Summary
The Drosophila tumour suppressor gene fat encodes a large cadherin that regulates growth and a form of tissue organization known as planar cell polarity (PCP). Fat regulates growth via the Hippo kinase pathway [1–4], which controls expression of genes promoting cell proliferation and inhibiting apoptosis (reviewed in [5–11]). The Hippo pathway is highly conserved and is implicated in the regulation of mammalian growth and cancer development [12–18]. Genetic studies suggest that Fat activity is regulated by binding to another large cadherin Dachsous (Ds) [19–25]. The tumour suppressor, discs overgrown (dco)/Casein Kinase I δ/ε, also regulates Hippo activity and PCP [1, 26, 27]. The biochemical nature of how Fat, Ds and Dco interact to regulate these pathways is poorly understood. Here we demonstrate that Fat is cleaved to generate 450kDa and 110kDa fragments (Fat450 and Fat110). Fat110 contains the cytoplasmic and transmembrane domain. The cytoplasmic domain of Fat binds Dco, and is phosphorylated by Dco at multiple sites. Importantly, we show Fat forms cis-dimers, and that Fat phosphorylation is regulated by Dachsous and Dco in vivo. We propose that Ds regulates Dco-dependent phosphorylation of Fat and Fat-associated proteins to control Fat signaling in growth and PCP.
Results and Discussion
Fat is proteolytically processed into 450kDa and 110kDa fragments
To characterize Fat, we generated antibodies against the cytoplasmic domain (α-Fat-IC) and analyzed larval extracts by Western blotting. Since Fat is predicted to encode a protein of ~ 560kDa, we separated Drosophila imaginal disc extracts on 2.5% acrylamide gels supported with agarose, to more clearly resolve large proteins. Western blotting demonstrated that α-Fat-IC was specific, as the 560kDa band expected for full-length Fat was present in wildtype extracts and absent from extracts of fat mutant larvae (Figure 1A). Surprisingly, this antibody also recognized a lower molecular weight band of ~110kDa that was absent in fat mutants, indicating that the 110kDa band was derived from the fat locus. Northern analysis and screening of EST databases indicated that Fat is not alternatively spliced, suggesting that the 110kDa protein may represent a cleavage product of Fat (data not shown).
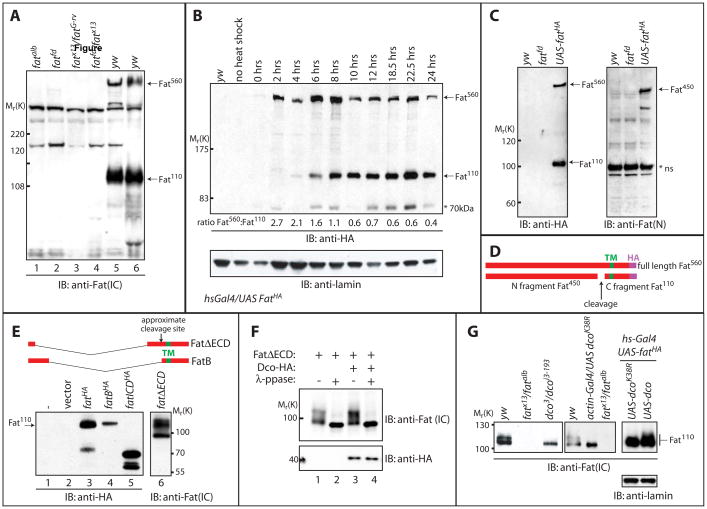
Figure 1. Fat processing generates a 110kDa form, which displays altered electrophoretic mobility in dco mutants.
A) Larval extracts from yw, fatalb, fatfd, fatfd/fatG-rv, fatfd/fatx13 were subjected to SDS-PAGE and analyzed by Western blotting with α-Fat (IC) antibody. 560kDa and 110kDa bands (indicated by arrows) recognized in yw (wildtype) extracts by the α-Fat (IC) antibody are absent from all fat extracts.
B) In vivo pulse chase analysis of FatHA. Larvae bearing a C terminally HA-tagged UAS-Fat transgene and hs-gal4 driver (hs-Gal4; UAS-FatHA) were subjected to a brief heat shock at 37°C and then returned to 18°C. Western blotting with α-HA of larval extracts from these animals at the indicated time points following heat shock revealed that Fat is first produced as a 560kDa protein (Fat560; top arrow). A band migrating at 110kDa (Fat110; lower arrow) however appears 4–6 hours post heat shock, and this band predominates after 10hrs (further emphasized by quantitation of the ratio of Fat560 to Fat110 below blot). Immunoblotting with α-lamin (lower panel) serves as a loading control. A 70kDa band (indicated with an asterisk) is also inconsistently detected with α-Fat (IC) antibody.
C) Larval extracts from yw, fatfd, and hs-Gal4;UAS-fatHA animals were subjected to SDS-PAGE and analyzed by Western blotting with α-Fat (N) antibody. This antibody detected a 450kDa band (Fat450; top arrow, right panel), while α-HA recognized a 560kDa band (Fat560; top arrow, left panel) and a 110kDa band (Fat110; bottom arrow, left panel). A non-specific band in all extracts (*ns) is also detected with α-Fat (N).
D) Schematic of Fat protein generated from expression of the UAS-fatHA transgene. The transmembrane domain (TM), C-terminal HA tag, and fragments generated upon cleavage of full length Fat (Fat560) are indicated.
E) Extracts from Drosophila S2 cells expressing HA-tagged full length Fat (lane 3), the Fat intracellular domain (ICD, lane 5), or versions of Fat encompassing both the transmembrane and intracellular domains (FatB and FatΔECD which vary in their extracellular sequence, lanes 4 and 6 respectively) were subjected to SDS-PAGE and analyzed by Western blotting with α-HA antibody. Expression of full-length Fat-HA generates Fat110, as well as Fat560 (not shown on this portion of the gel) while Fat ICD migrates at ~70kDa and FatB and FatΔECD at ~110kDa, suggesting Fat110 includes the transmembrane domain.
F) Lysates from Drosophila S2 cells expressing FatΔECD with or without HA-tagged Dco were divided. One half was treated with lambda phosphatase and the other mock treated. Samples were subjected to SDS-PAGE and Western blotting using α-HA or α-Fat (IC) antibodies.
G) Larval extracts from yw, dco3/dcoi3-193, fatAlb/fatx13, dcoK38R expressing, dco and fatHA co-expressing or dcoK38R and fatHA co-expressing larvae were analyzed by immunoblotting with α-Fat (IC) antibody. The Fat110 doublet is reduced to a single band in extracts from dco3/dcoi3-193 larvae and larvae expressing dcoK38A while overexpression of dco causes a decrease in the mobility of co-overexpressed Fat, visualized as an increase in the slower migrating form of the doublet. Immunoblotting with a-lamin (lower panel) serves as a loading control.
To determine if the 110kDa form derives from full length Fat, we conducted in vivo pulse-chase experiments, taking advantage of the temperature dependence of the UAS-Gal4 system. At 18°C, there is little Gal4-dependent transcription. Flies expressing UAS-fat-HA were raised at 18°C, and transferred to 37°C for 45 minutes, to allow a pulse of transcription, then returned to 18°C and samples taken at regular intervals for Western blotting (Figure 1B). Initially Fat-HA is produced as a 560kDa form – referred to herein as Fat560. The 110kDa form (Fat110) first appears after 2–4 hrs and is less abundant than Fat560 at these early time-points. At later time points Fat110 becomes more abundant (see ratio of Fat560:Fat110; Figure 1B). This suggests that Fat is synthesized as a 560kDa form, which undergoes proteolytic processing, yielding Fat110 and presumably an extracellular fragment of 450kDa (Fat450). We also detect a form of Fat at ~70kDa, but due to variability in detection (Figure 1B) we cannot exclude that this is a degradation product.
To detect the putative Fat450 N-terminal proteolytic fragment, we analyzed larval extracts with antibodies against the extracellular domain of Fat (α-Fat-N). Consistent with predicted extracellular cleavage, this antibody recognized a protein of ~450kDa (Figure 1C). This antibody also weakly detected Fat560 (data not shown). Together these data suggest Fat is synthesized as a transient 560kDa precursor, which is cleaved into 450kDa and 110kDa fragments (Figure 1D).
To determine if Fat110 and Fat450 form a stable complex, we generated a doubly-tagged Fat, tagged with Myc at the N terminus and HA at the C terminus. Immunoprecipitation of Fat450 (with Myc) or Fat110 (with HA) brought down Fat110 and Fat450 respectively, consistent with Fat110 and Fat450 stably associating (Figure S1). We note that the endogenous 560kDa band is not easily detected with α-Fat-IC when using standard chemoluminescence reagents (data not shown). In contrast the 110kDa band is readily detectable. The 560kDa band is thus likely transient, and endogenous Fat exists as 110kDa and 450kDa forms.
To determine if this processing is conserved, we analyzed mouse ES cell and embryonic extract using antibodies against the cytoplasmic domain of the vertebrate ortholog of Fat, Fat4 [28]. This antibody recognizes proteins of ~ 540kDa and ~140 kDa, suggesting that processing of the Fat family of cadherins is conserved (Figure S2). Significantly, an antibody against the extracellular domain of Fat4 recognizes both a ~540kDa band and a smaller N-terminal cleavage product of ~400kDa (Figure S2).
Fat110 is a transmembrane domain-containing protein
The cytoplasmic domain of Fat has a conceptual translation of 60kDa. Thus Fat110 likely includes the transmembrane domain and some of the extracellular domain. To test this, we expressed different fragments of Fat in Drosophila S2 cells. Expression of the cytoplasmic domain of Fat (ICD; Figure 1E, lane 5) yielded a protein of ~ 70kDa. Expression of either FatB or FatΔECD, which both contain between 150–160 amino acids of extracellular sequence, yielded a 110 kDa band comparable to that generated from processing of full length Fat (Figure 1E, lanes 4 and 6 compared to 3). This suggests that Fat110 is the product of cleavage of Fat560 ~150 amino acids N terminal of the transmembrane domain.
Post-translational modification of Fat110 is regulated by Discs overgrown (Dco)
Interestingly, FatΔECD runs as multiple bands that collapse with phosphatase treatment, indicating the cytoplasmic domain of Fat is phosphorylated in S2 cells (Figure 1F). Closer analysis indicated that Fat110 runs as a doublet in extracts from wildtype larvae (Figure 1G), suggesting that Fat is phosphorylated in vivo. Genetic studies indicate that the serine/threonine kinase Dco functions in the Fat growth pathway[1]. A specific hypomorphic dco allele, dco3, which harbors two point mutations in the N-terminal domain, leads to massive tissue overgrowth, phenocopying Hpo pathway mutants [29]. dco3 clones have elevated fj and Diap1 levels, indicative of increased Hpo pathway activity [1]. Interestingly, loss of dco results in reduced growth and decreased Diap1 levels, whereas overexpression of dco increases Diap1 protein levels [30]. Taken together this suggests that Dco has both positive and negative roles in tissue growth during development [29].
To assess whether Dco affects Fat phosphorylation in vivo, we isolated larval imaginal discs from dco3/dcoi3-193 transheterozygotes, and analyzed Fat electrophoretic mobility. Significantly, the Fat110 doublet ran largely as a single band in dco mutants, which co-migrated with the faster migrating band of the doublet (Figures 1G and Figure 4A). These data suggest that Dco regulates the phosphorylation of Fat. To confirm that Dco can alter Fat electrophoretic mobility in vivo, we ubiquitously expressed a dominant negative kinase-dead dco (DcoK38R) UAS transgene[27]. Similar to dco3/dcoi3-193 mutants, a single band at 110kDa was detected from larval extract of dcoK38R-expressing animals. Conversely, overexpression of wildtype dco decreased the mobility of co-expressed Fat110. Overexpression of dcoK38R failed to induce similar alterations in Fat mobility, suggesting that the slower migrating form of the doublet represents a hyperphosphorylated form of Fat110 (Figure 1G).
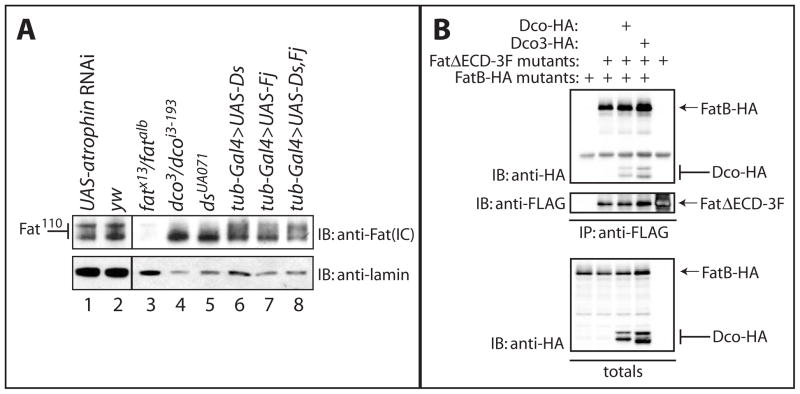
Figure 4. Fat110 is altered in ds mutants, and Fat can form dimers or oligomers.
A) Extracts from ey,GMR-gal4 UAS atrophin RNAi (lane 1), yw (lane 2), fat (lane 3), dco (lane 4), ds (lane 5) mutant and tub-gal4 UAS-ds (lane 6), tub-gal4 UAS-fj (lane 7), and tub-gal4 UAS-fj UAS-ds (lane 8) larvae were subjected to SDS-PAGE and analyzed by immunoblotting with α-Fat (IC) antibody. Fat110 mobility is altered in dco and ds mutants (lanes 4 and 5).
B) Lysates from HEK293T cells expressing HA-tagged FatB and/or 3FLAG-tagged FatΔECD together with Dco-HA or Dco3-HA were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. FatB was detected in immunoprecipitates of FatΔECD, along with Dco or Dco3, when co-expressed.
Dco/Casein kinase I δ/ε can bind and phosphorylate Fat
The alteration of Fat electophoretic mobility in dco mutants suggested that Dco might bind and phosphorylate Fat. Consistent with this hypothesis, co-expression of Fat with Dco in Drosophila S2 cells increased the proportion of phosphatase-susceptible Fat110 (Figure 1F, lanes 3–4). To examine whether Fat and Dco could interact, we expressed HA-tagged Dco and FLAG-tagged FatΔECD. Co-immunoprecipitation experiments in both HEK293T and Drosophila S2 cells revealed that Dco binds strongly to the cytoplasmic domain of Fat (Figures 2 and 3B). The cytoplasmic domain of vertebrate Fat4 [28], can specifically bind CKIε and CKIδ (Figure S4B), indicating this interaction is conserved.
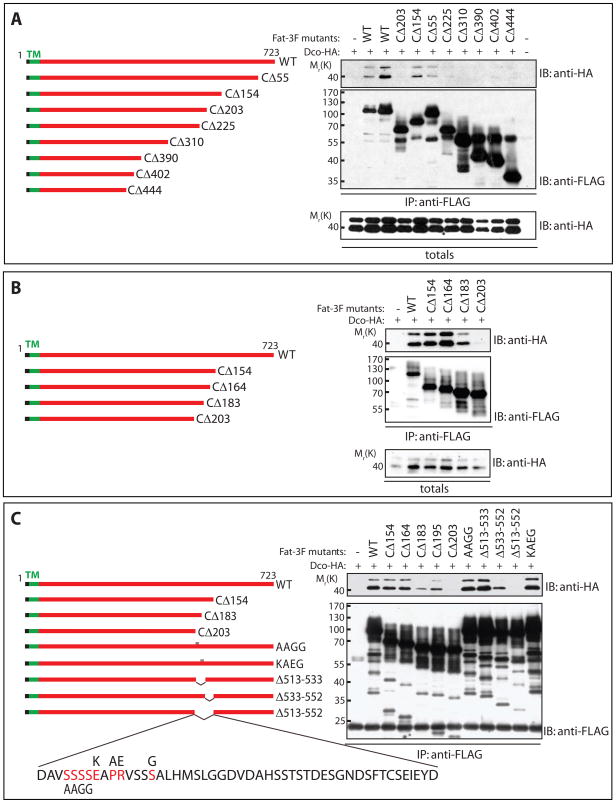
Figure 2. Dco interacts with the cytoplasmic domain of Fat.
Lysates from HEK293T cells expressing HA-tagged Dco together with 3FLAG-tagged variants of FatΔECD were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies.
A) Dco was detected in immunoprecipitates of full-length FatΔECD, comprised of the transmembrane and entire intracellular portion of Fat, and in immunoprecipitates from versions of FatΔECD lacking 55 and 154 C-terminal amino acids (CΔ55 and CΔ154). Versions of FatΔECD lacking more than 203 amino acids (CΔ203) failed to pull down Dco.
B) While FatΔECD lacking 154 C-terminal amino acids (CΔ154) can strongly interact with Dco, deletion of 183 amino acids (CΔ183) weakens the Dco interaction, and deletion of 203 amino acids (CΔ203) completely abolishes the interaction.
C) Internal deletions of 39 amino acids (Δ513–552) from the cytoplasmic portion of FatΔECD eliminates interaction with Dco, while removal of 19 of these internal residues (Δ533–552) weakens but does not abolish the interaction. Mutation of conserved residues within this region (AAGG or KAEG) does not block Fat interaction with Dco.
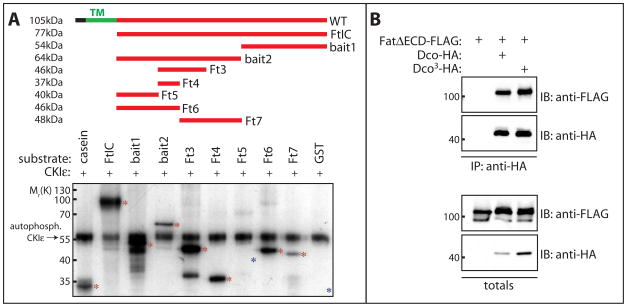
Figure 3. CKIε phosphorylates Fat and binding to Fat is not affected by the Dco3 mutation.
A) Recombinant GST-Fat fusion proteins (100ng) were incubated with human CKIε (50ng) and γ-32P-ATP. Casein was included as a positive control (lane 1). Phosphorylation of Fat fragments was analyzed by SDS-PAGE and autoradiography. The position of migration of input proteins is indicated with asterisks. Auto-phosphorylated CKIε migrates at ~55kDa. All fragments except Fat5 and GST (lanes 7 and 10 respectively, indicated with blue asterisks) showed robust phosphorylation, indicating multiple sites are phosphorylated by CKIε.
B) Both Dco and Dco3 interact with Fat. Lysates from Drosophila S2 cells expressing HA-tagged Dco or Dco3 together with FLAG-tagged FatΔECD were subjected to immunoprecipitation with anti-HA antibody and immunoprecipitates analyzed by immunoblotting with the indicated antibodies.
To define the Dco binding site, we generated a series of deletion mutants of Fat, and examined their ability to co-immunoprecipitate Dco. Deletions removing up to 164 amino acids from the C-terminus (CΔ164) had no effect on Fat-Dco binding. Further deletions weakened Dco binding. All detectable binding was lost when the last 203 residues were truncated from the C-terminus of Fat (CΔ203; Figure 2A and B). Internal deletions of 39 amino acids (Δ513–552) abolished Dco binding, while smaller deletions (Δ533–552) led to dramatically reduced binding (Figure 2C). Mutation of individual conserved residues within this region (AAGG or KAEG) had no detectable effect. These studies implicate a critical region for Dco binding that encompasses a 39 amino acid region, 164–203 residues from the C-terminus.
To test if Dco can phosphorylate the intracellular domain of Fat, we generated GST-fusion proteins and conducted in vitro phosphorylation assays. We incubated purified GST-Fat fragments with human CKIε (86% identity, 92% similarity to Drosophila Dco). Consistent with multiple CKI consensus sites in the Fat cytoplasmic domain (Figure S5), CKIε phosphorylated Fat at multiple sites (Figure 3A). Proteomic analysis has identified five phosphorylated serines in the cytoplasmic domain of Fat in Drosophila embryos[31] four of which conform to the CKI consensus. The finding that CKIε can phosphorylate Fat at multiple sites in vitro is consistent with the large alteration in electrophoretic mobility of Fat seen in dco3 mutants.
The dco3 mutation does not impede binding to the Fat cytoplasmic domain
As the electrophoretic mobility of Fat110 is altered in dco3 mutants we speculated that the dco3 mutation results in a loss of Fat binding. Unexpectedly, we found recombinant Dco3 bound strongly to the cytoplasmic domain of Fat in immunoprecipitation assays from Drosophila S2 and HEK293T cells (Figure 3B and Figure S4A respectively). Thus the alteration in Fat mobility seen in dco3 mutants is not due to a loss of Fat binding. We speculate that the dco3 mutation instead affects the ability of Dco to phosphorylate Fat and Fat-associated proteins.
These data suggest that loss of dco function, via the dco3 mutation, leads to loss of Fat signaling. Clones of dco3 exhibit increased expression of the well-characterized fat target gene fj [1]. To see if this reflected loss of dco function, or a neomorphic effect of the dco3 allele, we generated clones of dcoi3-193, a null allele for dco, and examined fj expression using a fj-lacZ enhancer construct. Importantly, clones of dcoi3-193 and dcoK38R-expressing clones both show strong upregulation of fj (Figure S3), similar to loss of fat.
Post-translational modification of Fat110 is altered in ds mutants
To explore whether Fat phosphorylation was altered in PCP or growth pathway regulating mutants, we examined extracts from wildtype, fat, fj, ex, sprouty, atrophin and ds mutant discs (Figure 4A and Figure S6). The Fat110 doublet was unaltered in fj, ex, sprouty, and atrophin mutants. Significantly however, mobility of Fat110 was altered in ds mutants. Like dco3/dcoi3-193 mutant and dcoK38R-expressing larvae, Fat110 resolves largely as a single band in ds mutants (Figure 4A and Table S1). Ds binds Fat and recruits it to cell contacts. The alteration of Fat110 mobility in ds mutants suggested Fat is differentially phosphorylated when not bound by Ds.
How might binding by Ds alter the phosphorylation of Fat by Dco? In ds mutants, Fat becomes diffusely localized, suggesting that Ds raises the local concentration of Fat at cell contacts [25, 32, 33]. We speculated that Fat molecule cis-associations, promoted by Ds, might promote trans-phosphorylation by Dco. We performed co-immunoprecipitation experiments with FLAG-tagged and HA-tagged Fat (Figure 4B and Figure S7). Significantly, we could co-immunoprecipitate HA-tagged Fat110 (or FatICD; Figure S7) with FLAG-tagged FatΔECD (Figure 4B). This indicates that Fat forms cis-dimers or oligomers, via the cytoplasmic region, and suggests a model whereby Fat activation involves receptor clustering. We propose that Ds binding increases the local concentration of Fat at cell contacts, favoring Fat cis-dimerization, thus promoting trans-phosphorylation of Fat by Dco.
Interestingly, borders of Ds expression promote Hippo activation [23] and uniform co-expression of fj and ds inhibits wing growth [22, 23]. However we find there is little, if any, alteration in the mobility of Fat when ds and fj are ubiquitously expressed (Figure 6A), nor when multiple clones are induced with eyFlp (not shown). Thus while ds is required for Fat phosphorylation, overexpression of ds is not sufficient to shift Fat110 entirely to the hyperphosphorylated form.
Loss of Ds leads to overgrowth and activation of the Hpo pathway, as does loss of Fat [34], suggesting that Ds binding enhances the ability of Fat to suppress growth. Ds binding promotes Fat phosphorylation. Together these observations suggest that phosphorylated Fat is the active form in growth repression. This is consistent with our observation that alleles of Dco that promote growth show reduced Fat phosphorylation. We suggest that Dco phosphorylates Fat. Dco may also phosphorylate other proteins recruited by Fat, modifying their activity in PCP or growth signaling. Significantly Dco not only regulates growth but also PCP [26, 27]. Fat has been proposed to recruit Expanded to control Hpo pathway activation, but not PCP [2–4, 35]. Conversely Atrophin binds to the Fat cytoplasmic domain, functioning with Fat to regulate PCP but not growth [36]. Phosphorylation of Expanded and/or Atrophin by Dco may provide a mechanism by which Dco and Fat can regulate growth and PCP.
By demonstrating that Dco binds and phosphorylates Fat, we have provided the first direct biochemical link between Fat and downstream components of the Hpo growth control pathway. Understanding how Fat coordinately regulates growth and planar polarity will require an understanding of the full complement of proteins that bind to Fat, and exploration of how Ds and Dco-dependent phosphorylation affects the activity of Fat in these diverse pathways.
Supplementary Material
Supplemental Figure 1. Processed forms of Fat interact in vivo.
Extracts from animals bearing an N terminal Myc and C terminal HA-tagged UAS-Fat transgene and daughterless-gal4 driver (da-Gal4; UAS-MYCFatHA) were subjected to immunoprecipitation with either α-Myc or α-HA antibody and analyzed by immunoblotting with the reciprocal antibody (α-Myc or α–HA). Immunoprecipitation of full length Fat (Fat560) or Fat450 with α-Myc brought down Fat560 and Fat110. Immunoprecipitation of full length Fat (Fat560) or Fat110 with α-HA brought down Fat560 and Fat450.
Supplemental Figure 2. Fat processing is conserved to vertebrates
Total extracts from Embryonic Stem cells (ES cells) and E14.5 embryos were analyzed by Western blotting with antibodies generated to the extracellular domain or to the intracellular domain of Fat4. Note both antibodies recognize proteins of ~540kDa, while a ~150kDa protein is seen only with the antibody to the intracellular domain, similar to the pattern of Drosophila Fat. The antibody to the extracellular domain also recognizes a protein of ~400kDa, which may represent the extracellular cleaved fragment.
Supplemental Figure 3. fourjointed is upregulated in dcoi3-193 null clones
A) Similar to that reported for dco3, clones of dcoi3-193 (MARCM clones, positively marked by GFP) in eye imaginal discs show elevated levels of fourjointed (fj-lacZ, red) near the morphogenetic furrow.
B) Clones of dcoK38R (MARCM clones, positively marked by GFP) in eye imaginal discs also show elevated levels of fourjointed (fj-lacZ, red) near the morphogenetic furrow.
Supplemental Figure 4. The interaction between Fat and Casein Kinase Iδ/ε is conserved
A) Lysates from HEK293T cells expressing HA-tagged FatB and/or 3FLAG-tagged FatΔECD together with Dco-HA or Dco3-HA were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. FatB was detected in immunoprecipitates of FatΔECD, along with Dco or Dco3, when co-expressed.
B) Lysates from HEK293T cells expressing a HA-tagged version of the Fat4 cytoplasmic domain fused to the Drosophila fat membrane targeting sequence together with 3FLAG-tagged CKIε (lane 2), CKIδ (lane 3), or Yap (lane 4), were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. Note the slower migrating forms of Fat4 in lanes 2 and 3 of the anti-HA immunoblot of totals (lower panel).
Supplemental Figure 5. Potential CKI phosphorylation sites in Fat
Amino acid sequence of Fat transmembrane domain (green) and Fat intracellular domain. Highlighted in red are regions adhering to the classical CKI consensus (S/T-X-X-S/T) and are potential CKI phosphorylation sites. Underlined and with asterisks are phosphorylated residues identified in vivo (Zhai et al, 2008).
Supplemental Figure 6. Fat110 is altered in ds mutants but not other growth or PCP mutants.
Extracts from yw (lane 1), fat (ft; lane 2), four-jointed (fj; lane 3), expanded (ex; lane 4), sprouty (sty; lane 5), and dachsous (ds; lane 6) mutant larvae were subjected to SDS-PAGE and analyzed by immunoblotting with α-Fat (IC) antibody. Fat110 mobility is altered in ds mutants.
Supplemental Figure 7. Fat can oligomerize.
Lysates from Drosophila S2 cells expressing HA-tagged full length Fat, FatB, or FatICD, and 3FLAG-tagged FatΔECD were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. Fat110 and FatICD were detected in immunoprecipitates of FatΔECD.
Acknowledgments
We thank Jeff Price, Markus Noll, Ken Irvine, David Strutt, DJ Pan and the DSHB for advice, antibodies, flies, and constructs. This work was funded by CIHR (FRN 84468) and CRS grants to HM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nature genetics. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 2.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 3.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Hariharan IK. Growth regulation: a beginning for the hippo pathway. Curr Biol. 2006;16:R1037–1039. doi: 10.1016/j.cub.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Bandura JL, Edgar BA. Yorkie and Scalloped: partners in growth activation. Developmental cell. 2008;14:315–316. doi: 10.1016/j.devcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 10.Pan D. Hippo signaling in organ size control. Genes & development. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 11.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development (Cambridge, England) 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 12.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. The Journal of biological chemistry. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 15.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer research. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 19.Casal J, Struhl G, Lawrence PA. Developmental compartments and planar polarity in Drosophila. Curr Biol. 2002;12:1189–1198. doi: 10.1016/s0960-9822(02)00974-0. [DOI] [PubMed] [Google Scholar]
- 20.Yang CH, Axelrod JD, Simon MA. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell. 2002;108:675–688. doi: 10.1016/s0092-8674(02)00658-x. [DOI] [PubMed] [Google Scholar]
- 21.Rawls AS, Guinto JB, Wolff T. The cadherins fat and dachsous regulate dorsal/ventral signaling in the Drosophila eye. Curr Biol. 2002;12:1021–1026. doi: 10.1016/s0960-9822(02)00893-x. [DOI] [PubMed] [Google Scholar]
- 22.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Developmental cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon MA. Planar cell polarity in the Drosophila eye is directed by graded Four-jointed and Dachsous expression. Development (Cambridge, England) 2004;131:6175–6184. doi: 10.1242/dev.01550. [DOI] [PubMed] [Google Scholar]
- 25.Matakatsu H, Blair SS. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development (Cambridge, England) 2004;131:3785–3794. doi: 10.1242/dev.01254. [DOI] [PubMed] [Google Scholar]
- 26.Klein TJ, Jenny A, Djiane A, Mlodzik M. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr Biol. 2006;16:1337–1343. doi: 10.1016/j.cub.2006.06.030. [DOI] [PubMed] [Google Scholar]
- 27.Strutt H, Price MA, Strutt D. Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila. Curr Biol. 2006;16:1329–1336. doi: 10.1016/j.cub.2006.04.041. [DOI] [PubMed] [Google Scholar]
- 28.Saburi S, Hester I, Fischer E, Pontoglio M, Eremina V, Gessler M, Quaggin SE, Harrison R, Mount R, McNeill H. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nature genetics. 2008;40:1010–1015. doi: 10.1038/ng.179. [DOI] [PubMed] [Google Scholar]
- 29.Zilian O, Frei E, Burke R, Brentrup D, Gutjahr T, Bryant PJ, Noll M. double-time is identical to discs overgrown, which is required for cell survival, proliferation and growth arrest in Drosophila imaginal discs. Development (Cambridge, England) 1999;126:5409–5420. doi: 10.1242/dev.126.23.5409. [DOI] [PubMed] [Google Scholar]
- 30.Guan J, Li H, Rogulja A, Axelrod JD, Cadigan KM. The Drosophila casein kinase Iepsilon/delta Discs overgrown promotes cell survival via activation of DIAP1 expression. Developmental biology. 2007;303:16–28. doi: 10.1016/j.ydbio.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai B, Villen J, Beausoleil S, Mintseris J, Gygi S. Phosphoproteome analysis of Drosophila melanogaster embryos. J Proteome Research. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma D, Yang CH, McNeill H, Simon MA, Axelrod JD. Fidelity in planar cell polarity signalling. Nature. 2003;421:543–547. doi: 10.1038/nature01366. [DOI] [PubMed] [Google Scholar]
- 33.Strutt H, Strutt D. Nonautonomous planar polarity patterning in Drosophila: dishevelled-independent functions of frizzled. Developmental cell. 2002;3:851–863. doi: 10.1016/s1534-5807(02)00363-5. [DOI] [PubMed] [Google Scholar]
- 34.Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development (Cambridge, England) 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- 35.Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Developmental cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Fanto M, Clayton L, Meredith J, Hardiman K, Charroux B, Kerridge S, McNeill H. The tumor-suppressor and cell adhesion molecule Fat controls planar polarity via physical interactions with Atrophin, a transcriptional co-repressor. Development (Cambridge, England) 2003;130:763–774. doi: 10.1242/dev.00304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Processed forms of Fat interact in vivo.
Extracts from animals bearing an N terminal Myc and C terminal HA-tagged UAS-Fat transgene and daughterless-gal4 driver (da-Gal4; UAS-MYCFatHA) were subjected to immunoprecipitation with either α-Myc or α-HA antibody and analyzed by immunoblotting with the reciprocal antibody (α-Myc or α–HA). Immunoprecipitation of full length Fat (Fat560) or Fat450 with α-Myc brought down Fat560 and Fat110. Immunoprecipitation of full length Fat (Fat560) or Fat110 with α-HA brought down Fat560 and Fat450.
Supplemental Figure 2. Fat processing is conserved to vertebrates
Total extracts from Embryonic Stem cells (ES cells) and E14.5 embryos were analyzed by Western blotting with antibodies generated to the extracellular domain or to the intracellular domain of Fat4. Note both antibodies recognize proteins of ~540kDa, while a ~150kDa protein is seen only with the antibody to the intracellular domain, similar to the pattern of Drosophila Fat. The antibody to the extracellular domain also recognizes a protein of ~400kDa, which may represent the extracellular cleaved fragment.
Supplemental Figure 3. fourjointed is upregulated in dcoi3-193 null clones
A) Similar to that reported for dco3, clones of dcoi3-193 (MARCM clones, positively marked by GFP) in eye imaginal discs show elevated levels of fourjointed (fj-lacZ, red) near the morphogenetic furrow.
B) Clones of dcoK38R (MARCM clones, positively marked by GFP) in eye imaginal discs also show elevated levels of fourjointed (fj-lacZ, red) near the morphogenetic furrow.
Supplemental Figure 4. The interaction between Fat and Casein Kinase Iδ/ε is conserved
A) Lysates from HEK293T cells expressing HA-tagged FatB and/or 3FLAG-tagged FatΔECD together with Dco-HA or Dco3-HA were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. FatB was detected in immunoprecipitates of FatΔECD, along with Dco or Dco3, when co-expressed.
B) Lysates from HEK293T cells expressing a HA-tagged version of the Fat4 cytoplasmic domain fused to the Drosophila fat membrane targeting sequence together with 3FLAG-tagged CKIε (lane 2), CKIδ (lane 3), or Yap (lane 4), were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. Note the slower migrating forms of Fat4 in lanes 2 and 3 of the anti-HA immunoblot of totals (lower panel).
Supplemental Figure 5. Potential CKI phosphorylation sites in Fat
Amino acid sequence of Fat transmembrane domain (green) and Fat intracellular domain. Highlighted in red are regions adhering to the classical CKI consensus (S/T-X-X-S/T) and are potential CKI phosphorylation sites. Underlined and with asterisks are phosphorylated residues identified in vivo (Zhai et al, 2008).
Supplemental Figure 6. Fat110 is altered in ds mutants but not other growth or PCP mutants.
Extracts from yw (lane 1), fat (ft; lane 2), four-jointed (fj; lane 3), expanded (ex; lane 4), sprouty (sty; lane 5), and dachsous (ds; lane 6) mutant larvae were subjected to SDS-PAGE and analyzed by immunoblotting with α-Fat (IC) antibody. Fat110 mobility is altered in ds mutants.
Supplemental Figure 7. Fat can oligomerize.
Lysates from Drosophila S2 cells expressing HA-tagged full length Fat, FatB, or FatICD, and 3FLAG-tagged FatΔECD were subjected to immunoprecipitation with α-FLAG antibody and analyzed by immunoblotting with the indicated antibodies. Fat110 and FatICD were detected in immunoprecipitates of FatΔECD.