Abstract
To better understand the molecular pathogenesis of neuroendocrine tumors (NET), we investigated the molecular and clinical characteristics of malignant poorly differentiated colorectal NET and compared these findings with sporadic CRC and well-differentiated benign and malignant fore-/midgut NET. Tumors were analyzed and correlated for microsatellite instability (MSI) and the CpG island methylator phenotype (CIMP). NET were scored for proliferation using Ki-67. A total of 34 malignant poorly differentiated colorectal NET, 38 well-differentiated benign and malignant fore-/midgut-NET and 150 sporadic colorectal cancers (CRC) with known MSI status were investigated. Among the sporadic CRC, CIMP was significantly correlated with MSI-high (MSI-H) (p < 0.001). Of the 34 colorectal NET, 0/1 of the MSI-H, 3/5 (60%) of the MSI-L and 13/19 (68%) of the MSS tumors were CIMP+ (p = 0.17). Of the fore-/midgut-NET, none was MSI-H. 20/34 (59%) colorectal NET vs. 11/38 (29%) fore-/midgut-NET were CIMP+ (p = 0.01). The Ki-67 index was significantly higher in poorly differentiated colorectal NET compared to the less malignant fore-/midgut-NET (p < 0.0001). Besides the location in the colon, Ki-67 predicted poor outcome in NET (p < 0.0001). CIMP status did not affect survival. In NET, p16 methylation predicted a poor outcome (p = 0.0004). We conclude that molecular pathogenesis in sporadic CRC and poorly differentiated colorectal NET is different despite some similarities. Main differences between malignant well-differentiated and poorly differentiated NET are the Ki-67 proliferation rate and differential methylation in tumor-associated genes. Predictors of a poor outcome in patients with NET are poor differentiation, a high Ki-67 index and p16 methylation.
Keywords: neuroendocrine tumors, fore-, and midgut tumors, hindgut tumors, microsatellite instability, loss of heterozygosity, CpG island methylation, promoter methylation
Gastroenteropancreatic neuroendocrine tumors (GEP-NET) represent heterogeneous tumors. The growth pattern ranges from very slow to fast growing aggressive types of tumors. Within these subgroups, the biological and clinical characteristics of the tumors may vary considerably.1 The classification currently used is the revised, second edition of the “WHO classification of the endocrine tumors of the gastroenteropancreatic tract.”2
It is well-known that tumor growth is the result of an uncontrolled cell proliferation and a defective cell death program.3 Deregulation of this balance results in tumor progression, resistance to therapy and poor prognosis. Genomic instability is a key mechanistic component of cancer progression.4,5 Three major mechanisms that increase the diversity of gene expression have been identified in most tumor types including colorectal cancer (CRC): microsatellite instability (MSI), chromosomal instability (CIN) and the CpG island methylator phenotype (CIMP).4,6–11 Current data indicate that CIMP is an important mechanism of gene inactivation in human carcinogenesis, and it has been shown that a number of tumor suppressor genes, including p16, p14, MGMT and hMLH1 are silenced by promoter methylation in CRC and many other tumor types.10–12
Several studies have been aimed to identify more precise predictors of prognosis of patients with GEP-NET. The current WHO classification attempted to define a more effective approach by introducing the concepts of cell differentiation and site-specific malignancy as well as specific criteria for carcinoma definition.2 WHO clinicopathological correlations embed the following prognostic features: degree of cell differentiation, angioinvasion, proliferation fraction as assessed by mitotic index and Ki-67, size and functional activity. NET are categorized into low or high proliferating tumors by the Ki-67 index. Following the WHO classification, NET in this study were categorized by the Ki-67 status into the subgroups with <2%, 2–19% and ≥20% positive cells. Other prognostic variables have been identified, most of which related to specific biological features of neuroendocrine cancer cells. Nonetheless, liver or distant metastases ultimately determine patients' survival and/or response to therapy. A recent proposal of tumor grading and tumor, nodes and metastases (TNM) staging aims at a simple and practical patients' stratification.13,14 In addition, recent studies addressed the prognostic value of immunohistochemical markers for a number of tumor-associated genes as well as clinical parameters such as age, gender and the functional activity in NET and its metastases. Only synaptophysin, cytokeratin-8 and Ki-67 had some though limited prognostic value.15 Age above 60 was the only clinical parameter of unfavorable prognostic significance. Another study tested Ki-67, expression of p53 and bcl-2 in patients with GEP-NET.16 Ki-67 proliferation rate and p53 were found to complement histological grading as prognostic indicators for metastatic disease, whereas bcl-2 appeared to be less useful. Recently, the role of nuclear survivin on tumor progression has been investigated in NET.17 Expression appeared to be upregulated during progression of GEP-NET. The analysis of nuclear survivin expression identified subgroups in metastatic disease (WHO class 2) with good or less favorable prognosis. It was proposed that the determination of nuclear survivin expression may be used to individualize therapeutic strategies.
In well-differentiated GEP-NET, methylation of cancer-associated genes has recently been shown.18,19 However, it has not been demonstrated that methylation of specific genes had a prognostic relevance. In patients with well-differentiated fore-/midgut NET, CIMP has been proposed as a prognostic marker, similar to a high Ki-67 proliferation index.18 The role of allelic losses has not been addressed in former studies studying well-differentiated and poorly differentiated NET.
In previous studies, NET from various origins, differentiation, tumor grade and functional activity had been studied but no clear separation between these variables had been made, mostly because of low case numbers. Our aim was to compare molecular characteristics of well-differentiated and poorly differentiated NET. For this reason, we selected a group of mostly malignant and well-differentiated NET from the fore- and midgut and of malignant mainly poorly differentiated NET from the colon and rectum. Poorly differentiated NET from the fore/midgut and well-differentiated hindgut tumors were not available at our institution. On the basis of these data, our aim was to correlate MSI, LOH, CIMP and the methylation patterns of well-differentiated NET (n = 38) with poorly differentiated NET (n = 34) in terms of tumor progression and survival. We furthermore compared mechanisms of genomic instability of poorly differentiated colonic NET with that of sporadic nonendocrine CRC.
Material and methods
Tumor tissues
Paraffin-embedded formalin-fixed tissue obtained by biopsy or surgical resection was available from 38 well-differentiated, mainly malignant fore- and midgut (WDEC, WHO class 1 and 2) and from 34 poorly differentiated colonic GEP-NET, respectively. Well-differentiated NET from the colon and rectum were not included into the study to be able to make a clear comparison between 2 cohorts of well-differentiated and poorly differentiated NET. Only sporadic GEP-NET were investigated. The MEN1-syndrome was excluded by a careful study of the personal and family history, and in cases of pancreatic NET by the chemical analysis for serum calcium and prolactin. Malignancy was scored according to WHO guidelines.2 In detail, poorly differentiated tumors with either metastases, invasion of the muscularis propria, angioinvasion and any tumor size were scored as malignant. Well-differentiated tumors with a tumor size of >2 cm (>3 cm for pancreatic endocrine tumors) and a Ki-67 proliferation index >2% were considered to be malignant similar to those with metastases or invasion of the muscularis propria. The fore-/midgut NET were either benign or low grade malignant [well-differentiated endocrine tumors (WDET) and well-differentiated endocrine cancers (WDEC)]. Patients' characteristics of fore-/midgut NET are shown in detail in Table I. All patients with NET from the colon and rectum were malignant and mainly poorly differentiated [WDEC and poorly differentiated endocrine cancers (PDEC), WHO class 2 and 3, respectively]. Malignant NET from the colon and rectum originated from the midpart of the transverse colon to the rectum. Patients' characteristics are also shown in detail in Table II. Follow-up data were available for most NET from time of diagnosis. In addition, 150 primary colon cancers were studied which were obtained from patients with sporadic CRC. They were selected by their MSI status, which had been investigated by standard PCR methods previously.6 Follow-up could not be obtained for these tumors.
TABLE I. Characteristics of Patients with Fore-/Midgut Net According to Functional Activity, Site and KI-67 Proliferation Index.
| Tumor no. | Diagnosis | Origin | Behavior | Ki-67 (%) | CIMP | MSI (marker) | LOH (marker) | Follow-up time (weeks) | Survival | Gender | Age at diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | Insulinoma | Pancreas | Malignant | <2 | − | MSS | − | 12 | tu | m | 79 |
| 14 | Insulinoma | Pancreas | Benign | <2 | − | MSS | − | 192 | Survived | f | 52 |
| 16 | Insulinoma | Pancreas | Benign | <2 | + | MSS | − | 60 | Survived | f | 77 |
| 19 | Insulinoma | Pancreas | Malignant | 2–20 | − | MSS | + (D5S346) | 12 | tu | f | 79 |
| 12 | Gastrinoma | Pancreas | Benign | 2–20 | − | MSS | na | 36 | tu | m | 81 |
| 2 | Vipoma | Pancreas | Malignant | 2–20 | − | MSS | − | 60 | Survived | m | 56 |
| 17 | Vipoma | Pancreas | Benign | 2–20 | + | MSS | − | 76 | Survived | m | 56 |
| 1 | Glucagonoma | Pancreas | Malignant | 2–20 | − | MSS | − | 8 | tu | f | 50 |
| 5 | Nonfunctional | Pancreas | Malignant | <2 | + | MSS | na | 84 | Survived | m | 61 |
| 4 | Nonfunctional | Pancreas | Malignant | 2–20 | − | MSS | na | 72 | Survived | f | 36 |
| 28 | Nonfunctional | Duodenum | Malignant | <2 | + | MSS | − | 96 | Survived | m | 71 |
| 34 | Nonfunctional | Duodenum | Malignant | <2 | − | MSI-L | na | 98 | Survived | f | 81 |
| 37 | Nonfunctional | Duodenum | Malignant | <2 | + | MSS | − | 96 | Survived | m | 70 |
| 9 | Nonfunctional | Duodenum | Malignant | 2–20 | − | MSS | na | 12 | tu | m | 78 |
| 30 | Nonfunctional | Duodenum | Malignant | 2–20 | − | MSS | na | 35 | tu | m | 70 |
| 6 | Gastrinoma | Duodenum | Malignant | <2 | − | MSI-L | na | 10 | tu | f | 49 |
| 15 | Gastrinoma | Duodenum | Benign | <2 | − | MSS | − | 192 | Survived | m | 48 |
| 18 | Gastrinoma | Duodenum | Benign | <2 | − | MSS | − | 50 | Survived | f | 49 |
| 3 | Gastrinoma | Duodenum | Benign | 2–20 | − | MSS | + (D2S123) | 48 | Survived | m | 66 |
| 23 | Carcinoid syndrome | Ileum | Malignant | <2 | − | MSS | − | 84 | Survived | m | 62 |
| 24 | Carcinoid syndrome | Ileum | Malignant | <2 | + | MSS | − | 72 | Survived | m | 66 |
| 25 | Carcinoid syndrome | Ileum | Malignant | <2 | − | MSS | − | 72 | Survived | m | 70 |
| 32 | Carcinoid syndrome | Ileum | Malignant | <2 | + | MSS | − | 120 | Survived | m | 63 |
| 35 | Carcinoid syndrome | Ileum | Malignant | <2 | + | MSS | − | 36 | tu | f | 56 |
| 38 | Carcinoid syndrome | Ileum | Malignant | <2 | − | MSS | na | 144 | Survived | f | 60 |
| 33 | Carcinoid syndrome | Jejunum | Malignant | <2 | − | MSS | − | 120 | Survived | m | 75 |
| 36 | Carcinoid syndrome | Jejunum | Malignant | <2 | − | MSS | na | 60 | Survived | m | 69 |
| 13 | Nonfunctional | Ileum | Malignant | <2 | + | MSS | − | 36 | tu | f | 81 |
| 21 | Nonfunctional | Ileum | Malignant | <2 | − | MSS | na | 84 | Survived | f | 61 |
| 22 | Nonfunctional | Ileum | Malignant | <2 | + | MSS | na | 84 | Survived | f | 61 |
| 20 | Nonfunctional | Ileum | Malignant | 2–20 | + | MSS | + (D17S250) | 36 | tu | m | 66 |
| 29 | Nonfunctional | Ileum | Malignant | 2–20 | − | MSI-L | na | 35 | tu | f | 38 |
| 31 | Nonfunctional | Ileum | Malignant | 2–20 | − | MSS | + (D17S250) | 24 | tu | m | 70 |
| 10 | Nonfunctional | Jejunum | Malignant | <2 | − | MSS | na | 12 | tu | m | 71 |
| 26 | Nonfunctional | Jejunum | Malignant | <2 | − | MSS | − | 84 | Survived | f | 49 |
| 27 | Nonfunctional | Jejunum | Malignant | <2 | + | MSS | − | 120 | Survived | m | 51 |
| 7 | Nonfunctional | Jejunum | Malignant | 2–20 | − | MSS | − | 10 | tu | m | 65 |
| 11 | Nonfunctional | Jejunum | Malignant | 2–20 | − | MSI-L | na | 12 | tu | f | 38 |
na, not available; tu, died from the tumor; f, female; m, male.
TABLE II. Characteristics of Patients with Net of the Colon and RECTUM According to KI-67 Proliferation Index.
| Tumor no. | Origin | Behavior | Ki-67 | CIMP | LOH (marker) | MSI (marker) | Follow-up time (weeks) | Survival | Gender | Age at diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|
| 17 | Colon | Malignant | <2 | − | − | MSI-H (BAT25, D17S250) | na | na | f | 70 |
| 14 | Colon | Malignant | 3 | + | + (D11S913) | MSI-L | 8 | Survived | f | 62 |
| 12 | Colon | Malignant | 5 | + | + (D2S123, PYGM, RH-93814) | MSS | 27 | Survived | f | 77 |
| 13 | Colon | Malignant | 5 | + | + (D17S250, PYGM) | MSI-L | 8 | tu | f | 54 |
| 18 | Colon | Malignant | 20 | + | + D11S913 | MSS | na | na | m | 70 |
| 23 | Colon | Malignant | 30 | + | + (D11S913) | MSS | 19 | tu | m | 32 |
| 11 | Colon | Malignant | 40 | − | − | MSI-L | 18 | Survived | m | 67 |
| 32 | Colon | Malignant | 45 | − | na | MSS | 62 | Survived | m | 54 |
| 33 | Colon | Malignant | 45 | + | − | Na | 46 | tu | f | 58 |
| 9 | Colon | Malignant | 50 | + | + (RH-93814) | Na | na | na | na | na |
| 10 | Colon | Malignant | 50 | + | − | Na | 15 | Survived | m | 71 |
| 16 | Colon | Malignant | 50 | + | − | MSS | na | na | f | 57 |
| 19 | Colon | Malignant | 50 | − | − | MSS | na | na | na | na |
| 21 | Colon | Malignant | 50 | + | − | MSS | 13 | tu | m | 69 |
| 27 | Colon | Malignant | 50 | + | − | MSS | 14 | tu | m | 32 |
| 5 | Colon | Malignant | 50 | + | − | Na | na | na | na | na |
| 6 | Colon | Malignant | 60 | − | − | Na | na | na | na | na |
| 25 | Colon | Malignant | 60 | + | − | MSS | 0 | tu | f | 63 |
| 8 | Colon | Malignant | 60 | + | − | MSS | 99 | Survived | m | 74 |
| 1 | Colon | Malignant | 65 | + | − | MSI-L | na | Na | na | na |
| 34 | Colon | Malignant | 65 | − | na | Na | 38 | Survived | m | 56 |
| 2 | Colon | Malignant | 70 | + | − | MSS | na | Na | na | na |
| 29 | Colon | Malignant | 75 | + | na | MSS | 2 | Tu | m | 64 |
| 30 | Colon | Malignant | 75 | − | na | MSS | 1 | Tu | f | 92 |
| 7 | Colon | Malignant | 80 | − | − | MSI-L | na | Na | na | na |
| 15 | Colon | Malignant | 80 | + | − | MSS | 2 | Tu | m | 53 |
| 3 | Colon | Malignant | 85 | − | na | na | na | na | na | na |
| 31 | Colon | Malignant | 85 | − | na | MSS | 68 | Survived | f | 58 |
| 4 | Colon | Malignant | 90 | − | na | na | na | na | na | na |
| 22 | Colon | Malignant | 90 | + | + (D5S346, D11S913) | MSS | 57 | tu | m | 49 |
| 28 | Rectum | Malignant | <2 | − | − | MSS | 6 | tu | m | 55 |
| 24 | Rectum | Malignant | 50 | + | − | MSS | 13 | tu | m | 29 |
| 26 | Rectum | Malignant | 50 | − | − | MSI-L | 7 | tu | m | 57 |
| 20 | Rectum | Malignant | 90 | − | + (D11S913) | MSS | 8 | tu | f | 81 |
na, not available; tu, died from the tumor; f, female; m, male.
Microdissection and DNA amplification
Serial sections from formalin-fixed paraffin-embedded matched normal and neoplastic primary tissues (5 μm) were stained with H&E, and representative normal and tumor regions were identified by microscopic examination. Normal control tissue (nontumor) was obtained from adjacent histological normal mucosa and/or normal lymph nodes.
Genomic DNA was isolated from the paraffin-embedded microdomains and removed from the slides by deparaffinization with multiple xylene washes. Subsequently, the tissues were hydrated, digested in Proteinase K and followed by DNA extraction using the QIAamp DNA mini kit (Qiagen, Valencia, CA), according to the manufacturer's instructions.
Loss of heterozygosity on chromosomes 2p16, 5q21 and 11q13
Sets of polymorphic microsatellite sequences that are tightly linked to known TSG and DNA mismatch repair (MMR) genes were used to identify significant allelic losses in the tumors.9 DNA was amplified by PCR using 32P-end-labeled primers at microsatellite loci linked to the hMSH2 locus on 2p16 (D2S123), APC locus on 5q21 (D5S346) and MEN1 locus on 11q13 (D11S913, PYGM, RH-93814) and D17S250. LOH was confirmed if a tumor allele showed at least a 50% reduction in the relative intensity of 1 allele in neoplastic tissue compared with the matched normal DNA. LOH was defined as allelic loss in at least 1 of 6 informative markers investigated. LOH analysis was performed for all NET.
MSI analysis
Microsatellite analysis of all matched normal and tumor tissues was performed by PCR amplification using a panel of 5 NCI-workshop recommended markers that included 2 mononucleotide (BAT25 and BAT26) and 3 dinucleotide repeat sequences (D2S123, D5S346 and D17S250).20 PCR was performed using 32P-labeled primers and subsequent electrophoresis on 8% polyacrylamide gels as described previously.18 Changes in the electrophoretic mobility of DNA amplified by PCR were used to assess MSI. Tumors showing a shift in at least 2 of the 5 markers were classified as MSI-H. MSI-L was defined as a shift in only 1 of the 5 markers. Tumors that did not show any allelic shifts were classified as microsatellite stable (MSS). MSI analysis was performed for CRC and NET.
Sodium bisulfite modification and methylation-specific PCR assays
The methylation status of DNA from human NET was determined by methylation-specific PCR (MSP). The specific primers for the methylated and unmethylated MSP for the hMLH1 (promoter region C), p16, APC, O6-MGMT, PTEN, HIC1, RASSF1A, TIMP3, MEN1 and RUNX3 genes were used as published.9,18
MSP was also performed on bisulfite-modified DNA templates obtained from human CRC to study the methylation status of 12 methylation targets. Among these, 9 methylation markers mapped to promoter regions of genes including hMLH1, APC, p16INK4a, p14ARF, TIMP3, RUNX3, HIC1, PTEN and RARβ2, and the remaining 3 markers amplified MINT (methylated in tumor) loci: MINT1, MINT2 and MINT31. All these markers were recently shown to be closely correlated with CIMP in CRC. The primer sequences, PCR conditions and product sizes for each of the methylation markers analyzed, and the specificity of the MSP assays have been described previously.6
Genomic DNA obtained from paraffin-embedded tissue sections was bisulfite modified to convert all unmethylated cytosine residues to uracil for subsequent detection of methylated cytosines using methylation specific primers. MSP assays were performed on the bisulfite-modified DNA using 2 sets of primers specific for amplification of methylated and unmethylated alleles as described previously.18 Briefly, 0.5–2.0 μg of genomic DNA were denatured with NaOH, treated with sodium bisulfite, and subsequently purified using the Wizard DNA Clean-up System (Promega, Madison, WI). PCR reactions were performed in a 25-μl reaction volume containing 1× PCR buffer (Invitrogen Life Technologies, Carlsbad, CA), 2.5 mM MgCl2, 200 μM dNTPs, 0.5 μM of each PCR primer, 0.75 U of AmpliTaq polymerase, and ∼25 ng of bisulfite-modified DNA. Reactions were hot-started at 95°C for 5 min. This was followed by 35–40 cycles at 95°C for 45 sec, 57–60°C for 30 sec and 72°C for 30 sec, followed by a 10-min extension at 72°C in a PTC 200 DNA Engine™ Thermocyler (MJ Research, Waltham, MA). The amplification products were separated on a 3% agarose gel and visualized by ethidium bromide staining and UV transillumination.
Human placental DNA (Sigma Chemical, St. Louis, MO) treated in vitro with SssI methylase (New England Biolabs, Beverly, MA) was used as a positive control for MSP of methylated alleles, whereas DNA from normal lymphocytes was used as a control for unmethylated alleles. Water was used as a negative PCR control to monitor for contamination.
Immunohistochemical detection of Ki-67
A standard avidin–biotin complex peroxidase method, using 3,30-diaminobenzidine as chromogen, and a monoclonal antibody (MIB-1; Dianova, Hamburg, Germany) were used for staining nuclear Ki-67. Paraffin sections were heated for 3 5-min periods, deparaffinized, rehydrated and immersed in 10 mmol/l sodium citrate buffer (pH 6.0). The primary antibody was diluted 1:30. The number of Ki-67 positive cells was assessed as percentage of about 2,000 tumor cells in areas of highest nuclear labeling (magnification 40×). A low Ki-67 index was defined as <2% positive cells, an intermediate index as 2–19% positive cells and a high proliferation index as ≥20% positive cells according to recent guidelines from the European Neuroendocrine Tumor Society (ENET) recommendations.13,14 Evaluation was performed independently by 2 investigators (CNA und IS). In case of disagreement, further evaluation was performed by AS-G.
Data analysis
The Fisher's exact test and the χ2 -test were used as appropriate to test the associations between tumor subgroups. Univariate associations of baseline characteristics of the tumor subgroups and prognostic variables were assessed using the Fisher's exact test or χ2 -test. Differences in the frequency of CIMP positive tumors and Ki-67 proliferation index between each subgroup were also analyzed with the χ2 test. Survival analyses were performed by the Kaplan-Meyer method. The risk ratio of methylation for survival was calculated using the effect likelihood test. All reported p-values are 2-sided and a p < 0.05 was considered statistically significant.
Results
Microsatellite instability in NET
Informative results were obtained for 25/34 NET from the colon and rectum and 34 of 38 fore- and midgut NET. Clinicopathological and genetic information are shown in Tables I and II. In all noninformative cases, nonneoplastic tissue was not available for comparison. Of the fore- and midgut NET, none was MSI-H, 4/38 (11%) were MSI-L and 30/38 (89%) were MSS. Of the NET from the colon and rectum, 1/26 (4%) was MSI-H (informative markers BAT25 and D17S250), 6/26 (23%) were MSI-L (informative markers BAT25 in 3 tumors, BAT26, D5S123 and D17S250 in 1 tumor, respectively) and 19 (73%) were MSS. MSI status was not different in fore-/midgut NET versus. NET from the colon and rectum (p = 0.12).
LOH in fore-/midgut NET versus NET of the colon and rectum
LOH is a common feature in sporadic CRC and NET of various origins.4,21,22 Of the 38 fore-/midgut NET LOH was seen in 4/24 informative tumors (17%) (tu # 3 D2S123; tu # 19 D5S346; tu # 20 D17S250; tu # 31 D17S250) (Table I). Of the 34 NET from the colon and rectum, LOH was found in 8/27 (30%) informative tumors (tu # 9 RH-93814; tu # 12 D2S123, PYGM, RH-93814; tu # 13 D17S250, PYGM; tu # 14 D11S913; tu # 18 D11S913; tu # 20 D11S913; tu # 22 D5S346, D11S913; tu # 23 D11S913) (Table II) (p = 0.02). The noninformative tumors were those without corresponding normal tissue. None of the LOH tumors were MSI-H.
CIMP and correlation with MSI in NET and CRC
In direct comparison, CIMP was much more frequent in poorly differentiated NET of the colon and rectum (20/34, 59%) than in well-differentiated fore-/midgut NET (11/38, 29%) (p = 0.01) (Table III). Comparing CRC subgrouped by MSI status, MSS NET from the colon and rectum were significantly more often CIMP positive than sporadic CRC (p = 0.0003) (Fig. 1a). Among all tumors, 24/94 (26%) MSS-, 10/43 (23%) MSI-L and 10/13 (77%) MSI-H sporadic CRC were CIMP positive (p < 0.0001), whereas 13/19 MSS informative NET from the colon and rectum (68%) and 3/6 (60%) MSI-L NET from the colon and rectum were CIMP positive (p = 0.17). Interestingly, the only MSI-H colon NET was CIMP negative, whereas CIMP was a predominant feature in MSI-H sporadic CRC.
TABLE III. Genomic Instability and Ki-67 Status in Colon/Rectum Net and Fore-/Midgut Net.
| Parameter | Colon and rectum NET | Fore-/midgut NET | p-value | |
|---|---|---|---|---|
| Age | Mean, SD | 61.2 (15.6) | 62.7 (12.6) | 0.691 |
| Gender | Female | 11 (41%) | 16 (42%) | 0.912 |
| No. of samples (%) | Male | 16 (59%) | 22 (58%) | |
| CIMP status | Positive (n = 31) | 20 (59%) | 11 (29%) | 0.012 |
| No. of samples (%) | Negative (n = 41) | 14 (41%) | 27 (71%) | |
| LOH status | Positive (n = 12) | 8 (30%) | 4 (17%) | 0.282 |
| No. of samples (%) | Negative (n = 39) | 19 (70%) | 20 (83%) | |
| MSI status | MSI-H (n = 1) | 1 (4%) | 0 (0%) | 0.172 |
| No. of samples (%) | MSI-L (n = 11) | 6 (23%) | 4 (11%) | |
| MSS (n = 52) | 19 (73%) | 34 (89%) | ||
| Ki-67 IHC score status | <2% (n = 16) | 2 (11%) | 14 (89%) | <0.00012 |
| No. of samples (%) | 2–19% (n = 27) | 3 (8%) | 24 (92%) | |
| >20% (n = 28) | 28 (100%) | 0 (0%) | ||
p-value is based on ANOVA.
p-values are based on X2 test.
Figure 1.
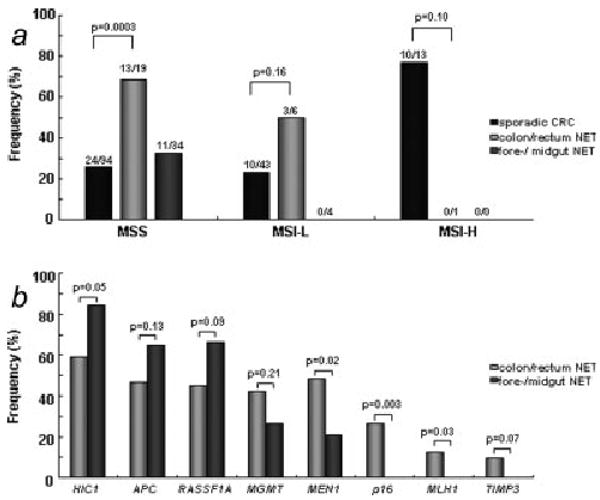
(a) CIMP in sporadic CRC, poorly differentiated NET from the colon and rectum and well differentiated fore-/midgut NET. Tumors are sorted by their MSI status. (b) Methylation rates in well differentiated fore-/midgut NET and poorly differentiated NET from the colon and rectum.
Distribution of NET by Ki-67 proliferation rate and CIMP
According to their poor differentiation, NET of the colon and rectum (29/34, 85%) revealed significantly more often a high-proliferation index ≥ 20% than the well-differentiated fore-/midgut NET (p < 0.0001) (Table III). Fore-/midgut NET had significantly more often a proliferation index of <2% (24/38, 63%) and 2–19% (14/38, 37%) than NET from the colon and rectum (2/34, 6% and 3/34, 9%, respectively) (Table III) (p < 0.0001).
Upon further subgroup analysis, we categorized NET by their proliferation index and CIMP status. We found that fore-/midgut NET did not differ significantly in CIMP status (Ki-67 <2% 13/23, 57%; Ki-67 2-19% 9/14, 64%) but in NET from the colon and rectum CIMP impressively correlated with a high Ki-67 index (Ki-67 2-19% 4/4, 100%; Ki-67 ≥ 20% 16/28, 57%) (p < 0.0001).
Difference in promoter methylation in fore-/midgut versus NET from the colon and rectum
Among 8 analyzed loci, HIC1, APC and RASSF1A were more often methylated in fore-/midgut-NET, whereas MEN1, MGMT, p16, hMLH1 and TIMP3 were more frequently methylated in NET from the colon and rectum (Tables IV and V, Fig. 1b). HIC1 was significantly more methylated in fore-/midgut NET (p = 0.05), whereas MEN1 (p = 0.02), p16 (p = 0.003) and hMLH1 (p = 0.03) were significantly methylated in NET from the colon and rectum. Interestingly, promoter methylation in p16, hMLH1 and TIMP3 were only detected in NET from the colon and rectum.
TABLE IV. Methylation Profile of Fore-/Midgut Net according to Site.
| Tumor no. | Diagnosis | hMLH1 | p16 | APC | MGMT | MEN1 | HIC1 | RASSF1a | TIMP3 |
|---|---|---|---|---|---|---|---|---|---|
| 3 | Gastrinoma | − | − | + | − | − | na | − | − |
| 6 | Gastrinoma | − | − | − | na | − | + | + | − |
| 12 | Gastrinoma | − | na | − | − | − | na | + | − |
| 15 | Gastrinoma | − | na | na | na | na | na | na | na |
| 18 | Gastrinoma | na | − | − | na | − | na | na | na |
| 8 | Insulinoma | − | − | − | + | − | na | + | − |
| 14 | Insulinoma | − | na | + | na | na | na | − | − |
| 16 | Insulinoma | − | − | + | + | − | + | + | − |
| 19 | Insulinoma | − | na | − | + | − | − | + | − |
| 1 | Glucagonoma | − | 2 | + | − | − | + | + | − |
| 2 | Vipoma | − | − | + | − | − | na | na | − |
| 17 | Vipoma | − | − | + | + | + | + | + | − |
| 23 | Carcinoid syndrome | − | − | + | − | − | − | − | − |
| 24 | Carcinoid syndrome | − | − | + | + | − | + | + | − |
| 25 | Carcinoid syndrome | − | − | − | − | − | + | + | − |
| 32 | Carcinoid syndrome | − | − | + | na | + | − | − | − |
| 33 | Carcinoid syndrome | − | − | − | − | − | + | + | − |
| 35 | Carcinoid syndrome | − | − | + | + | + | + | + | − |
| 36 | Carcinoid syndrome | − | na | − | − | − | na | − | − |
| 38 | Carcinoid syndrome | na | na | + | na | na | na | na | na |
| 4 | Nonfunctional pancreas | − | − | + | − | − | + | − | − |
| 5 | Nonfunctional pancreas | − | − | + | − | − | + | + | − |
| 7 | Nonfunctional jejunum | − | − | + | − | − | na | + | na |
| 9 | Nonfunctional duodenum | − | − | − | − | − | + | + | − |
| 28 | Nonfunctional duodenum | − | − | + | − | − | + | + | − |
| 30 | Nonfunctional duodenum | − | na | + | na | na | + | na | − |
| 34 | Nonfunctional duodenum | − | − | − | − | − | na | na | − |
| 37 | Nonfunctional duodenum | − | na | + | − | + | na | − | na |
| 10 | Nonfunctional jejunum | − | − | − | − | − | + | − | − |
| 11 | Nonfunctional jejunum | − | na | + | na | na | + | na | − |
| 26 | Nonfunctional jejunum | − | − | + | − | − | + | na | − |
| 27 | Nonfunctional jejunum | − | − | + | − | + | na | + | − |
| 13 | Nonfunctional ileum | − | − | + | + | + | + | + | − |
| 20 | Nonfunctional ileum | − | − | + | + | − | + | + | − |
| 21 | Nonfunctional ileum | − | − | − | − | − | + | + | − |
| 22 | Nonfunctional ileum | − | − | + | − | + | − | + | − |
| 29 | Nonfunctional ileum | − | − | − | − | − | + | − | − |
| 31 | Nonfunctional ileum | − | − | + | − | − | + | − | − |
Na, not available; +, methylated; −, not methylated.
TABLE V. Methylation Profile of Colon and Rectum Net According to Tumor Location.
| Tumor no. | Diagnosis | p16 | HIC 1 | APC | hMLH1 | RASSF1A | MEN 1 | TIMP3 | MGMT |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Colon | + | − | − | − | + | − | − | + |
| 2 | Colon | Na | + | + | − | + | − | + | − |
| 3 | Colon | − | − | − | − | − | − | − | − |
| 4 | Colon | − | na | + | − | na | − | − | na |
| 5 | Colon | − | + | − | − | − | + | − | + |
| 6 | Colon | − | − | − | − | − | − | − | − |
| 7 | Colon | na | na | − | − | − | − | − | − |
| 8 | Colon | na | + | + | + | − | − | − | − |
| 9 | Colon | − | + | + | − | + | − | − | − |
| 10 | Colon | + | − | + | − | − | − | − | + |
| 11 | Colon | − | − | + | − | + | − | − | − |
| 12 | Colon | − | + | + | − | − | + | − | + |
| 13 | Colon | + | + | + | − | − | + | − | − |
| 14 | Colon | − | + | − | − | + | + | − | − |
| 15 | Colon | − | − | − | − | + | + | − | + |
| 16 | Colon | − | + | − | − | − | + | − | + |
| 17 | Colon | − | na | − | − | na | + | − | − |
| 18 | Colon | + | + | − | − | − | + | − | + |
| 19 | Colon | − | − | na | − | − | + | − | − |
| 21 | Colon | − | + | + | − | + | + | − | − |
| 22 | Colon | − | − | + | − | + | + | − | + |
| 23 | Colon | − | − | + | − | − | + | − | + |
| 25 | Colon | + | + | − | + | + | − | − | − |
| 27 | Colon | + | + | − | − | + | + | − | − |
| 29 | Colon | + | + | + | − | − | − | − | + |
| 30 | Colon | − | na | − | − | − | na | − | − |
| 31 | Colon | na | na | − | na | na | na | − | − |
| 32 | Colon | − | na | na | na | na | − | na | na |
| 33 | Colon | − | + | + | + | + | + | + | + |
| 34 | Colon | − | − | − | + | − | − | − | + |
| 20 | Rectum | − | + | − | − | + | − | − | − |
| 24 | Rectum | − | + | + | − | + | + | + | + |
| 26 | Rectum | − | na | + | − | na | na | na | na |
| 28 | Rectum | + | − | − | − | − | − | − | − |
na, not available; +, methylated; −, not methylated.
The risk ratio of methylation for survival in all NET was calculated by proportional hazards analysis (Table VI). We did not find correlation with any of the investigated genes with the exception of p16. Methylation of p16 clearly associated with a poor survival and was only found in poorly differentiated NET from the colon and rectum and not in any case of the fore-/midgut NET (p = 0.01).
TABLE VI. Risk Ratio of Methylation for Survival.
| Gene | Risk ratio (confidence interval) | p-value |
|---|---|---|
| HIC1 | 1.46 (0.44–6.11) | 0.55 |
| MEN1 | 2.06 (0.72–5.99) | 0.18 |
| APC | 0.40 (0.11–1.40) | 0.15 |
| RASSF1A | 0.51 (0.16–1.66) | 0.25 |
| MGMT | 1.32 (0.38–5.15) | 0.66 |
| p16 | 5.21 (1.43–18.0) | 0.01 |
| hMLH1 | 0.58 (0.06–5.26) | 0.63 |
| TIMP3 | 3.00 (0.22–23.4) | 0.38 |
p-values were based on Effect Likelihood Tests.
Outcome by Ki-67 proliferation rate, CIMP and p16 methylation
Outcome data was available for most patients with NET. Unfortunately, follow-up was not available for patients with CRC. By this, direct outcome comparison between NET from the colon and rectum and sporadic CRC could not be performed in this study.
Our analyses demonstrated, however, that patients with poorly differentiated NET from the colon and rectum had a poorer outcome than patients with better differentiated fore-/midgut NET (p = 0.0045) (Fig. 2a). A three-year survival was 60% in patients with fore-/midgut NET as compared to 30% in patients with NET from the colon and rectum. The strongest predictor of survival was the Ki-67 proliferation index. Patients with NET with Ki-67 <2% had a significant longer survival than patients with a Ki-67 index of ≥ 2-19% or ≥ 20%, respectively (p < 0.0001) (Fig. 2b). After 2 years follow-up, 80% of the patients with Ki-67 <2% but only 30% of the patients with a Ki-67 index of ≥ 2–19% were alive (p = 0.001). Despite 5 patients with NET from the colon and rectum had a Ki-67 index of <5 % survival of these patients did not differ from those with a higher Ki-67 index (p = 0.26) (Fig. 2b). We have shown that CIMP was more frequent in poorly differentiated NET (colon and rectum) than in well-differentiated NET (fore-/midgut). However, CIMP was not a predictor for survival neither in patients with NET from the colon and rectum (p = 0.55) nor in patients with fore-/midgut NET (p = 0.66). It was further investigated if there was any influence in outcome by any of the molecular markers investigated. Interestingly, p16 methylation was a strong predictor of survival in all NET (p = 0.0004). In NET from the colon and rectum, we detected a trend toward worse outcome (p = 0.08) (Fig. 2c). Fore-/midgut NET were not methylated in p16.
Figure 2.
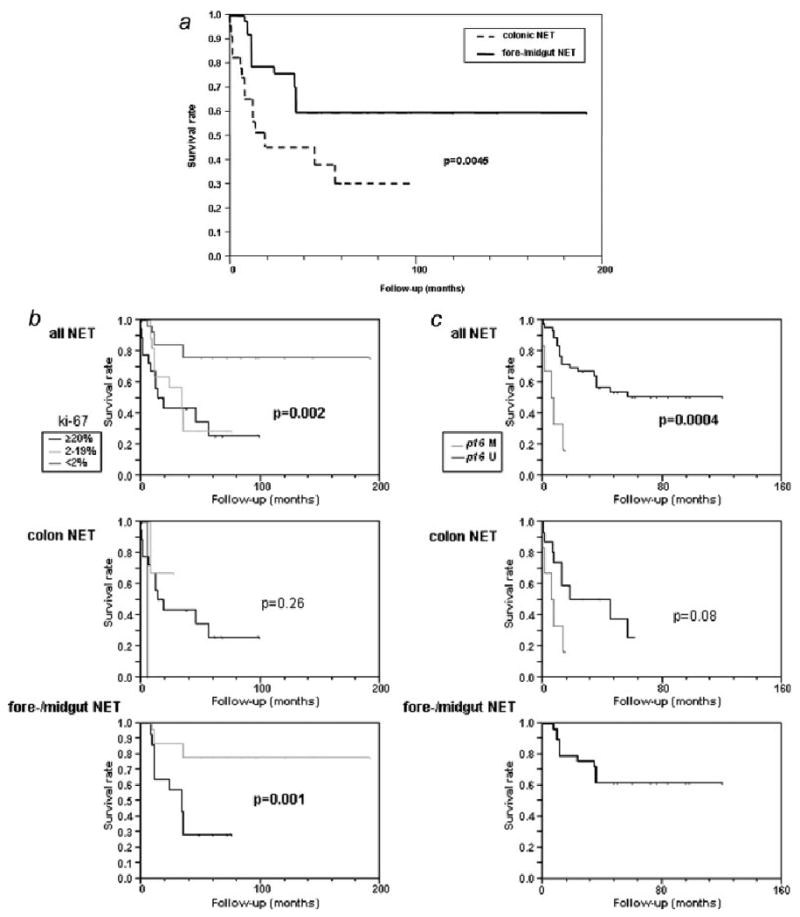
(a) Kaplan Meyer curves for survival in well differentiated fore-/midgut NET and poorly differentiated NET from the colon and rectum. (b) Kaplan Meyer survival analysis by Ki-67 proliferation rate in NET. (c) Kaplan Meyer survival analysis by p16 methylation in NET.
Discussion
In a previous study, we investigated the contribution of genetic and epigenetic alterations in molecular pathogenesis of sporadic well-differentiated fore-/midgut NET.18 It was shown that promoter methylation was a common event and a unique methylation profile for gastrointestinal NET was described. Interestingly, there were no significant differences in the promoter methylation profiles of any tumor subgroup. We only found slight differences for less frequent hypermethylation of the RUNX3 and the O6-MGMT genes in tumor subsets. Besides genetic alterations, CIMP appeared to play an important role in tumor pathogenesis of fore-/midgut NET. These results led us to hypothesize that CIMP is involved in the molecular pathogenesis also of NET of the colon and rectum, as has been shown for sporadic CRC.
In this study, we directly compared characteristic molecular events in well and poorly differentiated NET from different anatomical sites. We could demonstrate differences in molecular events for sporadic CRC, well-differentiated NET from fore-/midgut and poorly differentiated NET of the colon, which might influence growth characteristics and proliferation rates in both cancer types. We further demonstrated the influence of several predictors on outcome in well and poorly differentiated tumors. For the first group of tumors, the Ki-67 proliferation rate was confirmed as the most important known predictor of survival. However, that was not true for undifferentiated tumors (colon and rectum) in which only methylation of p16 appeared to influence survival. p16 methylation in general was a predictor of worse outcome in all NET. However, the numbers of poorly differentiated hindgut tumors with a low Ki-67 index was low. CIMP was not a predictor of survival in either group.
To date, only few outcome studies have been performed for NET. Most identified the Ki-67 proliferation index as the main predictor of outcome.23–26 Several molecular events have been described in NET including genetic or epigenetic alterations of p53, BAX, p16INK4a/CDKN2A, BRAF, bcl-2, PTEN, DPC4/Smad4, p27,16–18,21–23,27–36 allelic losses37 or differential expression of nuclear survivin and other proteins.17,38 Most of them did not correlate with patient survival or aggressiveness of the tumors. Tumors with frequent allelic losses correlate with a higher proliferation index and a poorer survival.22,25,37,39 Also retaining p27 expression was a predictor of a longer survival in some patients.40 This has been confirmed by a recent study (Grabowski 2008, submitted for publication) in which the loss of the cyclin-dependent kinase inhibitor p27 played a critical role for the aggressiveness of GEP-NET. This was explained by the increased cell cycle progression and proliferation by lack of inhibition of cyclin E in those tumors. Patients retaining p27 expression had a significantly longer outcome in that study. Further predictors of outcome are histological differentiation and the presence of malignancy (infiltration of adjacent structures or metastases).2 Our study confirmed the Ki-67 proliferation rate as a predictor of survival in GEP-NET. This was not consistent for poorly differentiated NET from the colon and rectum, however, in which none of the tumors had a low proliferation rate and the differentiation into <20% or ≥ 20% positive cells had no influence on survival. The aggressiveness of NET from the colon and rectum might be explained by the poor differentiation of the tumors with frequent allelic losses and loss of p27 expression.25,33,37,40 In our recent study, CIMP positive tumors had a better outcome than CIMP negative tumors in WDET and WDEC from the fore- and midgut.18 No other prognostic marker had been identified. CIMP did not influence survival in our study neither in well- and poorly differentiated NET. These discrepant results remain to be explained and might be due to the low tumor numbers investigated. Interestingly, methylation of the p16 promoter, which has been shown to correlate with expression in different studies,18,19 predicted a worse survival in NET and showed a trend toward a poorer outcome in poorly differentiated NET from the colon and rectum. However, the case number was too low to draw final conclusions from this finding. Methylation of p16 was not present in fore-/midgut NET and the number of NET from the colon and rectum was too low to yield significant data. Future studies, however, should address the role of p16 expression in NET on survival. Despite differences in the methylation rate of HIC1, MEN1, hMLH1 and TIMP3, these genes did not influence outcome in the respective tumor groups. The significance of MEN1 methylation is still unsolved. We have shown earlier that MEN1 expression and promoter methylation of the gene did not correlate.18 However, MEN1 mutations in sporadic foregut NET have been described before.41 It remains to be elucidated how these genes influence tumor growth and/or progression in sporadic NET. Recently, a new TNM classification of GEP-NET has been proposed.13,14 Future studies will clarify, whether this classification will represent a new outcome predictor and compliment the known predictors.
Apart from the low incidence of poorly differentiated NET of the colon and rectum compared to sporadic CRC, the molecular pathogenesis of both tumor types appears to be different although the growth characteristics of both tumors reveal some similarities. Other than in sporadic CRC, MSI is infrequent in poorly differentiated NET of the colon and rectum. MSI has also been shown to be an infrequent event in NET from different anatomic sites.18,42,43 CIMP is common in both sporadic CRC and poorly differentiated NET from the colon and rectum, but due to the different molecular pathogenesis of the tumors the relevant methylated genes have been shown to be different.6,9,11,18,19 Our study is the first to investigate the methylation status of tumor-associated genes in a relevant number of poorly differentiated NET from the colon and rectum.
In conclusion, our findings demonstrate that sporadic CRC and poorly differentiated NET of the colon and rectum differ in tumor pathogenesis despite the occurrence of CIMP in either. Until further studies are performed to directly compare methylation patterns in NET from the colon and rectum and sporadic CRC, we cannot know for certain whether CIMP is an acquired defect with a primary etiology or whether this abnormal pattern of promoter methylation is a random process that is selected for in tumor cells. MSI is common only in sporadic CRC and is not seen in GEP-NET from different anatomical sites and not dependent on tumor differentiation. In accordance with their poor differentiation, NET from the colon and rectum had a worse survival than well-differentiated fore-/midgut NET. Predictors of a poor outcome in this study were a high Ki-67 proliferation index and p16 methylation. Expression of p16 should be analyzed in future studies in a larger number of NET in order to define its value as predictor of survival that may contribute to individualize therapeutic strategies.
Acknowledgments
We thank Prof. Dr. G. Klöppel, Institute of Pathology, University of Kiel, Germany and Prof. Dr. A. Tannapfel, Institute of Pathology, University of Bochum, Germany, for providing tumor samples for the study. This work was supported by a grant from the Fritz-Thyssen Stiftung to Christian N. Arnold.
Grant sponsor: Fritz-Thyssen Stiftung.
References
- 1.Öberg K. Carcinoid tumors: molecular genetics, tumor biology, and update of diagnosis and treatment. Curr Opin Oncol. 2002;14:38–45. doi: 10.1097/00001622-200201000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Klöoppel G, Perren A, Heitz PU. The gastroenteropancreatic neuroendocrine cell system and its tumors: the WHO classification. Ann N Y Acad Sci. 2004;1014:13–27. doi: 10.1196/annals.1294.002. [DOI] [PubMed] [Google Scholar]
- 3.Hipfner DR, Cohen SM. Connecting proliferation and apoptosis in development and disease. Nat Rev Mol Cell Biol. 2004;5:805–15. doi: 10.1038/nrm1491. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–710. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 5.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–9. [PubMed] [Google Scholar]
- 6.Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM, Boland CR. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–38. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Issa JP, Baylin SB. Epigenetics and human disease. Nat Med. 1996;2:281–2. doi: 10.1038/nm0396-281. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, Kinzler KW, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci USA. 1997;94:2545–50. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–14. [PubMed] [Google Scholar]
- 10.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–96. [PubMed] [Google Scholar]
- 11.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 13.Rindi G, Kloppel G, Alhman H, Caplin M, Couvelard A, de Herder WW, Erikssson B, Falchetti A, Falconi M, Komminoth P, Korner M, Lopes JM, et al. TNM staging of foregut (neuro)endocrine tumors: a consensus proposal including a grading system. Virchows Arch. 2006;449:395–401. doi: 10.1007/s00428-006-0250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rindi G, Kloppel G, Couvelard A, Komminoth P, Korner M, Lopes JM, McNicol AM, Nilsson O, Perren A, Scarpa A, Scoazec JY, Wiedenmann B. TNM staging of midgut and hindgut (neuro) endocrine tumors: a consensus proposal including a grading system. Virchows Arch. 2007;451:757–62. doi: 10.1007/s00428-007-0452-1. [DOI] [PubMed] [Google Scholar]
- 15.Onaitis MW, Kirshbom PM, Hayward TZ, Quayle FJ, Feldman JM, Seigler HF, Tyler DS. Gastrointestinal carcinoids: characterization by site of origin and hormone production. Ann Surg. 2000;232:549–56. doi: 10.1097/00000658-200010000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyana TN, Xiang J, Senthilselvan A, Kulaga A. The spectrum of neuroendocrine differentiation among gastrointestinal carcinoids: importance of histologic grading. MIB-1, p53, and bcl-2 immunoreactivity. Arch Pathol Lab Med. 2000;124:570–6. doi: 10.5858/2000-124-0570-TSONDA. [DOI] [PubMed] [Google Scholar]
- 17.Grabowski P, Griss S, Arnold CN, Hörsch D, Göke R, Arnold R, Heine B, Stein H, Zeitz M, Scherübl H. Nuclear survivin is a powerful novel prognostic marker in gastroenteropancreatic neuroendocrine tumor disease. Neuroendocrinology. 2005;81:1–9. doi: 10.1159/000084892. [DOI] [PubMed] [Google Scholar]
- 18.Arnold CN, Sosnowski A, Schmitt-Graff A, Arnold R, Blum HE. Analysis of molecular pathways in sporadic neuroendocrine tumors of the gastro-entero-pancreatic system. Int J Cancer. 2007;120:2157–64. doi: 10.1002/ijc.22569. [DOI] [PubMed] [Google Scholar]
- 19.Chan AO, Kim SG, Bedeir A, Issa JP, Hamilton SR, Rashid A. CpG island methylation in carcinoid and pancreatic endocrine tumors. Oncogene. 2003;22:924–34. doi: 10.1038/sj.onc.1206123. [DOI] [PubMed] [Google Scholar]
- 20.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 21.Terris B, Meddeb M, Marchio A, Danglot G, Flejou JF, Belghiti J, Ruszniewski P, Bernheim A. Comparative genomic hybridization analysis of sporadic neuroendocrine tumors of the digestive system. Genes Chromosomes Cancer. 1998;22:50–6. doi: 10.1002/(sici)1098-2264(199805)22:1<50::aid-gcc7>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 22.Tonnies H, Toliat MR, Ramel C, Pape UF, Neitzel H, Berger W, Wiedenmann B. Analysis of sporadic neuroendocrine tumours of the enteropancreatic system by comparative genomic hybridisation. Gut. 2001;48:536–41. doi: 10.1136/gut.48.4.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amarapurkar AD, Davies A, Ramage JK, Stangou AJ, Wight DG, Portmann BC. Proliferation of antigen MIB-1 in metastatic carcinoid tumours removed at liver transplantation: relevance to prognosis. Eur J Gastroenterol Hepatol. 2003;15:139–43. doi: 10.1097/00042737-200302000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Arnold R, Rinke A, Klose KJ, Muller HH, Wied M, Zamzow K, Schmidt C, Schade-Brittinger C, Barth P, Moll R, Koller M, Unterhalt M, et al. Octreotide versus octreotide plus interferon-alpha in endocrine gastroenteropancreatic tumors: a randomized trial. Clin Gastroenterol Hepatol. 2005;3:761–71. doi: 10.1016/s1542-3565(05)00481-7. [DOI] [PubMed] [Google Scholar]
- 25.Rindi G, D'Adda T, Froio E, Fellegara G, Bordi C. Prognostic factors in gastrointestinal endocrine tumors. Endocr Pathol. 2007;18:145–9. doi: 10.1007/s12022-007-0020-x. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu T, Tanaka S, Haruma K, Kitadai Y, Yoshihara M, Sumii K, Kajiyama G, Shimamoto F. Growth characteristics of rectal carcinoid tumors. Oncology. 2000;59:229–37. doi: 10.1159/000012166. [DOI] [PubMed] [Google Scholar]
- 27.Grabowski P, Schindler I, Anagnostopoulos I, Foss HD, Riecken EO, Mansmann U, Stein H, Berger G, Buhr HJ, Scherübl H. Neuroendocrine differentiation is a relevant prognostic factor in stage III-IV colorectal cancer. Eur J Gastroenterol Hepatol. 2001;13:405–11. doi: 10.1097/00042737-200104000-00018. [DOI] [PubMed] [Google Scholar]
- 28.Grabowski P, Sturm I, Schelwies K, Maaser K, Buhr HJ, Dorken B, Zeitz M, Daniel PT, Scherübl H. Analysis of neuroendocrine differentiation and the p53/BAX pathway in UICC stage III colorectal carcinoma identifies patients with good prognosis. Int J Colorectal Dis. 2006;21:221–30. doi: 10.1007/s00384-005-0779-5. [DOI] [PubMed] [Google Scholar]
- 29.Lubomierski N, Kersting M, Bert T, Muench K, Wulbrand U, Schuermann M, Bartsch D, Simon B. Tumor suppressor genes in the 9p21 gene cluster are selective targets of inactivation in neuroendocrine gastroenteropancreatic tumors. Cancer Res. 2001;61:5905–10. [PubMed] [Google Scholar]
- 30.Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, Heitz PU, Eng C. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am J Pathol. 2000;157:1097–103. doi: 10.1016/S0002-9440(10)64624-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perren A, Saremaslani P, Schmid S, Bonvin C, Locher T, Roth J, Heitz PU, Komminoth P. DPC4/Smad4: no mutations, rare allelic imbalances, and retained protein expression in pancreatic endocrine tumors. Diagn Mol Pathol. 2003;12:181–6. doi: 10.1097/00019606-200312000-00001. [DOI] [PubMed] [Google Scholar]
- 32.Perren A, Schmid S, Locher T, Saremaslani P, Bonvin C, Heitz PU, Komminoth P. BRAF and endocrine tumors: mutations are frequent in papillary thyroid carcinomas, rare in endocrine tumors of the gastrointestinal tract and not detected in other endocrine tumors. Endocr Relat Cancer. 2004;11:855–60. doi: 10.1677/erc.1.00841. [DOI] [PubMed] [Google Scholar]
- 33.Perren A, Komminoth P, Heitz PU. Molecular genetics of gastroenteropancreatic endocrine tumors. Ann N Y Acad Sci. 2004;1014:208. doi: 10.1196/annals.1294.021. [DOI] [PubMed] [Google Scholar]
- 34.Serrano J, Goebel SU, Peghini PL, Lubensky IA, Gibril F, Jensen RT. Alterations in the p16INK4a/CDKN2A tumor suppressor gene in gastrinomas. J Clin Endocrinol Metab. 2000;85:4146–56. doi: 10.1210/jcem.85.11.6970. [DOI] [PubMed] [Google Scholar]
- 35.Tannapfel A, Vomschloss S, Karhoff D, Markwarth A, Hengge UR, Wittekind C, Arnold R, Horsch D. BRAF gene mutations are rare events in gastroenteropancreatic neuroendocrine tumors. Am J Clin Pathol. 2005;123:256–60. [PubMed] [Google Scholar]
- 36.Wild A, Ramaswamy A, Langer P, Celik I, Fendrich V, Chaloupka B, Simon B, Bartsch DK. Frequent methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene in pancreatic endocrine tumors. J Clin Endocrinol Metab. 2003;88:1367–73. doi: 10.1210/jc.2002-021027. [DOI] [PubMed] [Google Scholar]
- 37.Furlan D, Cerutti R, Uccella S, La RS, Rigoli E, Genasetti A, Capella C. Different molecular profiles characterize well-differentiated endocrine tumors and poorly differentiated endocrine carcinomas of the gastroenteropancreatic tract. Clin Cancer Res. 2004;10:947–57. doi: 10.1158/1078-0432.ccr-1068-3. [DOI] [PubMed] [Google Scholar]
- 38.Grabowski P, Schönfelder J, Ahnert-Hilger G, Foss HD, Heine B, Schindler I, Stein H, Berger G, Zeitz M, Scherübl H. Expression of neuroendocrine markers: a signature of human undifferentiated carcinoma of the colon and rectum. Virchows Arch. 2002;441:256–63. doi: 10.1007/s00428-002-0650-9. [DOI] [PubMed] [Google Scholar]
- 39.Modlin IM, Kidd M, Latich I, Zikusoka MN, Shapiro MD. Current status of gastrointestinal carcinoids. Gastroenterology. 2005;128:1717–51. doi: 10.1053/j.gastro.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 40.Hommura F, aka-Akita H, Mishina T, Nishi M, Kojima T, Hiroumi H, Ogura S, Shimizu M, Katoh H, Kawakami Y. Prognostic significance of p27KIP1 protein and ki-67 growth fraction in non-small cell lung cancers. Clin Cancer Res. 2000;6:4073–81. [PubMed] [Google Scholar]
- 41.Toliat MR, Berger W, Ropers HH, Neuhaus P, Wiedenmann B. Mutations in the MEN I gene in sporadic neuroendocrine tumours of gastroenteropancreatic system. Lancet. 1997;350:1223. doi: 10.1016/S0140-6736(05)63453-8. [DOI] [PubMed] [Google Scholar]
- 42.Ghimenti C, Tannergard P, Wahlberg S, Liu T, Giulianotti PG, Mosca F, Fornaciari G, Bevilacqua G, Lindblom A, Caligo MA. Microsatellite instability and mismatch repair gene inactivation in sporadic pancreatic and colon tumours. Br J Cancer. 1999;80:11–16. doi: 10.1038/sj.bjc.6690314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kidd M, Eick G, Shapiro MD, Camp RL, Mane SM, Modlin IM. Microsatellite instability and gene mutations in transforming growth factor-beta type II receptor are absent in small bowel carcinoid tumors. Cancer. 2005;103:229–36. doi: 10.1002/cncr.20750. [DOI] [PubMed] [Google Scholar]


