SUMMARY
The ribosome has an active site comprised of RNA that catalyzes peptide bond formation. To understand how RNA promotes this reaction requires a detailed understanding of the chemical transition state. Here, we report the Brønsted coefficient of the α-amino nucleophile (βnuc) using a series of puromycin derivatives. Both 50S subunit and 70S ribosome catalyzed reactions displayed linear free-energy relationships with slopes close to zero under conditions where chemistry is rate limiting. These results indicate that at the transition state the nucleophile is neutral in the ribosome catalyzed reaction, in contrast to the substantial positive charge reported for typical uncatalyzed aminolysis reactions. This suggests that the ribosomal transition state involves deprotonation to a degree commensurate with nitrogen-carbon bond formation. Such a transition state is significantly different from that of uncatalyzed aminolysis reactions in solution.
INTRODUCTION
The ribosome is the macromolecular machine that catalyzes cellular protein synthesis. The ribosome promotes amide bond formation within the peptidyl transferase center (PTC) of the 50S ribosomal subunit. Crystallographic studies have established that RNA surrounds the PTC, demonstrating that the ribosome is a ribozyme(Ban, Nissen et al., 2000). The two ribosomal substrates, the peptidyl-tRNA and the aminoacyl-tRNA bind to two adjacent sites within the PTC (P site and A site, respectively). In peptidyl transfer (PT), the α-amino group of the A-site aminoacyl-tRNA attacks the ester of the P-site peptidyl-tRNA. This results in the deacylation of the P-site tRNA and a peptide chain that is extended by one amino acid.
The ribosome enhances the PT reaction approximately 107-fold over the rate of comparable reactions in solution(Rodnina, Beringer et al., 2006). Several mechanisms have been proposed to account for this observed rate enhancement including general acid-base catalysis(Muth, Ortoleva-Donnelly et al., 2000; Nissen, Hansen et al., 2000), catalysis by substrate alignment(Sievers, Beringer et al., 2004) and substrate-assisted catalysis(Dorner, Panuschka et al., 2003; Weinger, Parnell et al., 2004; Trobro and Aqvist, 2005). Biochemical, mutational, and recent structural evidence suggest that the general acid-base mechanism as originally proposed is unlikely. The P-site A76 2′-OH is situated close to the attacking α-amine and O3′ leaving group, making it one of the few functional groups in a position to be directly involved in catalysis(Schmeing, Huang et al., 2005; Schmeing, Huang et al., 2005). Removing the A76 2′-OH decreases the rate of peptide bond formation by at least one million-fold(Weinger, Parnell et al., 2004). The catalytic importance and physical proximity of this key functional group has led to proposals that it is involved in a proton shuttle between the α-amine and the O3′ leaving group(Das, Bhattacharyya et al., 1999; Schmeing, Huang et al., 2005; Rodnina, Beringer et al., 2007).
A detailed knowledge of the transition state of the PT reaction will aid in understanding the catalytic mechanism of the ribosome. One method for defining the transition state is Brønsted analysis. The Brønsted plot is a linear free energy relationship that describes the correlation between the log of the reaction rate and the pKas of the respective nucleophile or leaving group. By using a series of modified substrates with different pKas, the magnitude and sign of the Brønsted coefficient (β) can be determined from the slope of the Brønsted plot. The Brønsted coefficient reflects the change in charge of an ionizing group between the ground state and the transition state. It is also indicative of the changes in bonding that occur in the transition state(Jencks, 1987; Fersht, 1999).
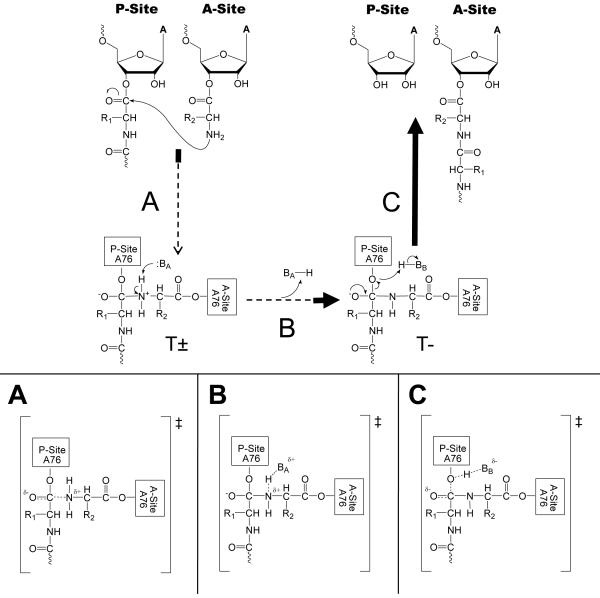
Except for highly activated nucleophiles or leaving groups, Brønsted coefficients for the nucleophile (βnuc) values in solution are typically between 0.8-0.9 for primary and secondary amines(Bruice and Lapinski, 1958; Jencks and Carriuolo, 1960; Jencks and Gilchrist, 1968; Satterthwait and Jencks, 1974). Positive values close to one suggest that the α-amine has a significant positive charge in the transition state, retaining the α-amine protons as the nitrogen-carbon (N-C) bond forms. Jencks proposed a stepwise reaction mechanism involving two tetrahedral intermediates (T± and T−). The α-amine in the first tetrahedral intermediate (T±) retains both protons and is deprotonated via a proton transfer to form the second intermediate (T−). This proton transfer limits the rate and a βnuc close to 1 indicates that the transition state closely resembles the T± intermediate(Satterthwait and Jencks, 1974).
Does PT on the ribosome go through the same transition state as uncatalyzed aminolysis in solution? Determining the Brønsted coefficient of the α-amino group of the A-site substrate would begin to address this question and help to define the nature of the catalytic contribution provided by the ribosome. A ribosomal Brønsted coefficient of ~0.8 would suggest that the transition state of both the catalyzed and uncatalyzed reactions are the same. A value that deviates significantly would indicate that the ribosome proceeds through an alternative transition state. Previously, we reported that β, β-difluorophenylalanyl- (pKa < 5.0) reacted 90-fold faster than the phenylalanyl derivative at acidic pH. At higher pH’s the rates of the phenylalanyl and fluorinated phenylalanyl deriviatives varied only 2-fold, suggesting an unexpectedly small βnuc value. However, the limited sample size and the use of only the 50S fragment assay were insufficient for mechanistic conclusions(Okuda, Seila et al., 2005). Here we apply Brønsted analysis to an expanded series of derivatives and establish that the transition states for the PT reaction catalyzed by both the 50S subunit and 70S ribosome substantially differ from that of the uncatalyzed reaction.
RESULTS
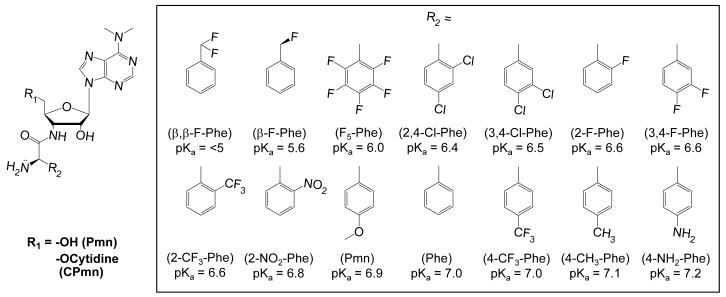
We prepared a series of puromycin derivatives (A-site substrates) (Figure 1) and used these compounds to determine the Brønsted coefficient for the nucleophilic α-amine. The substrates were designed to be structurally similar to the parental substrate with minor modifications to perturb the pKa of the nucleophilic amino group. A valid enzymatic Brønsted analysis requires that the modifications do not alter the binding of the substrate to the enzyme or the isostericity of the substrates, which significantly limits the diversity of modifications that can be made. We focused on the amino acid phenylalanine and its derivatives with either β-carbon or aromatic ring substitutions. The pKa of the α-amine was perturbed by introducing conservative electron withdrawing and donating substituents into the phenylalanine side chain. The result was a series of substrates whose pKas ranged from <5 to 7.2 (Figure 1).
Figure 1.
The series of puromycin derivatives used as A-site substrates.
βnuc Measurements on the 50S Ribosomal Subunit
The aminolysis rate for each puromycin (Pmn) derivative was determined under single turnover conditions using the modified 50S fragment assay(Schmeing, Seila et al., 2002; Okuda, Seila et al., 2005). The radiolabled P-site substrate, CCApcb (phenylalanine-caproic acid-biotin) was present in trace amounts in the presence of a saturating concentration of 50S subunits and A-site substrate(Seila, Okuda et al., 2005). The reactions were performed at pH 8.5, to ensure that all substrates were in the amino rather than the unreactive ammonium form (Supplemental Figure 1, Table 1). Eight substrates reacted efficiently [CPmn(β, β-F-Phe), CPmn(3, 4-Cl-Phe), CPmn(2-F-Phe), CPmn(3, 4-F-Phe), CPmn, CPmn(Phe), CPmn(4-CH3-Phe), and CPmn(4NH2-Phe)] while four of the substrates [CPmn(2, 4-Cl-Phe), CPmn(2-NO2-Phe), CPmn(2-CF3-Phe) and CPmn(F5-Phe)] were inactive. Three of the inactive and none of the active derivatives contained a bulky substitution at the ortho position of the benzyl ring. The inhibition constant for CPmn(F5-Phe) was comparable to the KM for CPmn (data not shown), suggesting that the derivative binds but is unreactive. Such steric effects are independent of the derivative pKa and preclude the use of these substrates in a Brønsted analysis.
Table 1.
50S modified fragment assay kinetic data (pH 8.5, 25 °C)
| pKa | kpep (s−1) | |
|---|---|---|
| CPmn(β,β-F-Phe) | <5.0 | 0.009 ± 0.0003 |
| CPmn(3,4-Cl-Phe) | 6.5 | 0.01 ± 0.01 |
| CPmn(2-F-Phe) | 6.6 | 0.01 ± 0.001 |
| CPmn(3,4-F-Phe) | 6.6 | 0.008 ± 0.0002 |
| CPmn | 6.9 | 0.02 ± 0.001 |
| CPmn(Phe) | 7.0 | 0.02 ± 0.002 |
| CPmn(4-CH3-Phe) | 7.1 | 0.01 ± 0.001 |
| CPmn(4-NH2-Phe) | 7.2 | 0.008 ± 0.001 |
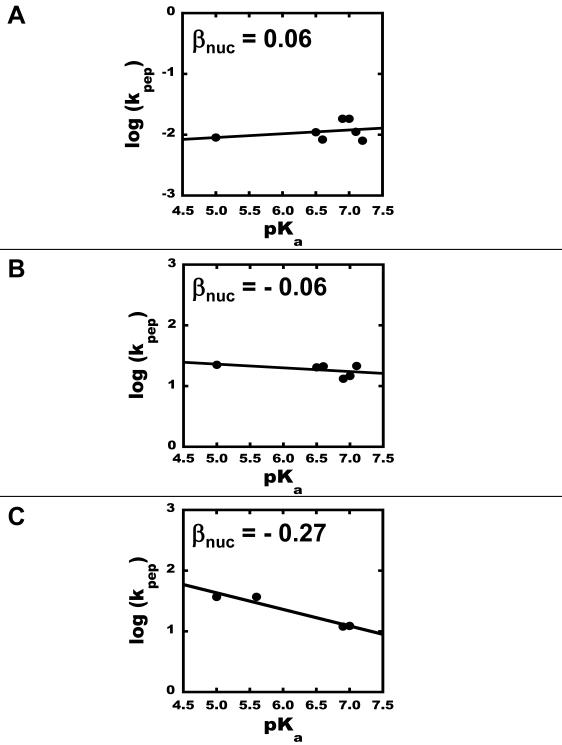
The rate of the PT reaction on 50S subunits was measured for each of the eight active compounds at substrate concentrations 3 to 4-fold above the KM of CPmn. Raising the concentrations 8-fold above the KM value did not change the rate, which implies that the substrate concentration was saturating and therefore the observed rates correspond to the chemical step, kpep. The PT rates for all A-site substrates tested varied less than 2.5-fold, ranging from 0.48 min−1 to 1.1 min−1. The βnuc for peptide bond formation on 50S subunits was determined from the slope of log kpep versus the pKa of the α-amino group of each aminoacyl substrate, resulting in a Brønsted coefficient of 0.06 (Figure 2A). The observed linearity for the plot indicates no change in the rate-determining step(Jencks, 1987).
Figure 2.
Brønsted plots for the PT reaction. (A) Brønsted plot of the PT reaction on 50S subunits. The line is the linear regression of the data, βnuc = 0.06. (B) Brønsted plot of the PT reaction on 70S initiation complexes in the presence of 60% DMSO, with βnuc = −0.06. (C) Brønsted plot of the PT reaction on 70S initiation complexes in the absence of DMSO, with βnuc = −0.27.
βnuc Measurements on the 70S Ribosome
Brønsted analysis was performed with the 70S initiation complex using a single-turnover kinetic assay(Katunin, Muth et al., 2002; Beringer, Bruell et al., 2005). f[3H]Met-tRNAfMet was preloaded into the P-site to form the 70S initiation complexes and the rate of dipeptide formation at pH 8.5 was measured for Pmn and its derivatives. Only four derivatives [Pmn(3,4-Cl-Phe), Pmn(3,4-F-Phe), Pmn(4-CH3-Phe), and Pmn(4-NH2-Phe)] were soluble at the concentrations required to reach substrate saturation. Previous reports found that 20% DMSO had no effect on the Pmn reaction rate(Katunin, Muth et al., 2002). We found that up to 60% DMSO was tolerated with the only consequence being a slight increase in the binding constant (KM) for Pmn (Supplemental Figures 2 and 3).
The rates of peptidyl transfer by the 70S ribosome for all of the puromycin derivatives were very similar. Data were collected for Pmn(β, β-F-Phe), Pmn(3, 4-Cl-Phe), Pmn(3, 4-F-Phe), Pmn, Pmn(Phe), and Pmn(4-CH3-Phe) in the presence of DMSO (Table 2). Although the pKa values of these derivatives range from <5 to 7.2, the rate of aminolysis by these six substrates varied less than 2-fold, from 13.2 s−1 to 22.4 s−1. Values for Pmn(β-F-Phe) and Pmn(4-NH2-Phe) are not obtained because an upper limit to solubility was reached before saturation occurred.
Table 2.
70S assay kinetic data (pH 8.5, 37 °C)
| 60% DMSO | No DMSO | ||||
|---|---|---|---|---|---|
| pKa | kpep (s−1) | KM (mM) | kpep (s−1) | KM (mM) | |
| Pmn(β,β-F-Phe) | <5.0 | 22.4 | 12.5 | 36.9 | 1.3 |
| Pmn(β-F-Phe) | 5.6 | - | - | 37.5 | 2.9 |
| Pmn(3,4-Cl-Phe) | 6.5 | 20.4 | 12.7 | - | - |
| Pmn(3,4-F-Phe) | 6.6 | 21.1 | 12.1 | - | - |
| Pmn | 6.9 | 13.2 | 6.1 | 12.0 | 2.5 |
| Pmn(Phe) | 7.0 | 14.7 | 8.3 | 12.3 | 3 |
| Pmn(4-CH3-Phe) | 7.1 | 21.5 | 17.9 | - | - |
A subset of analogs was sufficiently soluble in buffer B in the absence of DMSO to reach saturating concentrations. These substrates covered the full range of pKas in the collection. For the Pmn derivatives Pmn(β,β-F-Phe), Pmn(β-F-Phe), Pmn, and Pmn(Phe) (Table 2) all rates were no more than three fold different, ranging from 12 s−1 to 37 s−1.
The rate of PT reaction on 70S ribosomes, like that of the 50S subunits, is independent of the basicity of the nucleophile (Figure 2B & 2C). The Brønsted coefficients for the PT reaction on the 70S ribosomes were −0.06 and −0.27 in the presence and absence of DMSO, respectively. The linearity of the plot over the entire pKa range signifies that there is no change in the rate-determining step for the different analogues(Jencks, 1987).
DISCUSSION
The Brønsted coefficient of the nucleophilic α-amine (βnuc) in the PT reaction on the ribosome was determined for reactions catalyzed by both the 50S subunit and 70S ribosome. A series of puromycin-like substrates were synthesized with conservative, nearly isosteric substitutions of the phenylalanine side chain that resulted in pKas for the α-amino group ranging from <5 to 7.2. Using both the modified 50S fragment reaction and the 70S initiation complex reaction, the βnuc was found to be approximately zero, suggesting that the rate of peptidyl transfer is independent of the pKa of the nucleophilic amine. Since the magnitude and sign of the βnuc value reflects the change in charge on the attacking nucleophile between the ground state and the transition state, these near zero βnuc values suggest that the amine remains neutral in the PT transition state. Comparable βnuc values for reactions on the 50S subunit and the 70S ribosome support the similar intrinsic PT activity of the 50S subunits and 70S ribosomes(Wohlgemuth, Beringer et al., 2006).
In order to properly interpret the measured Brønsted coefficients, it is important to ensure that the assays are measuring the chemical rate and not another step in the pathway. If chemistry is not rate limiting, a near zero βnuc may arise because amine basicity would not affect the non-chemical steps of the assay. Several independent lines of evidence indicate that amide bond formation is rate limiting for both assays. In the 50S modified fragment assay: (i) there is a rapid-equilibrium between reactants and the bound complex (no forward commitment to catalysis), indicating that there is no rate limiting step prior to chemistry(Seila, Okuda et al., 2005); (ii) kinetic isotope effect (KIE) measured for the A-site substrate indicated that bond formation is at least partially rate limiting(Seila, Okuda et al., 2005); and (iii) a less-reactive nucleophile, hydroxypuromycin (the α-amino group is replaced with a hydroxyl) decreased the peptidyl transfer rate at least 20-fold(Seila, Okuda et al., 2005). For the 70S assay: (i) puromycin binding to the A site is not rate-limiting because peptidyl-tRNA substrates with different peptide lengths react at different rates. For example there is a 56-fold difference in the reaction rate of puromycin with fMet-tRNAfMet versus fMetAlaAsnMetPheAla-tRNAAla(Katunin, Muth et al., 2002). (ii) Substitution of puromycin with hydroxypuromycin decreases the rate of peptidyl transfer 200-fold(Katunin, Muth et al., 2002). (iii) Pmn binding into the PT center is not rate-limiting(Katunin, Muth et al., 2002; Sievers, Beringer et al., 2004). These observations argue that the near zero βnuc value does not result from monitoring a non-chemical step in the reaction pathway.
The βnuc values measured for the PT reaction on 50S subunits and 70S ribosomes differ significantly from those reported for most uncatalyzed aminolysis reactions. A βnuc near zero for peptide bond formation by the ribosome is inconsistent with formation of positive charge on the α-amine in the transition state. If this occurred, as in uncatalyzed aminolysis reactions(Bruice and Lapinski, 1958; Jencks and Carriuolo, 1960; Jencks and Gilchrist, 1968; Satterthwait and Jencks, 1974), then the addition of electron-donating groups to the amino acid side chain would stabilize the developing charge resulting in a positive correlation between reactivity and basicity(Zeeberg and Caplow, 1973). Also unlikely is the removal of the proton before nucleophilic attack, because the energetics associated with forming the amine anion are highly unfavorable(Blackburn and Jencks, 1968). The observed Brønsted coefficients indicate that the nucleophile remains uncharged in the ribosomal transition state, which suggests that the degree of deprotonation of the amine is similar to the degree of N-C bond formation. A transition state with a neutral nucleophile has implications for mechanistic models of the ribosomal reaction. Although the βnuc value does not fully define the transition state, it excludes several possibilities. Any mechanisms that require a transition state with significant positive charge development on the α-amine can be eliminated.
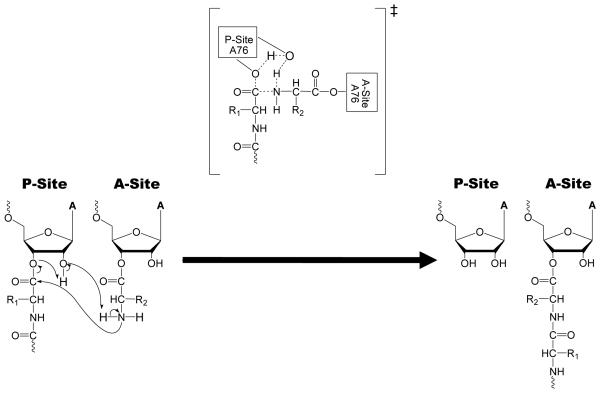
Several mechanisms have been proposed for the PT reaction. The first is equivalent to the solution aminolysis mechanism and proceeds stepwise through two intermediates (T± and T−)(Jencks and Gilchrist, 1968; Satterthwait and Jencks, 1974) (Figure 3). The second is also stepwise but involves only a single intermediate (T±) (Figure 4). This is one form of the “proton shuttle” mechanism and involves concerted proton transfer with C-O bond cleavage that resolves the T± intermediate directly to products(Das, Bhattacharyya et al., 1999; Schmeing, Huang et al., 2005; Trobro and Aqvist, 2005). The third mechanism is a fully concerted reaction without intermediates and is a second variant of the “proton shuttle” proposal (Figure 5). This mechanism requires that all bond formation, cleavage, and proton transfers occur simultaneously(Dorner, Polacek et al., 2002; Changalov, Ivanova et al., 2005). Each of these mechanisms can now be evaluated in light of the βnuc value determined for the ribosomal reaction.
Figure 3.
Stepwise mechanism with two intermediates (T± and T−). The relative energetics of intermediates and TS are arbitrary. The bolded portions of the arrows represent areas where a zero βnuc is consistent for each transition. (A) Possible TS if nucleophilic attack determines the reaction rate. (B) Possible TS if α-amine proton transfer following T± formation determines the reaction rate. (C) Possible TS if T- tetrahedral intermediate breakdown determines the reaction rate.
Figure 4.
Stepwise mechanism with one intermediate (T±). The relative energetics of intermediates and TS are arbitrary. The bolded portions of the arrows represent areas where a zero βnuc is consistent for each transition. (A) Possible TS if nucleophilic attack determines the reaction rate. (B) Possible TS if T± tetrahedral intermediate breakdown determines the reaction rate.
Figure 5.
Fully concerted reaction mechanism. Possible TS if nucleophilic attack, C-O bond cleavage, and proton transfer are concerted in the rate-determining step. The bolded portion of the arrow represents the area where the proposed mechanism is consistent with a zero βnuc.
Assuming the mechanism with two intermediates(Jencks and Gilchrist, 1968; Satterthwait and Jencks, 1974), (Figure 3), there are three transitions that could be rate limiting and each can be considered with regard to the experimentally determined βnuc value. If the rate-limiting step is nucleophilic attack and occurs upon the transition from substrates to the T± intermediate (Figure 3A), any degree of N-C bond formation without equivalent proton removal would increase the positive charge on the nucleophilic α-amine. The only transition state consistent with the observed βnuc would be an early one with minimal N-C bond formation. Low βnuc values of 0.1-0.2 were observed for model aminolysis reactions in solution for substrates with extreme pKas, which fell in the non-linear regions of reported Brønsted plots. The non-linearity of these plots was interpreted to denote a change in the rate-limiting step. Jencks interpreted the near zero βnuc values in these regions to indicate that nucleophilic attack was rate-limiting, resulting in an early transition state with little amide bond character and full retention of both amine protons(Jencks and Gilchrist, 1968; Satterthwait and Jencks, 1974).
A second possibility in this two-intermediate mechanism is that abstraction of the proton from T± to produce the T- intermediate is rate-limiting (Figure 3B). A near zero βnuc requires that the transition state must involve significant deprotonation, resulting in a transition state similar to the T- intermediate. If the rate-limiting step occurs after the T- intermediate, then C-O bond breaking would take place in the transition state. In this case, the βnuc value is not informative (Figure 3C) because inductive effects introduced by substitutions near the nucleophile are not conducted over the distance required to alter the leaving group. Thus, even assuming a stepwise reaction mechanism comparable to a typical aminolysis reaction in solution, the transition state is significantly different. Instead of resembling T±, the transition state either resembles substrates (Figure 3A), the T- intermediate (Figure 3B), or a transition state following the T- intermediate (Figure 3C).
The other mechanistic proposals that have been specifically suggested for the ribosome can also be evaluated in light of the Brønsted results. A stepwise mechanism with only one intermediate (T±) (Figure 4) has two transitions that could limit the overall rate(Das, Bhattacharyya et al., 1999; Schmeing, Huang et al., 2005; Trobro and Aqvist, 2005). This “proton shuttle” mechanism has been proposed based upon both in silico(Das, Bhattacharyya et al., 1999; Trobro and Aqvist, 2005) and crystallographic studies(Schmeing, Huang et al., 2005). In this mechanism, the 2′-OH of the P-site A76 shuttles protons between the α-amine and the 3′-O at the same time as the C-O bond cleaves. If the rate-limiting step is nucleophilic attack toward T± formation, the βnuc data are only consistent with a very early transition state resembling substrates for the same reasons as described above (Figure 4A). However, if resolution of the tetrahedral intermediate to products is rate-limiting (Figure 4B), then a zero βnuc value would only be possible in a transition state with significant C-O bond cleavage coupled to complete α-amine deprotonation. In this mechanism, the βnuc data constrain the transition state to one that is very late with a structure similar to products.
The fully concerted “proton shuttle” mechanism with no intermediates (Figure 5) (Dorner, Polacek et al., 2002; Changalov, Ivanova et al., 2005) requires that N-C bond formation, C-O bond cleavage, and proton-shuttling from the α-amine to the leaving group 3′-O all occur simultaneously. A near zero βnuc value is expected throughout the entire reaction coordinate as N-C bond formation occurs concurrently with α-amine proton abstraction. This coupling of bond formation and proton abstraction removes any relationship between the βnuc value and the extent of bond formation(Fersht, 1999). KIE and Brønsted analysis of the O3′ leaving group would be particularly useful for addressing the feasibility of this model.
A fourth model was proposed based upon a model system for the PT reaction. Changalov and Petkov argued that N-C bond formation is commensurate with α-amine deprotonation based on kinetic isotope effect (KIE) and Hammett correlation data for the transesterification of 2′/3′-O-p-substituted 2′-O-trityl adenosines and 2′-deoxyadenosines. Although these experiments were performed in solution, an organic base (DBU) was used as a catalyst and aprotic, apolar solvent conditions were chosen to mimic the interior of the ribosome. The KIEs indicate that a proton is “in flight” during the rate-determining transition state. The structure-activity relationship suggests that the rate-determining step for the model reaction is the formation of a tetrahedral intermediate that is similar to the T- intermediate, which they used as evidence for concerted nucleophilic attack and proton transfer(Changalov and Petkov, 2007). In this mechanism, the reaction proceeds from substrates to T- without forming the T± intermediate. This mechanism is also consistent with the βnuc value for the ribosome, but it requires deprotonation of the 2′-OH by a strong base to activate the 2′-O for deprotonation of the amine. There are no strong candidates for such an activating group within the ribosomal active site and chemical modification of the P-site A76 2′-OH indicates that it remains neutral in the transition state (Huang et al., submitted). Although the precise transition state cannot be fully defined from this βnuc value, the possible mechanisms that remain all differ significantly from the uncatalyzed reaction in solution.
There are examples of low βnuc coefficients for enzymes that have been interpreted to represent concerted or partially concerted reaction mechanisms. Bender investigated the serine protease-catalyzed cleavage of amides (a reaction resembling the reverse reaction of peptide bond formation) and proposed that the reaction proceeds through an ‘SN2-like’ rather than a step-wise mechanism because proton abstraction and nucleophilic attack are coupled(Komiyama and Bender, 1979). Inward and Jencks measured aminolysis rates on an acylenzyme, furoyl-chymotrypsin, and reported βnuc values of 0.13 – 0.19. They concluded that unassisted nucleophilic attack by the free amine could not occur and the proton must be removed either before or during the transition state(Inward and Jencks, 1965). Finally, a βnuc near zero was measured for amide bond synthesis by acetyltyrosylchymotrypsin. Deuterium isotope effects provided further support for the authors’ conclusion that proton abstraction is coupled with amine attack(Zeeberg and Caplow, 1973). These examples provide evidence that enzymes can perform aminolysis through different pathways than observed in solution and may be illustrative of the ribosomal reaction mechanism.
SIGNIFICANCE
This study explores a basic mechanistic question: Does the ribosomal peptidyl transfer reaction proceed through the same transition state as observed for a typical uncatalyzed aminolysis reaction in solution? To address this question, the Brønsted coefficient (βnuc) for the α-amino nucleophile was determined on 50S subunits and 70S ribosomes. The Brønsted coefficient reflects the change in charge between the ground state and the transition state of a reaction and thus provides valuable information regarding the reaction trajectory. βnuc was found to be approximately zero, suggesting that the rate of peptidyl transfer is independent of the pKa of the nucleophilic amine and that the amine remains neutral in the PT transition state. This value is significantly different than observed for uncatalyzed reactions in solution (βnuc = 0.8-0.9) where significant positive charge develops on the nucleophile in the transition state. A transition state with an uncharged amine suggests that the degree of N-C bond formation is commensurate with the degree of amine deprotonation. This value excludes mechanisms that require a transition state with significant positive charge development on the α-amine. The data suggest that the ribosome catalyzed reaction is facilitated by amine deprotonation, most likely through a proton shuttle mechanism.
METHODS
Reagents
Amino acids were purchased from CSPS Pharmaceuticals (San Diego, CA, USA). CPmn was purchased from Dharmacon (Lafayette, CO, USA). All other chemicals were purchased from Sigma (St. Louis, MO, USA). The cytidine-puromycin β, β -difluorophenylalanyl and the cytidine-puromycin phenylalanyl derivatives syntheses were previously reported(Okuda, Seila et al., 2005).
Puromycin-Phenylalanine and Cytidine-Puromycin-Phenylalanine Derivatives
Detailed descriptions of Pmn(3,4-Cl-Phe), Pmn(3,4-F-Phe), Pmn(4-CH3-Phe) synthesis and characterization is reported in the supplemental section. Amino acids were activated and then coupled to puromycin aminonucleoside (Pmn AN) then deprotected under basic conditions. The synthesis of Pmn(β-F-Phe) will be reported elsewhere. Detailed accounts of CPmn(β, β-F-Phe), CPmn(3,4-Cl-Phe), CPmn(2-F-Phe), CPmn(3,4-F-Phe), CPmn(Phe), CPmn(4-CF3-Phe), CPmn(4-CH3-Phe), CPmn(4-NH2-Phe) synthesis and characterization are reported in the supplemental section. The β-monofluorophenylalanyl (β-F-Phe) amino acid was incompatible with the solid phase chemistry described below due to β-elimination upon addition of base. Following coupling, a succinate linker was introduced at the 2′-oxygen. The derivatives were linked to solid support and a cytidine was added by solid phase chemical synthesis, followed by deprotection under basic conditions.
pKa Determination
Pmn, Pmn(β, β-F-Phe), and Pmn(Phe) pKa values were previously reported(Okuda, Seila et al., 2005). The pKas of the dihydrochloride salt of the puromycin derivatives were determined by monitoring the pH while titrating a 5 mM nucleoside solution in water:methanol:dichloromethane:conc. HCl (4:6:1:1) with 0.1 M NaOH (aq) at 25 °C. Methanol was added to improve the solubility of some derivatives. The measured pKa values for the synthesized compounds are reported in Figure 1.
Modified 50S Fragment Assay
Large ribosomal subunits were purified from E. coli strain MRE600(Seila, Okuda et al., 2005) and the aminolysis rates for the 50S modified fragment assay determined under single turnover conditions as previously described(Okuda, Seila et al., 2005). The activated 50S ribosomal subunits (final concentration 10 μM) were mixed with labeled CCApcb (<10 nM) and unlabeled aminoacyl substrates at concentrations more than 3-fold above the expected KM at 25°C. Buffer A (7 mM MgCl2, 7 mM KCl, 150 mM NH4Cl, 0.1 mM EDTA, 0.2 mM DTT, 25 mM MES, 25 mM MOPS, and 50 mM Tris-HCl pH 8.5 [25 °C]) was used for all 50S assays. Reaction rate (kpep) was determined by plotting the fraction of unreacted CCA-Pcb versus time using Kaleidagraph software and fit to the single exponential curve, fraction unreacted=M1*exp(-(kpep*t))+M2, where M1 is the reaction span, M2 is the endpoint, and t is time. Reactions using each A-site substrate were repeated 2-3 times, and the mean value and standard error were calculated. The log of the average maximum rate values log(kpep) were plotted versus the pKa for each cytidine puromycin derivative and βnuc value estimated from the slope of the plot by linear regression.
70S Rapid Kinetic Assay
The 70S ribosomes, initiation factors and f[3H]Met-tRNAfMet were prepared as previously described(Rodnina and Wintermeyer, 1995; Rodnina, Savelsbergh et al., 1999). MFT-mRNA (5′-GGCAAGGAGGUAAAUAAUGUUCACGAUU-3′, underlined sequence codes for fMet-Phe-Thr) was purchased from Dharmacon Research, Inc. (Boulder, CO, USA). Buffer B (50 mM Tris-HCl, 20 mM Bis-Tris, 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2 at pH 8.5 [37 °C]) was used throughout 70S protocols. Initiation complexes were prepared as previously discussed(Katunin, Muth et al., 2002; Beringer, Bruell et al., 2005). Time courses of PT reaction were measured in a quench-flow apparatus (KinTek, Austin, TX, USA). Reaction was started by mixing equal volumes (14 μl) of initiation complexes (0.2 μM) and Pmn(derivative) (1 – 40 mM) at 37 °C. The reaction was quenched by 25% formic acid and f[3H]MetPmn(derivative) was extracted and quantified as previously described(Beringer, Bruell et al., 2005). Pmn(3,4-Cl-Phe), Pmn(3,4-F-Phe), Pmn(4-CH3-Phe), and Pmn(4-NH2-Phe) were not soluble in the aqueous buffer, but all compounds were soluble in buffer B that contained 60% DMSO. Kinetic measurements were accomplished in the 60% DMSO buffer B using the entire Pmn derivative series. Saturation could not be achieved for Pmn(β-F-Phe) or Pmn(4-NH2-Phe). Pmn(β,β-F-Phe), Pmn(β-F-Phe), Pmn, and Pmn(Phe) were sufficiently soluble in buffer B without DMSO, so kinetics were measured with previously published conditions(Katunin, Muth et al., 2002). Single-exponential fitting was used for individual time courses to determine kobs at given concentration of Pmn derivative. Concentration dependencies of kobs were fit to a two-step model where a rapid, reversible binding step is followed by an irreversible reaction to yield kpep values(Katunin, Muth et al., 2002). The Brønsted coefficient was calculated from the two 70S data sets using log(kpep) and pKa as described above.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank W. Wintermeyer, M. Beringer, N. Carrasco, I. Suydam and D. Hiller for helpful kinetic discussions, J. Cochrane for comments on the manuscript, and C. Schillings, A. Böhm, S. Möbitz, and P. Striebeck for expert technical assistance. Funding for this work was provided by NIGMS training grant T32GM007223, a Beckman Scholar Award to RV, Ryobi Teien Memory Foundation grant to KO, NSF Travel Grant OISE-0339595, NIGMS grant 54839, and grants of the Deutsche Forschungsgemeinschaft to MVR.
REFERENCES
- Ban N, Nissen P, et al. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289(5481):905–20. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Beringer M, Bruell C, et al. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J Biol Chem. 2005;280(43):36065–72. doi: 10.1074/jbc.M507961200. [DOI] [PubMed] [Google Scholar]
- Blackburn GM, Jencks WP. The mechanism of the aminolysis of methyl formate. J. Am. Chem. Soc. 1968;90(10):2638–2645. [Google Scholar]
- Bruice TC, Lapinski R. Imidazole Catalysis. IV.1 The Reaction of General Bases with p-Nitrophenyl Acetate in Aqueous Solution. J. Am. Chem. Soc. 1958;80(9):2265–2267. [Google Scholar]
- Changalov MM, Ivanova GD, et al. 2′/3′-O-peptidyl adenosine as a general base catalyst of its own external peptidyl transfer: implications for the ribosome catalytic mechanism. Chembiochem. 2005;6(6):992–6. doi: 10.1002/cbic.200400349. [DOI] [PubMed] [Google Scholar]
- Changalov MM, Petkov DD. Linear free energy relationships and kinetic isotope effects reveal the chemistry of the Ado 2′-OH group. Tetrahedron Letters. 2007;48(13):2381–2384. [Google Scholar]
- Das GK, Bhattacharyya D, et al. A possible mechanism of peptide bond formation on ribosome without mediation of peptidyl transferase. J Theor Biol. 1999;200(2):193–205. doi: 10.1006/jtbi.1999.0987. [DOI] [PubMed] [Google Scholar]
- Dorner S, Panuschka C, et al. Mononucleotide derivatives as ribosomal P-site substrates reveal an important contribution of the 2′-OH to activity. Nucleic Acids Res. 2003;31(22):6536–42. doi: 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner S, Polacek N, et al. Molecular aspects of the ribosomal peptidyl transferase. Biochem Soc Trans. 2002;30(Pt 6):1131–6. doi: 10.1042/bst0301131. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W.H. Freeman; New York: 1999. [Google Scholar]
- Inward PW, Jencks WP. The Reactivity of Nucleophilic Reagents with Furoyl-chymotrypsin. J. Biol. Chem. 1965;240(5):1986–1996. [PubMed] [Google Scholar]
- Jencks WP. Catalysis in Chemistry and Enzymology. General Publishing Company; 1987. [Google Scholar]
- Jencks WP, Carriuolo J. General Base Catalysis of the Aminolysis of Phenyl Acetate. J. Am. Chem. Soc. 1960;82(3):675–681. [Google Scholar]
- Jencks WP, Carriuolo J. Reactivity of Nucleophilic Reagents toward Esters. J. Am. Chem. Soc. 1960;82(7):1778–1786. [Google Scholar]
- Jencks WP, Gilchrist M. Nonlinear structure-reactivity correlations. The reactivity of nucleophilic reagents toward esters. J. Am. Chem. Soc. 1968;90(10):2622–2637. [Google Scholar]
- Katunin VI, Muth GW, et al. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol Cell. 2002;10(2):339–46. doi: 10.1016/s1097-2765(02)00566-x. [DOI] [PubMed] [Google Scholar]
- Komiyama M, Bender ML. Do Cleavages of Amides by Serine Proteases Occur through a Stepwise Pathway Involving Tetrahedral Intermediates? PNAS. 1979;76(2):557–560. doi: 10.1073/pnas.76.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth GW, Ortoleva-Donnelly L, et al. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science. 2000;289(5481):947–50. doi: 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, et al. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289(5481):920–30. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Okuda K, Seila AC, et al. Uncovering the enzymatic pKa of the ribosomal peptidyl transferase reaction utilizing a fluorinated puromycin derivative. Biochemistry. 2005;44(17):6675–84. doi: 10.1021/bi047419c. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Beringer M, et al. Mechanism of peptide bond formation on the ribosome. Q Rev Biophys. 2006;39(3):203–25. doi: 10.1017/S003358350600429X. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Beringer M, et al. How ribosomes make peptide bonds. Trends Biochem Sci. 2007;32(1):20–6. doi: 10.1016/j.tibs.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, et al. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc Natl Acad Sci U S A. 1999;96(17):9586–90. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci U S A. 1995;92(6):1945–9. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwait AC, Jencks WP. Mechanism of the aminolysis of acetate esters. J. Am. Chem. Soc. 1974;96(22):7018–7031. doi: 10.1021/ja00829a034. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, et al. Structural insights into the roles of water and the 2′ hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell. 2005;20(3):437–48. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, et al. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438(7067):520–4. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Seila AC, et al. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat Struct Biol. 2002;9(3):225–30. doi: 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- Seila AC, Okuda K, et al. Kinetic isotope effect analysis of the ribosomal peptidyl transferase reaction. Biochemistry. 2005;44(10):4018–27. doi: 10.1021/bi047742f. [DOI] [PubMed] [Google Scholar]
- Sievers A, Beringer M, et al. The ribosome as an entropy trap. Proc Natl Acad Sci U S A. 2004;101(21):7897–901. doi: 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proc Natl Acad Sci U S A. 2005;102(35):12395–400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Parnell KM, et al. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol. 2004;11(11):1101–6. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth I, Beringer M, et al. Rapid peptide bond formation on isolated 50S ribosomal subunits. EMBO Rep. 2006;7(7):699–703. doi: 10.1038/sj.embor.7400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeberg B, Caplow M. Transition State Charge Distribution in Reactions of an Acetyltyrosylchymotrypsin Intermediate. J. Biol. Chem. 1973;248(16):5887–5891. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.