Abstract
Eukaryotic cell division is controlled by the activity of cyclin-dependent kinases (CDKs). Cdk1 and Cdk2, which function at different stages of the mammalian cell cycle, both require cyclin-binding and phosphorylation of the activation (T-) loop for full activity, but differ with respect to the order in which the two steps occur in vivo. To form stable complexes with either of its partners—cyclins A and B—Cdk1 must be phosphorylated on its T-loop, but that phosphorylation in turn depends on the presence of cyclin. Cdk2 can follow a kinetically distinct path to activation in which T-loop phosphorylation precedes cyclin-binding, and thereby out-compete the more abundant Cdk1 for limiting amounts of cyclin A. Mathematical modeling suggests this could be a principal basis for the temporal ordering of CDK activation during S phase, which may dictate the sequence in which replication origins fire. Still to be determined are how: 1) the activation machinery discriminates between closely related CDKs, and 2) coordination of the cell cycle is affected when this mechanism of pathway insulation breaks down.
Introduction
Two decades of investigation into the mechanisms of eukaryotic cell division control have produced a unified model, in which cyclin-dependent kinase (CDK) activity initiates both the DNA synthesis (S) phase and mitosis [reviewed in 1]. There are both quantitative and qualitative differences between the CDK requirements at the G1/S and G2/M boundaries: the threshold of activity needed to trigger mitosis is set higher than the one for S phase 2, 3; and the association with different cyclins confers distinct biochemical properties, such as substrate preferences, on the S- and M-phase forms of CDK in yeast 4, 5. Recently, we uncovered another way in which functions of closely related CDKs can be differentiated and, perhaps, insulated: activation by distinct mechanisms. Specifically, human Cdk1 and Cdk2, which are ~65% identical in aminoacid sequence 6–8, follow different kinetic paths to activation in vivo, even though they share a CDK-activating kinase (CAK) and at least one cyclin partner 9, 10. Here we discuss the possible sources of this difference, and its potential consequences for the proper functioning of the CDK network during cell division.
Cell cycle control by CDKs: revisionism begets reductionism
In both budding and fission yeast, a single CDK catalytic subunit pairs with multiple cyclin partners that accumulate with distinct cell-cycle timing. In metazoans, there has been expansion of the CDK family, such that different catalytic subunits are activated by different cyclins. There are clear examples of specialization in yeast and mammalian systems—in the propensities of CDKs to bind individual cyclins 11 and phosphorylate specific substrates in vitro 5, 12, and in the relative abilities of different CDK/cyclin complexes to trigger discrete cell-cycle events in vivo 1, 13—but there is also a surprising degree of plasticity or redundancy. The fission yeast Schizosaccharomyces pombe, for example, can complete both S phase and mitosis with only a single cyclin partner for the cell-cycle CDK 14. In the budding yeast Saccharomyces cerevisiae, loss of the cyclins normally responsible for triggering DNA replication does not block entry to S phase, but delays it until the next cyclins to be expressed can accumulate to sufficient levels 15. Finally, the mammalian cell cycle machinery can withstand loss of one or more of the catalytic subunits normally activated during interphase and still achieve faithful cell duplication [reviewed in 16].
Based on their activation timing and cyclin-binding preferences in mammalian cells, Cdk2 and Cdk1 were suspected of promoting S phase and mitosis, respectively. Overexpression of either CDK as a dominant negative (DN) version—a catalytically inactive mutant protein that retains normal cyclin-binding ability—produced discrete cell-cycle arrest phenotypes in human U2OS osteosarcoma cells: a G2/M arrest in the case of DN Cdk1; and arrest or delay in G1, S or G2 phase in cells expressing DN Cdk2 under different conditions 17, 18. In other cell lines, however, decreasing Cdk2 activity with DN Cdk2 expression, RNA interference (RNAi), or antisense DNA oligonucleotides produced no clear cell-cycle phenotypes 19. Mice lacking Cdk2 because of homozygous gene disruption, moreover, were viable 20, 21. The sequential activation of different CDKs that had seemed central to mammalian cell cycle control was now proposed to be no more than fine tuning, needed only in specialized cases such as the meiotic cell cycle—Cdk2−/− mice are infertile 20, 21—or in specific tissues or cell types 16. Mouse embryonic fibroblasts (MEFs) lacking all “interphase” CDKs—Cdk2, Cdk4 and Cdk6—are capable of faithful completion and alternation of S phase and mitosis, suggesting that Cdk1 might be the only CDK a mammalian cell needs for the basic task of cell division, and seeming to consign the other CDKs to limited, supporting roles 22.
Cdk2 stays in the picture
The survival of Cdk2−/− mice 20, 21, and the proliferation of cells lacking Cdk2, Cdk4 and Cdk6 22, leave little doubt that Cdk1 is sufficient for the essential functions previously ascribed to other CDKs. However, in chicken cells expressing a mutant Cdk1 sensitized to inhibition by bulky ATP analogs (analog-sensitive, or AS), Cdk1 activity was not necessary for S phase unless Cdk2 was removed by homozygous gene disruption 23. Therefore Cdk2 can support S-phase completion in the absence of active Cdk1, just as Cdk1 can do when Cdk2 protein is absent, leaving unclear the precise role of either CDK—when both are present—in executing a “normal” S phase.
Defining the division of labor between Cdk1 and Cdk2 is important both for understanding the regulation of cell division and for efforts to target its core machinery in cancer treatment 16, 24. Because each CDK is able to substitute for the other to varying degrees, gene disruption or silencing may not uncover the functions that either one performs, perhaps exclusively, in wild-type cells. Recently, two independent investigations into CDK-regulatory mechanisms in vivo yielded new evidence that Cdk2 might execute essential functions at discrete points in the mammalian cell cycle.
CDK activation: the same steps for different dances?
All CDKs involved directly in cell cycle control have the same minimal requirements for full activation: binding to a cyclin and phosphorylation within the activation segment (T-loop) of the kinase domain by a CAK (reviewed in 25, 26). In the best-studied case of human Cdk2, the two modifications collaborate to remodel the active site for efficient catalysis and protein substrate recognition; although cyclin-binding alone confers enzymatic activity on an intrinsically inert Cdk2 monomer 27, T-loop phosphorylation results in a further, ~300-fold stimulation of activity towards a model substrate 28. For Cdk1 activation, both cyclin-binding and T-loop phosphorylation are strictly required; unphosphorylated Cdk1/cyclin complexes have no measurable activity 11, 29, 30. Even the combined effects of cyclin and CAK might not be sufficient in all cases; a recent crystal structure of Cdk4 bound to cyclin D and phosphorylated on the T-loop revealed catalytic residues that were not properly oriented for phosphotransfer 31 and suggested an additional requirement—perhaps for the binding of protein substrate—in the conformational maturation of Cdk4 31, 32.
In vivo, both Cdk1 and Cdk2 bind cyclin A 33, but unlike Cdk2, Cdk1 cannot form stable complexes with cyclin A in vitro without T-loop phosphorylation 11, 29. Conversely, phosphorylation of Cdk1 by the Cdk7 complex—the sole CAK identified in mammalian cells—requires the presence of a cyclin 11, 34, whereas Cdk7 can efficiently phosphorylate monomeric Cdk2 34, 35. Consistent with the more stringent requirements for assembly into stable complexes, mutant forms of Cdk1 with the Thr residue phosphorylated by CAK mutated to Ala or Val were defective for cyclin-binding in Xenopus extracts 36, 37, and inactivation of Drosophila Cdk7 by a temperature-sensitive mutation specifically depleted Cdk1/cyclin A complexes in vivo 38. These results suggested that Cdk1 and Cdk2 might have different modes of regulation in vivo, despite their extensive homology and shared activation machinery.
Tracing the steps to CDK activation in vivo: order out of AS
The ATP-binding sites of eukaryotic protein kinases nearly invariably contain a bulky amino-acid residue—the so-called gatekeeper—that can be mutated in the majority of cases to a less-bulky Gly or Ala residue without inactivating the enzyme 39. The expanded pocket can accommodate derivatized ATP analogs—substrates or non-hydrolyzable inhibitors—that bind poorly to the wild-type kinase, allowing both monospecific inhibition in vivo and labeling of specific substrates by a single kinase in crude extracts 2, 40. Once an AS kinase is introduced into cells in place of the wild-type version, its activity can be controlled specifically in vivo with the speed and reversibility uniquely afforded by small molecules. Although this chemical-genetic manipulation of the cell-cycle machinery is still most easily accomplished in yeast, advances in gene-targeting with recombinant adeno-associated virus (rAAV) 41 have facilitated the creation of AS kinase-dependent mammalian cell lines 9, 42.
Human colon carcinoma HCT116 cells with both copies of the Cdk7 gene replaced by Cdk7as alleles divided at near-normal rates in the absence of drugs, but ceased proliferation when treated with inhibitory analogs specific for the mutated enzyme 9. Inhibition of Cdk7as impeded G1/S and G2/M progression, and led to loss of T-loop phosphorylation on both Cdk1 and Cdk2, providing genetic validation of Cdk7 as a general CAK in mammalian cells 9. Precise control over CAK activity, moreover, enabled dissection of the assembly and activation mechanisms of the two major cell-cycle CDKs in vivo. Inhibition of Cdk7 within the first six hours after release from a block at the G1/S boundary prevented subsequent passage into mitosis. Although cyclin B (the partner of Cdk1 needed for the G2/M transition) accumulated, it failed to assemble with Cdk1 when CAK was inhibited 9. That dependency could be recapitulated in extracts of Cdk7as/as cells; treating extracts with the inhibitor 1-NM-PP1 blocked the ability of added cyclin B to assemble with endogenous Cdk1, whereas addition of a 1-NM-PP1-insensitive CAK rescued binding 9. Therefore, although purified Cdk1 and cyclin B can form complexes in vitro in the absence of CAK, phosphorylation of the Cdk1 T-loop is required for stable binding to cyclin B in vivo 11, 29. Conversely, phosphorylation of the Cdk1 T-loop by Cdk7 depends on the presence of cyclin 11, 34. The two steps in Cdk1 activation are thus mutually dependent in vivo and must occur in concert. (Intrinsic instability of Cdk1/cyclin complexes, relative to Cdk2/cyclin complexes, might also explain why only the latter show activity independent of T-loop phosphorylation 11, 27, 29.)
Inhibition of CAK had no apparent effect on the ability of Cdk2 to form complexes with its cyclin partners (E and A) 9, 10. In fact, inhibiting Cdk7 increased binding of Cdk2 to an unnatural partner, cyclin B, presumably because of impaired Cdk1/cyclin B assembly and the resulting liberation of cyclin B 10. The ability to shut off CAK activity selectively also allowed us to measure turnover of phospho-Thr160—the T-loop residue phosphorylated by Cdk7—in different populations of Cdk2. Surprisingly, there was little or no decay of T-loop phosphorylation in the cyclin-bound fraction after 12 hours in the absence of CAK activity, whereas phosphorylated Cdk2 monomer disappeared within ~2 hours 10. These results implied that, in vivo: 1) cyclin-bound Cdk2 is resistant to attack by phosphatases, and active CAK is not needed to maintain T-loop phosphorylation of pre-formed and activated Cdk2/cyclin complexes; 2) monomeric Cdk2 is phosphorylated by Cdk7, in opposition to the known T-loop phosphatases that are specific for the cyclin-free form 43, 44; and 3) Cdk2 is not dependent on T-loop phosphorylation for cyclin-binding, unlike Cdk1.
Kinetic measurements with purified enzymes and substrates support the idea that phosphorylation of monomeric Cdk2, followed by cyclin-binding to generate active complexes, is a physiologic pathway of Cdk2 activation. The Cdk7 complex phosphorylated Cdk2 monomers and Cdk2/cyclin A complexes with nearly equal efficiency (kcat/Km), meaning that the relative frequencies of the two reactions in vivo should be determined by the relative concentrations of their respective substrates. In asynchronous HCT116 cells, monomeric Cdk2 appears to be in slight excess over cyclin-bound forms. At steady state, however, most cyclin-bound Cdk2 is phosphorylated on the T-loop (and not a substrate for CAK), whereas the majority of monomeric Cdk2 is not (and therefore available for phosphorylation) 10, suggesting that Cdk2 is likely to be phosphorylated before it binds cyclin in vivo.
CDK-cyclin pairing: activating kinase as molecular matchmaker
The distinct activation pathways could be reconstituted with monomeric Cdk1 and Cdk2 recovered from Cdk7as/as human cells that had been treated with inhibitory analogs to deplete the phosphorylated isoforms 10. Human extracts also recapitulated the preferential binding of cyclin A to Cdk2 observed in vivo, despite having an ~tenfold greater abundance of Cdk1 10, 45. This selectivity appears to be a direct consequence of the different activation mechanisms available to the two CDKs; when extracts were supplemented with Csk1, a fission yeast CAK that can phosphorylate monomeric human Cdk1 46–48, cyclin A bound Cdk1 and Cdk2 roughly in proportion to their initial concentrations 10.
This switch in cyclin A-binding preference, upon redirection of Cdk1 into an activation pathway normally exclusive to Cdk2, suggests a role for Cdk7 as a chaperone (in the colloquial sense) for CDK-cyclin pairs. By virtue of its substrate preferences, Cdk7 helps determine which CDKs bind which cyclins—and when those couplings occur—during the cell cycle. For example, CAK normally prevents cyclin B pairing with Cdk2 by stabilizing intrinsically weak Cdk1/cyclin B complexes 9, 10. Conversely, CAK actively promotes pairing between Cdk2 and cyclin A; when Csk1 was added to extracts from untreated cells, cyclin A-binding to Cdk1 came at the expense of binding to unposphorylated Cdk2, while the Thr160-phosphorylated form of endogenous Cdk2 still bound normally to added cyclin A 10. This indicates that both the ability of Cdk7 to phosphorylate Cdk2 monomers and its inability to phosphorylate monomeric Cdk1 contribute to a kinetic barrier preventing Cdk1/cyclin A assembly. Consistent with this notion, cyclin A bound nearly exclusively to Cdk2 in G1 and early S phase, and began to assemble with Cdk1 only after Cdk2/cyclin A levels reached a plateau in mid- to late-S phase 10.
Sequential CDK assembly during S phase: a quantitative model
To address whether this kinetic difference is sufficient to account for the observed specificity and timing of CDK/cyclin pairings in vivo, we built a mathematical model based on experimentally determined or calculated rate constants and intracellular protein concentrations 49. Reliable affinity measurements for CDK-cyclin binding are unavailable, so in the initial simulations we uniformly set Kd at 10 nM for complexes that form readily in vitro (Cdk2/cyclin A, Cdk2-P/cyclin A and Cdk1-P/cyclin A) and at 10 μM for those that do not (Cdk1/cyclin A). The model recapitulates the preference of cyclin A for Cdk2 observed in human cells 10; in a simulation of normal cell-cycle progression, Cdk1/cyclin A began to accumulate only after Cdk2/cyclin A complex formation was saturated (Figure 1A). This transition occurred around the time free Cdk2 was depleted, perhaps suggestive of a passive mechanism rather than an active switch. In addition, when phosphorylation of monomeric Cdk1 was permitted (i.e., when Km and Vmax values for monomeric Cdk2 were applied to Cdk1), the model predicted a switch in cyclin A-binding preference from Cdk2 to Cdk1, such that the temporal sequence of Cdk2- and Cdk1-binding is obliterated (Figure 1B). These results fit our experimental findings in whole-cell extracts without or with added Csk1, respectively 10, and support the notion that distinct pathways of activation could by themselves account for the order of cyclin A-CDK binding events in vivo.
Figure 1. A mathematical model of CDK activation recapitulates CDK-cyclin binding preferences observed in vivo.
A model of ordinary differential equations was solved in Virtual Cell 49 using the values indicated in the reaction schematic. We modeled accumulation of cyclin A to occur in linear, time-dependent fashion. The intracellular concentrations of Cdk1, Cdk2, and Cdk7 were calculated based on experimentally determined amounts of each CDK per gram of cellular protein 45, 57, and the kinetic parameters of Cdk2 phosphorylation were derived from experimentally determined values 10. Models (A) and (B) are identical, except that (B) is modified to allow Cdk1 monomer to be phosphorylated by Cdk7 and dephosphorylated by a phosphatase with kinetics identical to those measured or estimated for Cdk2. The kinetics of phosphorylation of Cdk1 (which occurs only in model B) and Cdk1/cyclin A have not been determined in vitro, so they were assumed to be equivalent to those determined for Cdk2 and Cdk2/cyclin A, respectively. As the values of Kd have not been determined, they were arbitrarily set to 10 nM for those complexes that readily form in vitro (Cdk2/CycA, Cdk2-P/CycA, and Cdk1-P/CycA) and to 10 μM for those that do not (Cdk1/CycA). The kinetic parameters used for CDK monomer dephosphorylation are estimates based on values typical of phosphatases 72. We did not include phosphatases acting on CDK/cyclin complexes in the model, because cyclin-binding generally blocks CDK T-loop dephosphorylation 43, 44.
We next tested robustness of the model in predicting the behavior of CDKs and cyclins in mammalian cells, by varying Vmax of monomeric Cdk1 phosphorylation from 0 (as in the first simulation) to 50% of Vmax of Cdk2 monomer phosphorylation (Figure 2A). The model predicted a strong preference by cyclin A for Cdk2 even when the rate of Cdk1 phosphorylation was set at 10% that of Cdk2 phosphorylation—almost certainly an overestimate, because we have not detected phosphorylation of monomeric Cdk1 by human CAK under any conditions 10, 34. We also evaluated the potential importance of the equilibrium binding constant Kd and its component on- and off-rates (kforward and kreverse, respectively) in determining the predicted specificity of CDK-cyclin binding. Upon varying either the on- or off-rate of Cdk1/cyclin A complex formation while holding the other parameter constant, we observed that the preference of cyclin A for Cdk2 was maintained as long as we enforced at least a 100-fold difference in the rate of association, but was relatively insensitive to changes in dissociation rate (Figure 2B). In an extreme example, a simulation in which Kd for Cdk1 was only 10-fold higher than that for Cdk2 still produced “normal” temporal profiles of Cdk2 and Cdk1 activation if we maintained a 1,000-fold faster association rate for Cdk2 relative to Cdk1 while setting the rate of Cdk2/cyclin A dissociation 100 times faster than that of Cdk1/cyclin A. This makes intuitive sense—a high on-rate for Cdk1-cyclin A interaction would increase the likelihood that Cdk7 can phosphorylate Cdk1 and thereby stabilize the complex (i.e., diminish the off-rate)—but will require experimental validation.
Figure 2. Modeling contributions of enzymologic parameters and binding affinities to proper CDK-cyclin pairing in vivo.
Simulations were performed in which (A) Vmax of the Cdk1 monomer phosphorylation reaction or (B) binding affinities of cyclin A for unphosphorylated Cdk1 were varied from the limit values set in Figure 1. In (B), the equilibrium binding constant Kd was varied by changes in either on-rate kforward (kf, top row) or off-rate kreverse (kr, bottom row), while holding the other parameter constant.
We need not invoke a similar mechanism to account for the behavior of cyclins B and E, which bind nearly exclusively to Cdk1 and Cdk2, respectively, in vivo 10. Despite their scarcity in wild-type cells, Cdk2/cyclin B complexes form readily from purified components in vitro, and can be detected in vivo when Cdk1 protein levels are decreased by RNAi or when Cdk1 complex formation is disrupted by CAK inhibition 10, 50. This indicates that endogenous Cdk2 is competent to bind cyclin B when Cdk1 is unavailable or unable to form stable complexes. Moreover, it suggests that the high ratio of Cdk1-to-Cdk2 monomer concentration observed in multiple human cell lines 10, 45 is likely to be important for normal cyclin B pairing fidelity. The restriction of cyclin B expression to later times in the cell cycle, when most Cdk2 is already bound to cyclin E or A, would also serve to reinforce “selectivity” for Cdk1 in vivo.
The nearly exclusive binding of cyclin E to Cdk2, on the other hand, might be a simple function of relative affinities. A comparison of Cdk2 co-crystal structures with cyclin E or A revealed similar interfaces, but more extensive contacts with cyclin E (~14% increase in buried surface area relative to Cdk2/cyclin A), consistent with a tighter complex 51. Only relatively minor steric clashes occurred when a modeled structure of Cdk1 was substituted for Cdk2 in the co-crystal with cyclin E, and these could be relieved by small adjustments of Cdk1 conformation 51. Cdk1 and cyclin E do interact productively, but transiently, in vitro: cyclin E can facilitate phosphorylation of Cdk1 by Cdk7 but cannot form a stable binary complex with Cdk1, even after Cdk1 T-loop phosphorylation 11. Nevertheless, Cdk1/cyclin E complexes can accumulate in cells lacking Cdk2 22, 52, by an unknown mechanism. One possibility is that removal of Cdk2 permits Cdk1/cyclin E assembly assisted by an accessory factor such as p21Cip1 or p27Kip1, both of which can promote assembly of intrinsically unstable Cdk4/cyclin D pairs 53. This puzzle may remain unsolved at least as long as no actual structure of Cdk1 is available.
Just-in-time CDK assembly and activation: keys to a normal S phase?
The ability of Cdk2 to exclude Cdk1 from complexes with cyclins E and A helps establish a temporal hierarchy of CDK assembly and activation in vivo. Recently, another mechanism for delaying Cdk1 function was uncovered in MEFs, where ablation of Chk1—an essential kinase with roles in normal cell cycle progression and the DNA damage checkpoint—led to premature activation of Cdk1/cyclin A during S phase. Complex assembly was not advanced in time, arguing against an effect mediated through CAK or Cdk2. Instead, inhibitory phosphorylation of Cdk1 on Tyr15 was lost prematurely, apparently due to increased levels of the Tyr15-specific phosphatase Cdc25A, which is destabilized during a normal S phase by Chk1-mediated phosphorylation 54.
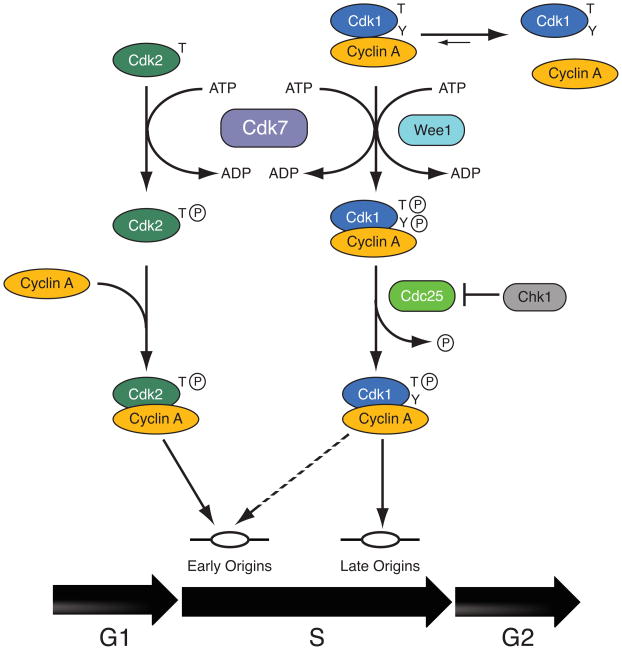
Therefore, reinforcing mechanisms ensure that activation of Cdk2/cyclin A precedes that of Cdk1/cyclin A 10, 54. The order of CDK activation may be important for S-phase coordination; normally late-firing origins initiated replication early in S phase when Chk1 was inactivated in MEFs and in cells engineered to express a constitutively active Cdk1-cyclin A fusion protein 54. The authors proposed a model in which Cdk2/cyclin A triggers initiation only from the early-firing subset of origins, whereas Cdk1/cyclin A can activate most or all origins, regardless of their normal replication timing (Figure 3) 54. This resembles the division of labor between budding yeast S-phase cyclins; both Clb5 and Clb6 can activate Cdk1 to trigger replication at early-firing origins, but Clb5 is specifically required at late-firing origins 55 as Cdk1 appears to be in mammalian cells. It remains to be determined what effects, if any, the loss of this coordination will have on long-term cell survival and genomic integrity.
Figure 3. Sequential activation of Cdk2 and Cdk1: a way to order S phase?
A model of S-phase coordination by timed activation of Cdk2/cyclin A and Cdk1/cyclin A. Cdk2 has a competitive advantage in binding cyclin A conferred by its ability to be recognized by Cdk7 as a monomer and to bind cyclin A when unphosphorylated. This largely excludes Cdk1 from active complexes until Cdk2 is saturated with cyclin in mid- to lateS-phase 10. Another, potentially reinforcing mechanism for delaying Cdk1 activation was recently described: Chk1-mediated down-regulation of the CDK-activating phosphatase Cdc25, which removes phosphates from the Tyr15 and Thr14 residues added by the inhibitory kinase Wee1. Loss of Chk1 led to premature activation of late-firing replication origins 54. We hypothesize that advancing Cdk1/cyclin A assembly might have the same effect, and propose that the normal temporal sequence, of Cdk2- followed by Cdk1-binding to cyclin A, is important for orderly S phase progression.
Customized CDK activation pathways: one size does not fit all, but why not?
Cdk1 and Cdk2 defy the naïve expectation that they would follow the same path to full activation 9, 10. The structures of Cdk4 complexes suggest requirements beyond cyclin D-binding and T-loop phosphorylation for maximal activity 31, 32. Indeed, it is beginning to look as if each CDK/cyclin pair might be handled in its own unique way by the activation machinery—versatility accomplished, in the case of Cdk1 and Cdk2, without evolving separate CAKs. Cdk4 regulation might represent another variation on the same theme; although the kinase responsible for activating Cdk4 in vivo has not been identified conclusively, Cdk7 is capable of phosphorylating Cdk4/cyclin D complexes in vitro, and removal of Cdk7 from mouse cell extracts abolished their ability to activate added Cdk456. Cdk4 was also a target of Cdk7as identified by radiolabeling with a substrate analog in HeLa nuclear extracts 57. Recent work suggests that Cdk4 T-loop phosphorylation may be coupled to mitogenic signaling through the CDK inhibitor (CKI) p27Kip1 58–60. In response to growth-promoting signals, Cdk4-bound p27Kip1 is phosphorylated at Tyr residues in its 3–10 helix motif, neutralizing its inhibitory activity both directly, by dislodging the helix from the active site of the kinase 59; and indirectly, by reversing a CKI-mediated block to Cdk4 T-loop phosphorylation 61. Only a mammalian CAK, the Cdk7 complex, was sensitive to this modification, whereas a heterologous CAK from S. pombe, Csk1, could phosphorylate the T-loop of Cdk4 bound to phosphorylated or unphosphorylated p27Kip1 equally well 61.
Thus, mechanistic studies of CDK regulation continue to elucidate how CDKs and their activators co-evolved to coordinate mammalian cell division. Seemingly subtle distinctions between Cdk1 and Cdk2—in their affinities for different cyclins, recognition by CAK and dependence on T-loop phosphorylation for cyclin-binding 11, 29, 34—have turned out to be important determinants of their cyclin-specificity and activation timing in vivo 9, 10. All the more reason, then, to dig deeper and ask why two CDKs so similar in primary structure are processed so differently inside the cell. There are still gaps in our basic knowledge of these enzymes, the most glaring perhaps being a structure of Cdk1—either active or inactive—from any organism. A series of Cdk1 structures, analogous to the available snapshots of Cdk2 in monomeric 62 and cyclin A-bound forms 28, 63, could explain why T-loop phosphorylation is required for complex stability. The structure of a Cdk1 monomer could also reveal why its T-loop is incompetent for phosphorylation by Cdk7. Then again it might not, given that the T-loop of monomeric Cdk2 is accessible to CAK in solution but not solvent-exposed in the crystal 62.
Insight into the differential regulation of Cdk1 and Cdk2 is also likely to come from a better understanding of CAK-CDK interactions. How Cdk7 finds its CDK substrates is still an open question. We know how it doesn’t work—through direct recognition of protein sequence surrounding the phosphorylation site—based on T-loop swaps between a CDK that is a natural Cdk7 target (Cdk2) and one that is not (Cdk7 itself) 64. In vitro, CAK appears to bind Cdk2/cyclin A stably and turn it over slowly to yield free, phosphorylated Cdk2/cyclin A 28, 57, 65. A structure of the complex between Cdk7/cyclin H/Mat1 and Cdk2/cyclin A would help define the determinants of their interaction, but our recent data suggest this might not be the most physiologically relevant intermediate. We should now add a complex of CAK with monomeric Cdk2 to the structural wish list—a tall order, perhaps, because of the likely contribution by cyclin A to CAK-Cdk2 binding affinity 10. To date the only Cdk7 structure available is of an inactive monomer 66. That was the basis for a model of Cdk2 docked to Cdk7 67, which might illustrate how the two CDKs are able to phosphorylate one another 64, but clearly more actual structures—preferably of active complexes—are needed to validate any proposed model of the Cdk2-Cdk7 interaction.
Concluding thoughts
Chemical-genetic dissection of CDK activation in mammalian cells yielded a confirmation and a surprise, both of which have important implications for our understanding of cell-cycle control. Selective inhibition of Cdk7 in vivo showed it to be a CAK for both Cdk1 and Cdk2, needed at both G1/S and G2/M transitions, as suggested by biochemical data and genetic evidence in lower organisms 25. Nonetheless, comparison of Cdk1- and Cdk2-activating mechanisms in vivo uncovered two distinct pathways 9, 10. We expect deeper exploration of CDK-regulatory pathways—both activating and inhibitory—to reveal more complexity of the kind described here. For example, in both yeast and human cells, different CDK/cyclin pairs are phosphorylated on inhibitory Tyr residues with different efficiencies by the same kinase 68, 69. It is likely that no two CDK complexes are regulated in quite the same way in vivo. This need not involve different regulators, however, if the biochemical output of a pathway can be modulated merely by re-ordering the steps. As in the example illustrated here, we should not reflexively posit additional CAKs, just because two CDKs appear to obey different rules when it comes to T-loop phosphorylation in vivo.
A second lesson is that the multiple catalytic subunits of metazoan CDKs are likely to perform exclusive functions in vivo, even if Cdk1 can take over essential functions when other CDKs are lost. Cdk1 is by far the most abundant CDK in mammalian cells 10, 45, and can complement the essential functions of Cdk1 in yeast 70, 71, so its latent ability to drive S phase by itself should not come as a surprise. In wild-type cells, however, Cdk1 is excluded from complexes with essential cyclins at decisive points in the cell cycle, by mechanisms that depend on the presence of Cdk2 10 (and, perhaps, other CDKs). Inevitably, Cdk gene knockouts and knockdowns disrupt normal pairing, allowing the unnatural formation of some complexes (e.g. Cdk1/cyclin E), and the premature or excessive activation of others (e.g. Cdk1/cyclin A). Future chemical-genetic studies in cells expressing the normal complement of CDKs and cyclins should deepen our understanding of cell-cycle coordination, and provide more accurate models for discovery of anti-cancer drug targets.
Acknowledgments
We thank R. Iyengar and S. Larochelle for critical review of the manuscript, and members of the Fisher lab for helpful discussions. This work was supported by NIH grant GM056985 to R.P.F.
References
- 1.Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press Ltd; 2007. [Google Scholar]
- 2.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 3.Stern B, Nurse P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996;12:345–50. [PubMed] [Google Scholar]
- 4.Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–60. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- 5.Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- 6.Elledge SJ, Spottswood MR. A new human p34 protein kinase, CDK2, identified by complementation of a cdc28 mutation in Saccharomyces cerevisiae, is a homolog of Xenopus Eg1. EMBO J. 1991;10:2653–9. doi: 10.1002/j.1460-2075.1991.tb07808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ninomiya-Tsuji J, Nomoto S, Yasuda H, Reed SI, Matsumoto K. Cloning of a human cDNA encoding a CDC2-related kinase by complementation of a budding yeast cdc28 mutation. Proc Natl Acad Sci USA. 1991;88:9006–10. doi: 10.1073/pnas.88.20.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai L-H, Harlow E, Meyerson M. Isolation of the human cdk2 gene that encodes the cyclin A- and adenovirus E1A-associated p33 kinase. Nature. 1991;353:174–7. doi: 10.1038/353174a0. [DOI] [PubMed] [Google Scholar]
- 9.Larochelle S, Merrick KA, Terret ME, Wohlbold L, Barboza NM, Zhang C, Shokat KM, Jallepalli PV, Fisher RP. Requirements for Cdk7 in the assembly of Cdk1/cyclin B and activation of Cdk2 revealed by chemical genetics in human cells. Mol Cell. 2007;25:839–50. doi: 10.1016/j.molcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merrick KA, Larochelle S, Zhang C, Allen JJ, Shokat KM, Fisher RP. Distinct activation pathways confer cyclin binding selectivity on Cdk1 and Cdk2 in human cells. Mol Cell. 2008;32:662–72. doi: 10.1016/j.molcel.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai D, Wessling HC, Fisher RP, Morgan DO. The effect of phosphorylation by CAK on cyclin binding by CDC2 and CDK2. Mol Cell Biol. 1995;15:345–50. doi: 10.1128/mcb.15.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem. 1997;272:12738–46. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
- 13.Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–69. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 14.Fisher DL, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. Embo J. 1996;15:850–60. [PMC free article] [PubMed] [Google Scholar]
- 15.Piatti S, Bohm T, Cocker JH, Diffley JF, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–31. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 16.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 17.Hu B, Mitra J, van den Heuvel S, Enders GH. S and G2 phase roles for Cdk2 revealed by inducible expression of a dominant-negative mutant in human cells. Mol Cell Biol. 2001;21:2755–66. doi: 10.1128/MCB.21.8.2755-2766.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–4. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 19.Tetsu O, McCormick F. Proliferation of cancer cells despite CDK2 inhibition. Cancer Cell. 2003;3:233–45. doi: 10.1016/s1535-6108(03)00053-9. [DOI] [PubMed] [Google Scholar]
- 20.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–85. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 21.Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 22.Santamaria D, Barriere C, Cerqueira A, Hunt S, Tardy C, Newton K, Caceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 23.Hochegger H, Dejsuphong D, Sonoda E, Saberi A, Rajendra E, Kirk J, Hunt T, Takeda S. An essential role for Cdk1 in S phase control is revealed via chemical genetics in vertebrate cells. J Cell Biol. 2007;178:257–68. doi: 10.1083/jcb.200702034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol. 2006;24:1770–83. doi: 10.1200/JCO.2005.03.7689. [DOI] [PubMed] [Google Scholar]
- 25.Fisher RP. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J Cell Sci. 2005;118:5171–80. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- 26.Morgan DO. The dynamics of cyclin dependent kinase structure. Curr Opin Cell Biol. 1996;8:767–72. doi: 10.1016/s0955-0674(96)80076-7. [DOI] [PubMed] [Google Scholar]
- 27.Connell-Crowley L, Solomon MJ, Wei N, Harper JW. Phosphorylation-independent activation of human cyclin-dependent kinase 2 by cyclin A in vitro. Mol Biol Cell. 1993;4:79–92. doi: 10.1091/mbc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo AA, Jeffrey PD, Pavletich NP. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nature Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 29.Desai D, Gu Y, Morgan DO. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–82. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solomon MJ, Lee T, Kirschner MW. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day PJ, Cleasby A, Tickle IJ, O’Reilly M, Coyle JE, Holding FP, McMenamin RL, Yon J, Chopra R, Lengauer C, Jhoti H. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci U S A. 2009;106:4166–70. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takaki T, Echalier A, Brown NR, Hunt T, Endicott JA, Noble ME. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci U S A. 2009;106:4171–6. doi: 10.1073/pnas.0809674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–71. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher RP, Morgan DO. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–24. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 35.Wohlbold L, Larochelle S, Liao JC, Livshits G, Singer J, Shokat KM, Fisher RP. The cyclin-dependent kinase (CDK) family member PNQALRE/CCRK supports cell proliferation but has no intrinsic CDK-activating kinase (CAK) activity. Cell Cycle. 2006;5:546–54. doi: 10.4161/cc.5.5.2541. [DOI] [PubMed] [Google Scholar]
- 36.Ducommun B, Brambilla P, Felix M-A, Franza BR, Karsenti E, Draetta G. cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 1991;10:3311–9. doi: 10.1002/j.1460-2075.1991.tb04895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee TH, Turck C, Kirschner MW. Inhibition of cdc2 activation by INH/PP2A. Mol Biol Cell. 1994;5:323–38. doi: 10.1091/mbc.5.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larochelle S, Pandur J, Fisher RP, Salz HK, Suter B. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 1998;12:370–81. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–30. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 40.Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–64. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 41.Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burkard ME, Randall CL, Larochelle S, Zhang C, Shokat KM, Fisher RP, Jallepalli PV. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc Natl Acad Sci U S A. 2007;104:4383–8. doi: 10.1073/pnas.0701140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng A, Ross KE, Kaldis P, Solomon MJ. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 1999;13:2946–57. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poon RYC, Hunter T. Dephosphorylation of Cdk2 Thr160 by the cyclin-dependent kinase-interacting phosphatase KAP in the absence of cyclin. Science. 1995;270:90–3. doi: 10.1126/science.270.5233.90. [DOI] [PubMed] [Google Scholar]
- 45.Arooz T, Yam CH, Siu WY, Lau A, Li KK, Poon RY. On the concentrations of cyclins and cyclin-dependent kinases in extracts of cultured human cells. Biochemistry. 2000;39:9494–501. doi: 10.1021/bi0009643. [DOI] [PubMed] [Google Scholar]
- 46.Lee KM, Saiz JE, Barton WA, Fisher RP. Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases CAKs) Curr Biol. 1999;9:441–4. doi: 10.1016/s0960-9822(99)80194-8. [DOI] [PubMed] [Google Scholar]
- 47.Saiz JE, Fisher RP. A CDK-activating kinase network is required in cell cycle control and transcription in fission yeast. Curr Biol. 2002;12:1100–5. doi: 10.1016/s0960-9822(02)00903-x. [DOI] [PubMed] [Google Scholar]
- 48.Tsakraklides V, Solomon MJ. Comparison of Cak1p-like cyclin-dependent kinase-activating kinases. J Biol Chem. 2002;277:33482–9. doi: 10.1074/jbc.M205537200. [DOI] [PubMed] [Google Scholar]
- 49.Moraru II, Schaff JC, Slepchenko BM, Blinov ML, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM. Virtual Cell modelling and simulation software environment. IET Syst Biol. 2008;2:352–62. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.L’Italien L, Tanudji M, Russell L, Schebye XM. Unmasking the redundancy between Cdk1 and Cdk2 at G2 phase in human cancer cell lines. Cell Cycle. 2006;5:984–93. doi: 10.4161/cc.5.9.2721. [DOI] [PubMed] [Google Scholar]
- 51.Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, Brown NR, Johnson LN. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. Embo J. 2005;24:452–63. doi: 10.1038/sj.emboj.7600554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–6. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 53.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 1997;11:847–62. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 54.Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:3184–9. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae Mol Cell. 1998;2:173–82. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 56.Matsuoka M, Kato J, Fisher RP, Morgan DO, Sherr CJ. Activation of cyclin-dependent kinase-4 (CDK4) by mouse MO15-associated kinase. Mol Cell Biol. 1994;14:7265–75. doi: 10.1128/mcb.14.11.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larochelle S, Batliner J, Gamble MJ, Barboza NM, Kraybill BC, Blethrow JD, Shokat KM, Fisher RP. Dichotomous but stringent substrate selection by the dual-function Cdk7 complex revealed by chemical genetics. Nat Struct Mol Biol. 2006;13:55–62. doi: 10.1038/nsmb1028. [DOI] [PubMed] [Google Scholar]
- 58.Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, Sun P, Tan CK, Hengst L, Slingerland J. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–94. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128:269–80. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 60.James MK, Ray A, Leznova D, Blain SW. Differential modification of p27Kip1 controls its cyclin D-cdk4 inhibitory activity. Mol Cell Biol. 2008;28:498–510. doi: 10.1128/MCB.02171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ray A, James MK, Larochelle S, Fisher RP, Blain SW. p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol Cell Biol. 2009;29:986–99. doi: 10.1128/MCB.00898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De Bondt HL, Rosenblatt J, Jancarik J, Jones HD, Morgan DO, Kim S-H. Crystal structure of cyclin-dependent kinase 2. Nature. 1993;363:595–602. doi: 10.1038/363595a0. [DOI] [PubMed] [Google Scholar]
- 63.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–20. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 64.Garrett S, Barton WA, Knights R, Jin P, Morgan DO, Fisher RP. Reciprocal activation by cyclin-dependent kinases 2 and 7 is directed by substrate specificity determinants outside the T-loop. Mol Cell Biol. 2001;21:88–99. doi: 10.1128/MCB.21.1.88-99.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larochelle S, Chen J, Knights R, Pandur J, Morcillo P, Erdjument-Bromage H, Tempst P, Suter B, Fisher RP. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. EMBO J. 2001;20:3749–59. doi: 10.1093/emboj/20.14.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure (Camb) 2004;12:2067–79. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 67.Lolli G, Johnson LN. Recognition of Cdk2 by Cdk7. Proteins. 2007;67:1048–59. doi: 10.1002/prot.21370. [DOI] [PubMed] [Google Scholar]
- 68.Chow JP, Siu WY, Ho HT, Ma KH, Ho CC, Poon RY. Differential contribution of inhibitory phosphorylation of CDC2 and CDK2 for unperturbed cell cycle control and DNA integrity checkpoints. J Biol Chem. 2003;278:40815–28. doi: 10.1074/jbc.M306683200. [DOI] [PubMed] [Google Scholar]
- 69.Keaton MA, Bardes ES, Marquitz AR, Freel CD, Zyla TR, Rudolph J, Lew DJ. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr Biol. 2007;17:1181–9. doi: 10.1016/j.cub.2007.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–5. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 71.Wittenberg C, Reed SI. Conservation of function and regulation within the Cdc28/cdc2 protein kinase family: characterization of the human Cdc2Hs protein kinase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:4064–8. doi: 10.1128/mcb.9.9.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–82. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]