G encapsidation into exosomes in a Gag-independent manner. Since A3G is able to interact with diverse retrotransposon Gag proteins (Bogerd et al., 2006a; Dutko et al., 2005), we cannot dismiss the possibility that A3G interacts with endogenous retroviral Gag and cellular RNA (including 7SL RNA) complexes to gain access to exosomes. The mechanism by which other A3 proteins gain access into exosomes is presently unknown.
The biological significance of exosomes bearing encapsidated A3 proteins and respective mRNAs is unknown. One possibility is that encapsidation of A3 in exosomes may serve to dispose of excess cytidine deaminases that might otherwise lead to genomic instability (Anant and Davidson, 2003; Pham, Bransteitter, and Goodman, 2005). However, we speculate that exosomes with encapsidated A3 proteins may serve to fine tune the response against transiently expressed retroelements in germ cells or during the early stages of human embryogenesis (Garcia-Perez et al., 2007; Kazazian, 2004; Sawyer, Emerman, and Malik, 2004). In addition, exosomes with encapsidated A3 proteins may control retrotransposition which may occasionally occur in somatic cells (Kubo et al., 2006; Schulz, 2006).
In conclusion, the findings reported here show for the first time that exosomes packaging A3G and A3F potently restrict mobilization of L1 and Alu in recipient cells in vitro. Whether exosomes contribute to restriction of mobilization of retroelements in vivo will require further investigation.
Acknowledgments
This work was supported by NIH grants from NCRR (U54RR019192) and NIAID (P30AI054999).
We thank T. Heidmann for the neoTet-marked Alu and p220.CMV-L1RP constructs, and E. T. L. Prak and H. H. Kazazian for L1RP-EGFP and EFO5J. A3H-V5-His was provided by Y. H. Zheng and HA-tagged A3 expression vectors were obtained from B. R. Cullen.
Vectors pDON/EGFP (coding for GFP) and pDON/EGFP-CEM15 (coding for GFP-A3G) were obtained from A. Takaori-Kondo (Kyoto, Japan). We thank K. Strebel for anti-A3G antibodies. We thank Mr. Jared Elzey for his assistance in manuscript editing and preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279(33):34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Anant S, Davidson NO. Hydrolytic nucleoside and nucleotide deamination, and genetic instability: a possible link between RNA-editing enzymes and cancer? Trends Mol Med. 2003;9(4):147–152. doi: 10.1016/s1471-4914(03)00032-7. [DOI] [PubMed] [Google Scholar]
- Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum Mutat. 2007;28(6):527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34(5):1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80(17):8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006a;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006b;103(23):8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100(9):5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387(2):313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno S, Grange C, Deregibus MC, Calogero RA, Saviozzi S, Collino F, Morando L, Busca A, Falda M, Bussolati B, Tetta C, Camussi G. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol. 2009;20(5):1053–1067. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callinan PA, Batzer MA. Retrotransposable elements and human disease. Genome Dyn. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem. 2004;279(32):33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16(5):480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435(7038):108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, Heidmann T, Greene WC. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103(42):15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem. 2008;283(17):11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gassart A, Geminard C, Hoekstra D, Vidal M. Exosome secretion: the art of reutilizing nonrecycled proteins? Traffic. 2004;5(11):896–903. doi: 10.1111/j.1600-0854.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B, Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110(7):2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35(1):41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Douaisi M, Dussart S, Courcoul M, Bessou G, Vigne R, Decroly E. HIV-1 and MLV Gag proteins are sufficient to recruit APOBEC3G into virus-like particles. Biochem Biophys Res Commun. 2004;321(3):566–573. doi: 10.1016/j.bbrc.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Dutko JA, Schafer A, Kenny AE, Cullen BR, Curcio MJ. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr Biol. 2005;15(7):661–666. doi: 10.1016/j.cub.2005.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433(7024):430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
- Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34(5):1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Priet S, Ribet D, Heidmann O, Heidmann T. Restriction by APOBEC3 proteins of endogenous retroviruses with an extracellular life cycle: ex vivo effects and in vivo "traces" on the murine IAPE and human HERV-K elements. Retrovirology. 2008;5:75. doi: 10.1186/1742-4690-5-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T, Singer M. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 1987;15(5):2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash EA, Kao GD, Horman SR, Prak ET. Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 2006;34(4):1196–1204. doi: 10.1093/nar/gkj522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Holmes RK, Swanson CM, Fernandez-Ocana M, Byers HL, Ward MA, Malim MH. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol. 2008;82(11):5636–5642. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16(13):1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282(4):2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- Holmes SE, Singer MF, Swergold GD. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J Biol Chem. 1992;267(28):19765–19768. [PubMed] [Google Scholar]
- Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390(1–2):199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303(5664):1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatua AK, Taylor HE, Hildreth JE, Popik W. Exosomes packaging APOBEC3G confer human immunodeficiency virus resistance to recipient cells. J Virol. 2009;83(2):512–521. doi: 10.1128/JVI.01658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet. 1999;8(8):1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, Sata T, Tokunaga K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35(9):2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosha VO, Martin SL. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1) J Biol Chem. 2003;278(10):8112–8117. doi: 10.1074/jbc.M210487200. [DOI] [PubMed] [Google Scholar]
- Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281(39):29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Jr, Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A. 2006;103(21):8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173(6):949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol. 2004;78(21):11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack M, Kleinschmidt A, Bruhl H, Klier C, Nelson PJ, Cihak J, Plachy J, Stangassinger M, Erfle V, Schlondorff D. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000;6(7):769–775. doi: 10.1038/77498. [DOI] [PubMed] [Google Scholar]
- Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J Mol Biol. 2000;304(1):11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254(5039):1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81(24):13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281(31):22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333(2):374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15(2):166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Niewiadomska AM, Tian C, Tan L, Wang T, Sarkis PT, Yu XF. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol. 2007;81(17):9577–9583. doi: 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28(6):1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35(4):363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Goodman MF. Reward versus risk: DNA cytidine deaminases triggering immunity and disease. Biochemistry. 2005;44(8):2703–2715. doi: 10.1021/bi047481+. [DOI] [PubMed] [Google Scholar]
- Pion M, Granelli-Piperno A, Mangeat B, Stalder R, Correa R, Steinman RM, Piguet V. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J Exp Med. 2006;203(13):2887–2893. doi: 10.1084/jem.20061519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prak ET, Dodson AW, Farkash EA, Kazazian HH., Jr Tracking an embryonic L1 retrotransposition event. Proc Natl Acad Sci U S A. 2003;100(4):1832–1837. doi: 10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006a;20(5):847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006b;20(9):1487–1495. doi: 10.1038/sj.leu.2404296. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J, Jimenez-Velasco A, Agirre X, Cervantes F, Sanchez J, Garate L, Barrios M, Castillejo JA, Navarro G, Colomer D, Prosper F, Heiniger A, Torres A. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24(48):7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2(9):E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328(2):163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schulz WA. L1 retrotransposons in human cancers. J Biomed Biotechnol. 2006;(1):83672. doi: 10.1155/JBB/2006/83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82(6):2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35(Pt 3):637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418(6898):646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281(25):16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3(5):321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, Freed EO, Hu WS, Pathak VK. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;279(34):35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 2009;23(1):279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J Biol Chem. 2004;279(42):43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, Hoefsloot LH, Sistermans EA, de Wijs IJ, Mukhopadhyay A, Plomp AS, de Jong PT, Kazazian HH, Cremers FP. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16(13):1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- Vetter ML, D'Aquila RT. Cytoplasmic APOBEC3G restricts incoming Vif-positive human immunodeficiency virus type 1 and increases two-long terminal repeat circle formation in activated T-helper-subtype cells. J Virol. 2009;83(17):8646–8654. doi: 10.1128/JVI.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene. 2008;419(1–2):75–81. doi: 10.1016/j.gene.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FX, Huang J, Zhang H, Ma X, Zhang H. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008a;89(Pt 3):722–730. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- Wang JH, Janas AM, Olson WJ, Wu L. Functionally distinct transmission of human immunodeficiency virus type 1 mediated by immature and mature dendritic cells. J Virol. 2007a;81(17):8933–8943. doi: 10.1128/JVI.00878-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007b;81(23):13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J Mol Biol. 2008b;375(4):1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Wang T, Zhang W, Tian C, Liu B, Yu Y, Ding L, Spearman P, Yu XF. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008c;377(1):71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Morrish TA, Alisch RS, Moran JV. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem. 2000;284(2):435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13(9):763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78(21):12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]