Abstract
Astrocytomas are the leading cause of solid tumor-related death in children. Progress toward developing effective therapies for these tumors has been modest, in part because of a limited understanding of the genetic alterations responsible for their development. To identify genetic changes that occur in pediatric astrocytomas, we examined genome-wide DNA copy number aberrations (CNAs) in a panel of 33 tumors representing pilocytic astrocytomas (PAs, WHO grade I), diffuse infiltrating low-grade astrocytomas (DLGA, WHO grade II), anaplastic astrocytomas (AA, WHO grade III), and glioblastoma multiforme (GBM, WHO grade IV). We found high-level genomic amplifications (>10 copies) restricted to AAs and GBMs, which involved the MDM4 (1q32), PDGFRA (4q12), MET (7q21), CMYC (8q24), PVT1 (8q24), WNT5B (12p13), and IGF1R (15q26) genes. Homozygous deletion (HD) of CDKN2A (9p21), PTEN (10q26), and TP53 (17p3.1) were evident among grade II-IV tumors. CNAs suggestive of BRAF gene rearrangement were observed in 3 tumors, and in two of these, both PAs, RT-PCR analysis revealed expression of KIAA1549-BRAF fusion transcripts. Examination of eight additional PAs also revealed BRAF fusion transcripts. In total, 10/10 PAs showed expression of KIAA1549-BRAF mRNAs; however, none of the grade II-IV tumors expressed these gene products. In contrast, oncogenic missense BRAF mutation (BRAFV600E) was detected in 2/11 DLGAs (18%), 3/9 AAs (33%), and 2/11 GBMs (18%), but in none of the PAs. Five of seven grade II-IV astrocytomas with BRAFV600E had concomitant CDKN2A HD, suggesting that these alterations define a subset of pediatric malignant astrocytomas.
Keywords: pediatric astrocytoma, genomic profiling, gene alterations
Introduction
Targeted molecular therapeutics holds great promise for the development of less toxic and more effective personalized treatment strategies for cancer (1). This approach, however, requires a detailed understanding of the molecular alterations that drive tumor formation and malignant progression. The identification of seminal changes in cancer can be used to inform the development of therapeutic agents that specifically inhibit the gene products and pathways that are deregulated in association with these changes. This strategy has been used successfully in adult high-grade astrocytomas, leading to clinical trials that target the epidermal growth factor receptor (EGFR), for example. However, astrocytomas in children are distinct clinical entities from those seen in adults, and do not harbor many of the critical genetic alterations found in their adult counterparts (2–5). It is therefore important that the molecular alterations unique to pediatric astrocytomas are identified and characterized.
Recently, it was shown that the majority of pilocytic astrocytomas (PAs, WHO grade I) harbor 7q34 duplications (6), which result in gene fusions between KIAA1549 and BRAF (7), and concomitant expression of KIAA1549:BRAF fusion transcripts (7–9). While this discovery suggests that targeted inhibition of BRAF or relevant downstream signaling mediators may be especially effective for the treatment of PA, identification of therapeutically informative molecular alterations in pediatric malignant astrocytomas (WHO II, III, and IV) has been an elusive goal.
To provide a more detailed characterization of the spectrum of genetic alterations in all malignancy grades of pediatric astrocytoma, we report a genome-wide analysis of DNA copy number alterations for a series of these tumors, presented in combination with the sequence analysis of five relevant genes, including three frequently mutated in adult astrocytomas (TP53, PTEN, and IDH1), and two involved in MAP kinase signaling (BRAF and KRAS). Our results confirm that genes targeted for sequence alteration in adult astrocytomas are less frequent mutation targets in pediatric astrocytomas. In addition, we show that BRAF fusion is a signature event in PA, but not in the malignant astrocytomas, whereas BRAF activating mutation is common in grade II-IV tumors. Moreover, we found that the majority of grade II-IV tumors with BRAF activating mutation have concomitant CDKN2A homozygous deletion. Collectively, these findings suggest that the combination of BRAF activation and CDKN2A inactivation may be unique drivers of malignancy in a subset of pediatric astrocytomas, and whose alteration suggest targets for future therapeutic drug design.
Materials and Methods
Clinical specimens
A total of 41 astrocytomas were included in this study (Table 1). Non-neoplastic brain samples derived from epileptic surgeries from pediatric (n=2) and adult (n=6) patients were also included. All samples were subject to detailed histopathological review (SRV) and approved for use according to CHR guidelines. Sufficient RNA was obtained from all cases for determination of presence or absence of KIAA1549-BRAF fusion transcripts, and sufficient DNA was obtained from 40 cases for gene sequence analysis. DNA from 33 of the cases was examined for copy number alterations.
Table 1.
Summary of Pediatric Astrocytoma Gene and Genomic Alterations
| Tumor | Class | Age | Location | FGA | BRAF | Other | |
|---|---|---|---|---|---|---|---|
| 1 | SF2085 | PA (I) | 8 | Cerebellum | ND | K16-B9 | None |
| 2 | SF2415 | PA (I) | 9 | Cerebellum | ND | K16-B9 | None |
| 3 | SF2420 | PA (I) | 9 | Brain Stem | 0.000 | K15-B9 | None |
| 4 | SF2974 | PA (I) | 4 | Cerebellum | ND | K16-B11 | None |
| 5 | SF3526 | PA (I) | 5 | Cerebellum | ND | K16-B9 | ND |
| 6 | SF3663 | PA (I) | 14 | Optic nerve | ND | K15-B9 | None |
| 7 | SF3975 | PA (I) | 2 | Temporal | ND | K15-B9 | None |
| 8 | SF4035 | PA (I) | 12 | Spinal Cord | ND | K16-B9 | None |
| 9 | SF4282 | PA (I) | 1 | Post. Fossa | 0.000 | K16-B9 | None |
| 10 | SF4283 | PA (I) | 5 | Cerebellum | ND | K15-B9 | None |
| 11 | SF2652 | DLGA (II) | 10 | Hypothalamus | 0.014 | N | None |
| 12 | SF2995 | DLGA (II) | 17 | Unspecified | 0.000 | ND | ND |
| 13 | SF3094 | DLGA (II) | 15 | Temporal | 0.010 | N | None |
| 14 | SF3310 | DLGA (II) | 17 | Intraventricular | 0.000 | N | None |
| 15 | SF4762 | DLGA (II) | 21 | Unspecified | 0.001 | N | None |
| 16 | SF4825 | DLGA (II) | 10 | Parietal | 0.000 | N | None |
| 17 | WU12516 | DLGA (II) | 19 | Brain Stem | 0.096 | V600E | CDKN2A (HD) |
| 18 | WU12517 | DLGA (II) | 20 | Occipital Lobe | 0.056 | V600E | CDKN2A (HD) |
| 19 | WU108305 | DLGA (II) | 13 | Thalamus/Pineal | 0.000 | N | None |
| 20 | WU108309 | DLGA (II) | 3 | Cerebellar Vermis | 0.000 | N | None |
| 21 | WU108312 | DLGA (II) | 1 | Frontal | 0.001 | N | None |
| 22 | SF1692 | AA (III) | <1 | Frontal | 0.003 | N | None |
| 23 | SF1734 | AA (III) | 13 | Temporal | 0.025 | V600E | None |
| 24 | SF1752 | AA (III) | 5 | Frontal | 0.000 | V600E | CDKN2A (HD) |
| 25 | SF1762 | AA (III) | 14 | Intraventricular | 1.158 | N | TP53, 273:R>H |
| 26 | SF2007a | AA (III) | 13 | Cerebellum | 0.131 | V600E | None |
| 27 | SF2390 | AA (III) | 11 | Temporal | 7.198 | N |
CDKN2A (HD) CCND2/WNT5B (A) |
| 28 | SF2570 | AA (III) | 15 | Brain Stem | 3.860 | N | TP53, 272:V>L |
| 29 | SF7269 | AA (III) | 7 | Thalamus | 0.000 | N | None |
| 30 | SF7432 | AA (III) | 11 | Brain Stem | 0.000 | N | None |
| 31 | SF1983 | GBM (IV) | 9 | Posterior Fossa | 5.137 | N | TP53, 179:H>Y |
| 32 | SF2975 | GBM (IV) | 7 | Frontal | 8.001 | N |
CDKN2A (HD) PTEN(HD) TP53 (HD) |
| 33 | SF3343 | GBM (IV) | 2 | Basal Ganglia | 0.001 | N | None |
| 34 | SF4212 | GBM (IV) | 14 | Parietal | 0.984 | N | KRAS: 12 G>V |
| 35 | SF4532 | GBM (IV) | 17 | Parietal | 0.722 | V600E | CDKN2A (HD) |
| 36 | SF4635 | GBM (IV) | 17 | Parietal | 0.262 | V600E | CDKN2A (HD) |
| 37 | SF4761 | GBM (IV) | 17 | Parietal | 5.562 | N | TP53, 273:R>H PDGFRA/KIT (A) |
| 38 | SF4870 | GBM (IV) | 6 | Parietal | 4.296 | N | PDGFRA/KIT (A) MET (A) IGFR1 (A) |
| 39 | SF6751 | GBM (IV) | 13 | Parietal | 4.113 | N | None |
| 40 | SF6906 | GBM (IV) | 16 | Parietal | 1.736 | N | PTEN: 214 T>STOP CHIC2 (A) |
| 41 | SF7124 | GBM (IV) | 9 | Temporal | 3.790 | N | TP53, 172:V>F PIK3C2B/MDM4 (A) MYC/PVT1 (A) |
Key for types of gene alterations: A = high level amplification; HD = homozygous deletion; missense mutations resulting in amino acid changes are indicated by amino acid sequence number and resulting amino acid change. FGA = fraction genome altered. No IDH1 mutation was identified in this tumor series.
Recurrent tumor initially diagnosed as grade III astrocytoma.
Identification of Copy Number Aberrations
To identify copy number gains and losses, we used the custom-designed Molecular Inversion Probe (MIP) cancer panel consisting of 24,037 SNPs (Affymetrix, Santa Clara, CA). The MIP assay and analysis of resultant data were performed as described (10, 11), using 37 ng of genomic DNA as starting material. The fraction of the genome altered (FGA) for each tumor was calculated as the total length of gained and deleted loci divided by the total genome length (2866 Mb; NCBI Build 35).
Assessment of KIAA1549-BRAF fusion transcripts
Total RNA was extracted using the miR-Vana RNA isolation kit (Applied Biosystems, Foster City, CA), and first strand cDNA was generated using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). PCR was conducted using 60 ng of cDNA and final concentrations of 2.5mM MgCl2, 25nM dNTPs, 400nM primers and 0.05U/ul AmpliTaq Gold (Applied Biosystems). Cycling conditions were: 95°C 10min, 35 cycles of 95°C 15sec, 55°C 30sec, 72°C 1min, followed by 72°C 10min & 4°C hold. The forward primer, 5′-CGGAAACACCAGGTCAACGG-3′ (KIAA1549 exon 15), and reverse primer, 5′-GTTCCAAATGATCCAGATCCAATTC-3′ (BRAF exon 11), used in this study detect 3 fusion transcripts: KIAA1549-exon-15/BRAF-exon-9, KIAA1549-exon-16/BRAF-exon-11, and KIAA1549-exon-16/BRAF-exon-9 (7). Wild type BRAF primers were: forward primer 5′-TTGTGACTTTTGTCGAAAGCTGC-3′, and reverse primer 5′-AAGGGGATGATCCAGATGTTAGG-3′.
Gene Sequence Analysis
For BRAFV600E mutation analysis, genomic DNA was PCR amplified using BRAF exon 15 primers with M13 tails: forward primer 5′- TGTAAAACGACGGCCAGTCATAATGCTTGCTCTGATAGGA-3′, reverse primer 5′-AGCGGATAACAATTTCACACAGGCCAAAAATTTAATCAGTGGA-3′. PCR final concentrations (25ul volume) were 2.0mM MgCl2, 250nM dNTPs, 0.02U/ul AmpliTaq Gold (Applied Biosystems), 0.2mg/ml BSA, 500nM primers and 10ng of genomic DNA. Cycling conditions were: 95°C 10min, 35 cycles at 95°C 15sec, 64°C 45sec, 72°C 1min, followed by 72°C 10min, 4°C hold. PCR products were treated with Exo-SAP (USB, Cleveland, OH) per manufacturer’s protocol, and sequenced (ELIM Biopharmaceuticals, Hayward, CA).
For analysis of IDH1 sequence alterations we used the protocol described by Balss et al (12). Briefly, a fragment of 129bp length spanning the catalytic domain of IDH1 including codon 132 was amplified using the sense primer IDH1f 5′-CGGTCTTCAGAGAAGCCATT-3′ and the antisense primer IDH1r 5′-GCAAAATCACATTATTGCCAAC-3′. PCR conditions, using 20ng of DNA, were for 35 cycles with denaturing at 95°C for 30s, annealing at 56°C for 40s and extension at 72°C for 50s. Sequencing was with sense primer IDH1f 5′-CGGTCTTCAGAGAAGCCATT-3′.
KRAS mutations were identified as described by Lievre et al (13). Specific probes for mutated and non-mutated alleles labeled with fluorescence reporter dyes were included in 5 ul reactions mixtures containing 10 ng of DNA, 1 ug of specific primers and probes, and 1 ul TaqMan Universal PCR Master Mix (Applied Biosystems). Reaction mixtures were subjected to the following cycle conditions: 95°C for 15 minutes; 40 cycles, 95°C for 15 seconds; and 60°C for 1 minute. The single mutation detected by allelic discrimination was confirmed by direct sequencing of exon 2 of the KRAS gene.
Phospho-MAP Kinase Immunohistochemistry
All tissue was routinely fixed in either phosphate buffered 4% formalin, dehydrated by graded ethanols and embedded in wax (Paraplast Plus, McCormick Scientific, St. Louis, MO) using routine techniques. Immunohistochemistry was performed using the Ventana Medical Systems Benchmark XT (Ventana Medical Systems, Tuscon, AZ) and the Ultraview (multimer) detection system. All sections were cut at 5μm and mounted on Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA). Epitope retrieval was performed for 30 minutes in Tris buffer pH 8 at 90° prior to application of the primary antibodies. Rabbit polyclonal antibodies to phospho-MAP kinase (ERK1+2, Invitrogen, Carlsbad, CA) were used at a dilution of 1:200 with an incubation time of 2 h at 37°C.
Results
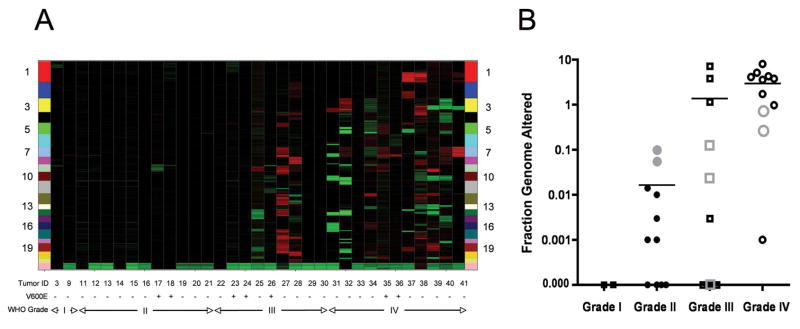
To identify regions of genomic copy number gain and loss, we analyzed DNA from 33 astrocytomas (Table 1) using a panel of ~24,000 Molecular Inversion Probes (MIPs) enriched for sequences that directly interrogate allele-specific copy number of ~1,000 known cancer genes. Included in this analysis were two grade I tumors, eleven grade II tumors, nine grade III tumors, and eleven grade IV tumors. Calculation of the mean fraction genome altered (FGA) for each grade of tumor revealed increasing FGA with increasing malignancy grade (grade I < 0.000; grade II = 0.016; grade III = 1.375; grade IV = 2.964), with the mean FGA for combined grade III + grade IV tumors significantly higher than for grade I + grade II tumors (p < 0.003, 2-sided Wilcoxon rank-sum test: Table 1 and Figure 1).
Figure 1. Copy number changes in pediatric astrocytoma.
A, Heat map showing genome-wide copy number gains (red) and losses (green) in 33 pediatric astrocytomas. Individual astrocytomas are arranged left to right on the X-axis from low-to-high grade. Tumors with (+) and without (−) BRAFV600E mutations are indicated. Chromosome landmarks are listed vertically on both sides of the map. Note the minor number of red and green blocks among grade I and grade II tumors. B, Vertical scatter plot showing individual and mean fractional genome alteration values for each malignancy grade of tumor. Gray colored data points represent cases with BRAFV600E mutations.
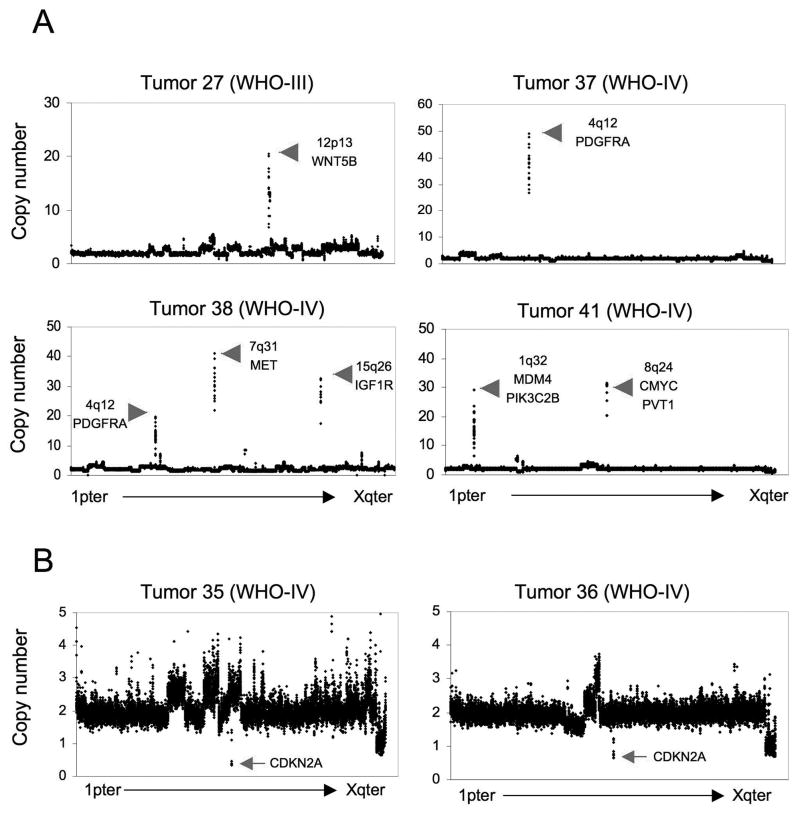
Several locations of copy number alteration were observed among grade II-IV tumors. Among these, high level amplifications (>10 copies) were observed exclusively in grade III and grade IV tumors, and included known oncogenes MDM4 (1q32), PDGFRA (4q12), MET (7q21), CMYC (8q24), PVT1 (8q24), WNT5B (12p13), and IGF1R (15q26) (Figure 2A). In contrast to adult grade III and grade IV astrocytomas, no high level or focal copy number gains for EGFR were observed. Regions of homozygous deletion (HD: <1 copy) were observed in grade II-IV astrocytomas, involving CDKN2A (7 tumors: Figure 2B), PTEN (1 tumor), and TP53 (1 tumor) (Table 2 and supplemental Table S1).
Figure 2. High level amplifications and homozygous deletions in pediatric astrocytoma.
Examples of genome copy number plots for tumors with high-level (>10 copies) genomic amplifications (A), and for tumors showing focal homozygous deletions of the CDKN2A locus (B).
Table 2.
Recurrent Gene Alterations in Pediatric Astrocytomas
| Gene Alteration | # Occurrences | Tumor Malignancy Grade Distribution |
|---|---|---|
| KIAA1549-BRAF: fusion | 10 | 10 Grade I |
| BRAF: V600E | 7 | 2 Grade II, 3 Grade III, and 2 Grade IV |
| CDKN2A: HD | 7 | 2 Grade II, 2 Grade III, and 3 Grade IV |
| TP53: mutation or HD | 6 | 2 Grade III and 4 Grade IV |
| PTEN: mutation or HD | 2 | 2 Grade IV |
| PDGFRA: amplification | 2 | 2 Grade IV |
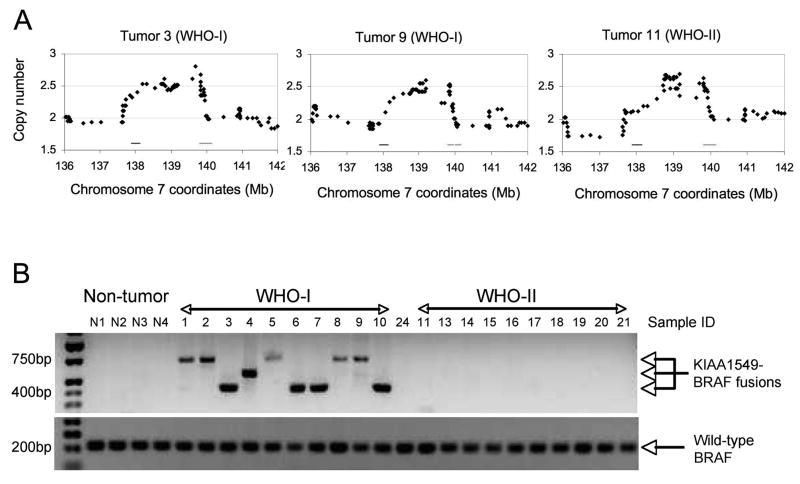
As revealed by the CNA results (Figure 1 and Table 1), genomic alterations in grade I and grade II tumors are exceedingly rare. Nonetheless, a single recurrent alteration was identified in two grade I tumors and a single grade II tumor: a 2 Mb duplication at 7q34, with breakpoints encompassing the KIAA1549 and BRAF genes (Figure 3A). Duplications of this region have been associated with KIAA159-BRAF rearrangements, resulting in the production of fusion mRNAs detectable by RT-PCR (7–9). To determine whether the 7q34 duplications represented KIAA1549-BRAF fusion transcripts, mRNA was isolated from each of the initial cohort of 33 tumors, and examined for these products. Both of the grade I tumors expressed BRAF fusion transcripts, but not the grade II tumor with a similar duplication of 7q34 (Figure 3B). Eight additional grade I astrocytomas subsequently examined by RT-PCR all revealed expression of KIAA1549-BRAF fusion transcripts. In total, 10 of 10 PAs expressed one of the three common KIAA1549-BRAF fusion transcripts previously described (7), whereas none of 31 grade II, grade III, and grade IV tumors expressed these fusion transcripts (Figure 3B).
Figure 3. BRAF fusion alterations are common in PA tumors.
A, High resolution view of copy number duplications at 7q34 and juxtaposition of BRAF (left horizontal line at position 138) and KIAA1549 (right horizontal line at positioin 140) sequences in grade I and grade II tumors. B, RT-PCR analysis of KIAA1549-BRAF fusion transcripts in non-neoplastic brain (N1-N4), grade I astrocytoma (n=10), grade II astrocytoma (n=10), and one grade III astrocytoma (tumor 24) reveals selective expression of fusion transcripts in grade I tumors. RT-PCR analysis of wild-type BRAF serves as a positive control. DNA sequencing of the RT-PCR fusion products revealed expression of KIAA1549-exon-15/BRAF-exon-9 transcripts (highest mobility fragment: 4 cases), KIAA1549-exon-16/BRAF-exon-11 transcripts (intermediate fragment: 1 case), and KIAA1549-exon-16/BRAF-exon-9 transcripts (lowest mobility fragment; 5 cases). Note that one grade II astrocytoma displayed a 7q34 duplication (tumor 11), but no KIAA1549-BRAF fusion transcript was detected.
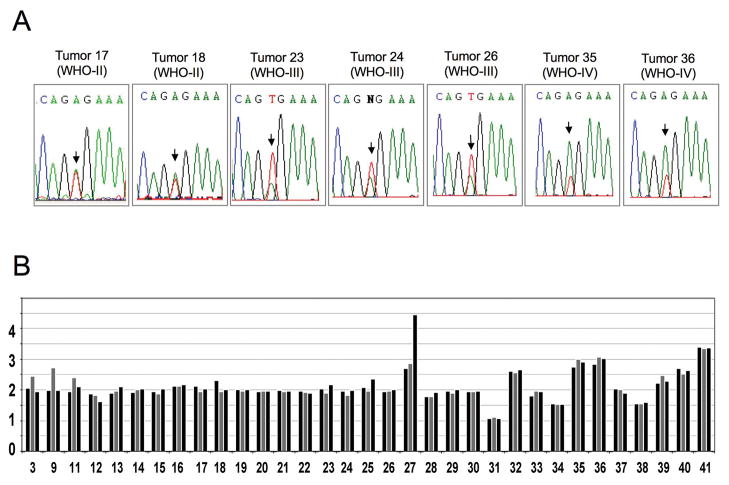
In addition to the gene rearrangements of BRAF resulting in the synthesis of KIAA1549-BRAF fusion transcripts, a missense activating BRAF mutation (BRAFV600E) has been previously reported in grade I and grade II astrocytomas at low incidence (6, 7). Here, DNA sequence analysis revealed BRAFV600E mutation in 0/10 grade I, 2/11 (18%) grade II, 3/9 (33%) grade III, and 2/11 (18%) grade IV astrocytomas (Figure 4A; Tables 1 and 2). MIP analysis indicated that both GBM tumors with BRAFV600E had 7q34 copy number gains (Figure 4B), consistent with the relative peak areas of the BRAF DNA sequence tracings seen in these tumors (Figure 4A). BRAF copy number gains were not evident in the three grade III tumors or two grade II tumors with BRAFV600E mutations.
Figure 4. BRAF mutation in pediatric astrocytomas.
A, Sequence traces from grade II, III, and IV astrocytomas showing presence of both BRAF wild-type alleles (T) and mutant (A) BRAFV600E alleles in all instances where the V600E mutation has occurred. B, Average copy number variation across 7q34. For each tumor, copy number was calculated for: a ~ 2Mb region inclusive of KIAA1549 and BRAF (gray bar); a 5Mb region proximal to KIAA1549 and BRAF (left black bar); and a 5Mb distal to KIAA1549 and BRAF (right black bar). This data representation highlights tumors with duplications limited to KIAA1549 and BRAF (tumors 3, 9, 11) and tumors with duplications of a larger 7q34 chromosomal region that includes KIAA1549 and BRAF (tumors 32, 35, 36, 39, 40, 41). Tumor 27 contains a 7q34 duplication and further 7q34 amplification distal to BRAF.
Genes that are frequent mutagenic targets in the development of adult astrocytoma include TP53, PTEN, and IDH1 (4, 5, 12). To address the incidence of these alterations in pediatric astrocytomas, we sequenced relevant coding regions of each of these genes in the current set of tumors. Sequence alterations of TP53 were identified in 5 cases (2 grade III and 3 grade IV tumors), PTEN in 1 case (1 grade IV tumor), and IDH1 in none of the tumors (Table 1). Activating mutation of KRAS, an infrequent occurrence in both adult and pediatric astrocytoma, was found in one grade IV astrocytoma lacking the BRAFV600E alteration (Table 1). Together, these data demonstrate that BRAF is the most common mutagenic target in pediatric astrocytomas (Table 2). Specifically, seventeen of 41 tumors had BRAF alterations, followed by CDKN2A HD (7 instances) and TP53 sequence alteration or HD (6 instances). Inactivating mutation and/or deletion of PTEN and amplification of PDGFRA were the only other specific gene alterations observed in multiple tumors (2 tumors each).
We next examined gene alterations that occur concomitant with BRAF mutation, and found that CDKN2A HD was observed in 5 of the 7 tumors with the BRAFV600E alteration (Table 1). Among grade II-IV tumors, the association between CDKN2A HD and BRAFV600E mutation was significant: Whereas 71% of grade II-IV tumors with BRAFV600E have CDKN2A HD (5 of 7), only 8% (2 of 24) of the remaining grade II-IV astrocytomas lacking the BRAFV600E mutation harbored CDKN2A HD (p=0.0016, 2-sided Fishers exact test). In fact, no additional specific gene alterations, including those resulting from high-level genomic amplifications, were evident in the two remaining grade III tumors with BRAFV600E mutation (Table 1).
Since increased MAPK signaling accompanies BRAF activation, we next examined phospho-MAP kinase (pMAPK) expression in astrocytomas (14, 15). Interestingly, all pediatric astrocytomas examined (40 cases), irrespective of malignancy grade or BRAF mutation status, showed strong immunoreactivity for pMAPK (supplemental Figure S1).
Discussion
Grade II-IV pediatric astrocytomas share histopathologic similarities with their corresponding adult counterparts, and, in general, pathologic examination does not distinguish pediatric and adult tumors of the same malignancy grade. Numerous studies have identified signature genetic mutations in high-grade adult astrocytomas, including the recent TCGA analysis of GBM tumors (4). The TCGA study demonstrated that EGFR, PTEN, and TP53 changes are among the most frequently observed alterations in adult GBM, whereas recent studies using parallel genomic approaches showed that mutations in the IDH1 gene predominate in adult gliomas (5, 16). With the possible exception of TP53 mutation, the frequent genetic alterations seen in adult astrocytomas have been identified at lower frequencies in pediatric astrocytomas (2, 3, 17). This lack of signature genetic mutations in pediatric astrocytomas is unfortunate, as these serve as important reference points with which to test therapeutic hypotheses.
The results of the current study further emphasize the distinct genetic etiologies of pediatric and adult astrocytomas. In our series, the only common adult astrocytoma gene alterations observed at appreciable frequencies were CDKN2A HD (7 of 31 cases: 23%), and alterations effecting p53 function (6 of 31 cases: 19%). The incidence of these alterations, however, is much lower than that reported by the TCGA project for adult GBM (4). Although a negative observation, the lack of IDH1 mutation in the current series of astrocytomas is further evidence of the distinct genetic nature of the pediatric tumors. In this regard, a recent study that included 42 pediatric gliomas examined for IDH1 mutation, also concluded that these alterations are rare in childhood astrocytomas (18).
The approach taken here to identify common alterations in pediatric astrocytoma involved microarray analysis for high-resolution whole genome examination. The SNP platform used has particularly high density coverage of 1000 genes associated with human cancer, yet has sufficient coverage for identifying focal gains and deletions across the majority of the genome (median distance between probes, across the entire genome, is 43 kb), and certainly has sufficient density to identify copy number alterations affecting all large chromosomal regions. Whereas recurrent copy number alterations indicating low level gains or hemizygous deletions are of importance, and are presented in supplemental Table S2, it is the regions of high level amplification and homozygous deletion that are of particular interest in revealing specific oncogene and tumor suppressor gene targets, respectively (Tables 1, 2, and S1). These array data highlight PDGFRA and CDKN2A as important oncogene and tumor suppressor gene targets, respectively, in the development of pediatric malignant astrocytoma.
The identification of focal 7q34 copy number gains with rearrangement in 3 tumors (Figure 3A) prompted an analysis of BRAF alterations, revealing KIAA1549-BRAF fusion transcript expression in ten grade I tumors, but not in a single grade II tumor with this genomic alteration. While it is possible that there are additional KIAA1549-BRAF alterations not detected with the primer pairs we used, it appears that BRAF fusion transcripts are highly diagnostic of pediatric grade I astrocytoma. This is particularly important in clinical practice, as biopsy material from these tumors arising in the brainstem or optic nerve may not be representative of the entire tumor, and pilocytic astrocytoma may be misdiagnosed as grade II or grade IV astrocytoma. The development of reagents for immunohistochemical detection of the junction sequences in paraffin-embedded tissues would therefore be of great value.
In light of prior reports indicating a few instances of BRAFV600E activating mutations in grade I and grade II pediatric astrocytomas (6, 7), we determined the presence or absence of this alteration in the current series of tumors to address the overall incidence of BRAF alterations. Unexpectedly, 7 tumors with BRAFV600E mutation were identified among the 31 grade II-IV tumors (23%) suggesting this alteration is one of the most common gene alterations in pediatric malignant astrocytomas (Table 2). Observation of KRAS activating mutation (Table 1), high level PDGFRA or MET amplification (Table 1), and low-level copy number increases of PTK2 (Table S2) in seven additional grade III and grade IV tumors that lack BRAFV600E, indicate the occurrence of BRAF or surrogate genetic alterations for achieving heightened BRAF activity in most, and perhaps all (as implied by Figure S1 results) pediatric astrocytomas.
Even more surprising than the number of instances of BRAFV600E in pediatric grade II-IV astrocytomas was the occurrence of CDKN2A HD in 5 of the 7 tumors with BRAFV600E: a frequency of co-occurrence that is highly significant. This association is underscored by the lack of other specific gene alterations occurring in combination with BRAFV600E. BRAFV600E in combination with CDKN2A inactivation has been described in other cancers, especially melanoma (19), and in a recent report, this combination was identified in 2 of 18 grade II pediatric astrocytomas (8). Our results reinforce the important of coincident BRAFV600E with CDKN2A HD in pediatric malignant astrocytomas, and suggest this combination of gene alterations occurs at similar frequencies across grade II-IV tumors.
Finally, there are several BRAF signaling pathway inhibitors currently in clinical trial (20), including inhibitors of MAP kinase, a key downstream effector of BRAF. Interestingly, our immunohistochemical results indicate that MAP kinase activity is not differentially elevated in tumors with BRAF alterations (Figure S1), and sugget that additional activators of MAP kinase may be deregulated in pediatric astrocytoma Studies are underway to determine how BRAF controls astrocyte cell growth as well as to assess the effect of MAP kinase inhibitors on deregulated BRAF-mediated astrocytoma growth. Moreover, the new pediatric astrocytoma-associated genetic mutations identified in this report should inform the development of more relevant animal models of pediatric astrocytoma.
Supplementary Material
Acknowledgments
Our thanks to Cynthia Cowdrey for her outstanding assistance in tissue acquisition and processing.
Financial Support: Funded in part by grants from the Pediatric Low Grade Astrocytoma Foundation (J.G.H., D.H.R., C.D.J.), Brain Tumor Society (D.H.G.), Pediatric Brain Tumor Foundation (J.G.H., S.R.V., D.H.R., M.S.B., C.D.J.), Harriet H Samuelsson Foundation (J.D.S.), Center for Children’s Brain Tumors at LPCH (J.D.S., P.G.F., J.M.F., H.J.), and the NIH (CA097257: M.S.B., C.D.J.; CA101777: J.G.H.; CA121940 and HG000205: P.F.). D.H.R. is an HHMI Investigator.
Footnotes
Potential Conflicts of Interest: None.
References
- 1.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–70. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 2.Raffel C, Frederick L, O’Fallon JR, Atherton-Skaff P, Perry A, Jenkins RB, James CD. Analysis of oncogene and tumor suppressor gene alterations in pediatric malignant astrocytomas reveals reduced survival for patients with PTEN mutations. Clin Cancer Res. 1999;5:4085–90. [PubMed] [Google Scholar]
- 3.Pollack IF, Hamilton RL, James CD, et al. Rarity of PTEN deletions and EGFR amplification in malignant gliomas of childhood: results from the Children’s Cancer Group 945 cohort. J Neurosurg. 2006;105:418–24. doi: 10.3171/ped.2006.105.5.418. [DOI] [PubMed] [Google Scholar]
- 4.The Cancer Genome Atlas Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfister S, Janzarik WG, Remke M, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. 2008;118:1739–49. doi: 10.1172/JCI33656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forshew T, Tatevossian RG, Lawson AR, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J Pathol. 2009;218:172–81. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Deshmukh H, Gutmann RJ, Emnett RJ, Rodriguez FJ, Watson MA, Nagarajan R, Gutmann DH. Alterations of BRAF and HIPK2 loci predominate in sporadic pilocytic astrocytoma. Neurology. 2009 doi: 10.1212/WNL.0b013e3181c0664a. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Moorhead M, Karlin-Neumann G, et al. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33:e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Moorhead M, Karlin-Neumann G, et al. Analysis of molecular inversion probe performance for allele copy number determination. Genome Biol. 2007;8:R246. doi: 10.1186/gb-2007-8-11-r246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 13.Lièvre A, Bachet JB, Boige V, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 14.Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG. Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol. 2008 Sep;67(9):878–87. doi: 10.1097/NEN.0b013e3181845622. [DOI] [PubMed] [Google Scholar]
- 15.Sievert AJ, Jackson EM, Gai X, Hakonarson H, Judkins AR, Resnick AC, Sutton LN, Storm PB, Shaikh TH, Biegel JA. Duplication of 7q34 in pediatric low-grade astrocytomas detected by high-density single-nucleotide polymorphism-based genotype arrays results in a novel BRAF fusion gene. Brain Pathol. 2009;19:449–58. doi: 10.1111/j.1750-3639.2008.00225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;19(360):765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pollack IF, Finkelstein SD, Burnham J, Holmes EJ, Hamilton RL, Yates AJ, Finlay JL, Sposto R Children’s Cancer Group. Age and TP53 mutation frequency in childhood malignant gliomas: results in a multi-institutional cohort. Cancer Res. 2001;61:7404–7. [PubMed] [Google Scholar]
- 18.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–74. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 19.Haluska FG, Tsao H, Wu H, Haluska FS, Lazar A, Goel V. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res. 2006;12:2301s–2307s. doi: 10.1158/1078-0432.CCR-05-2518. [DOI] [PubMed] [Google Scholar]
- 20.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–34. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.