Abstract
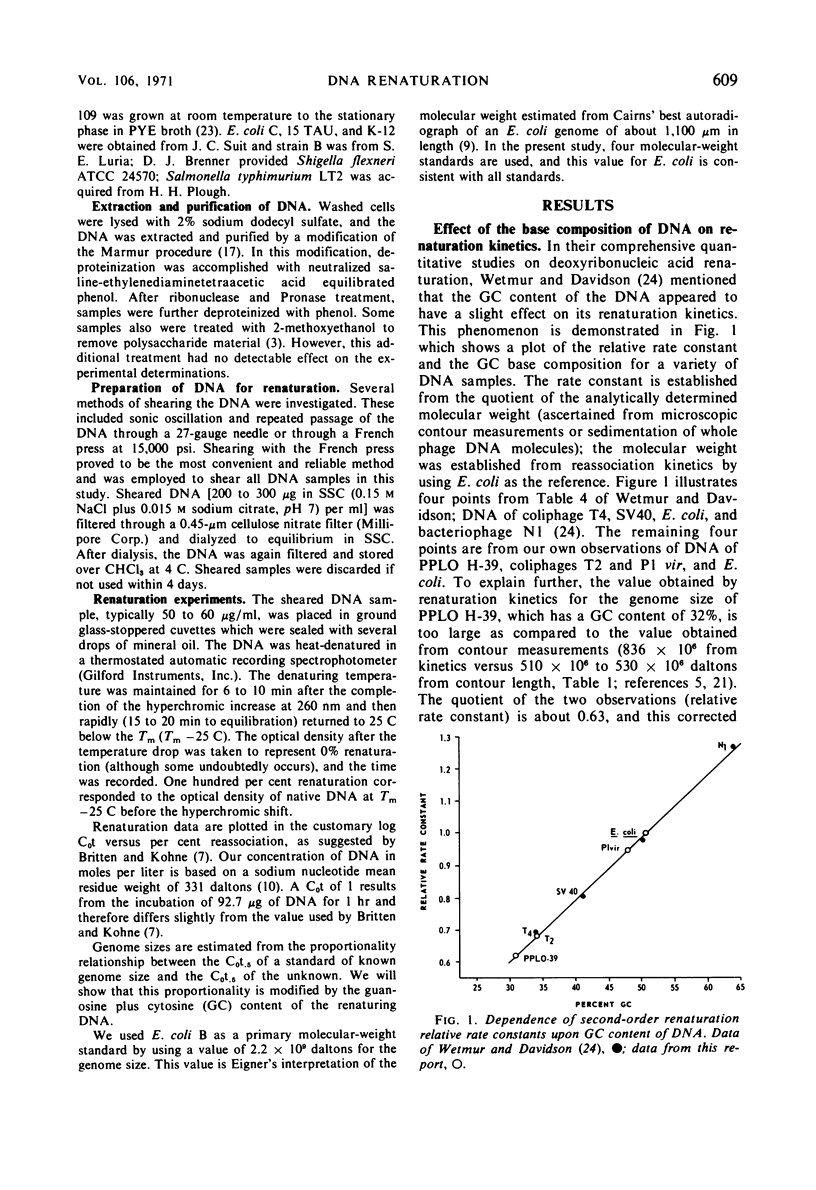
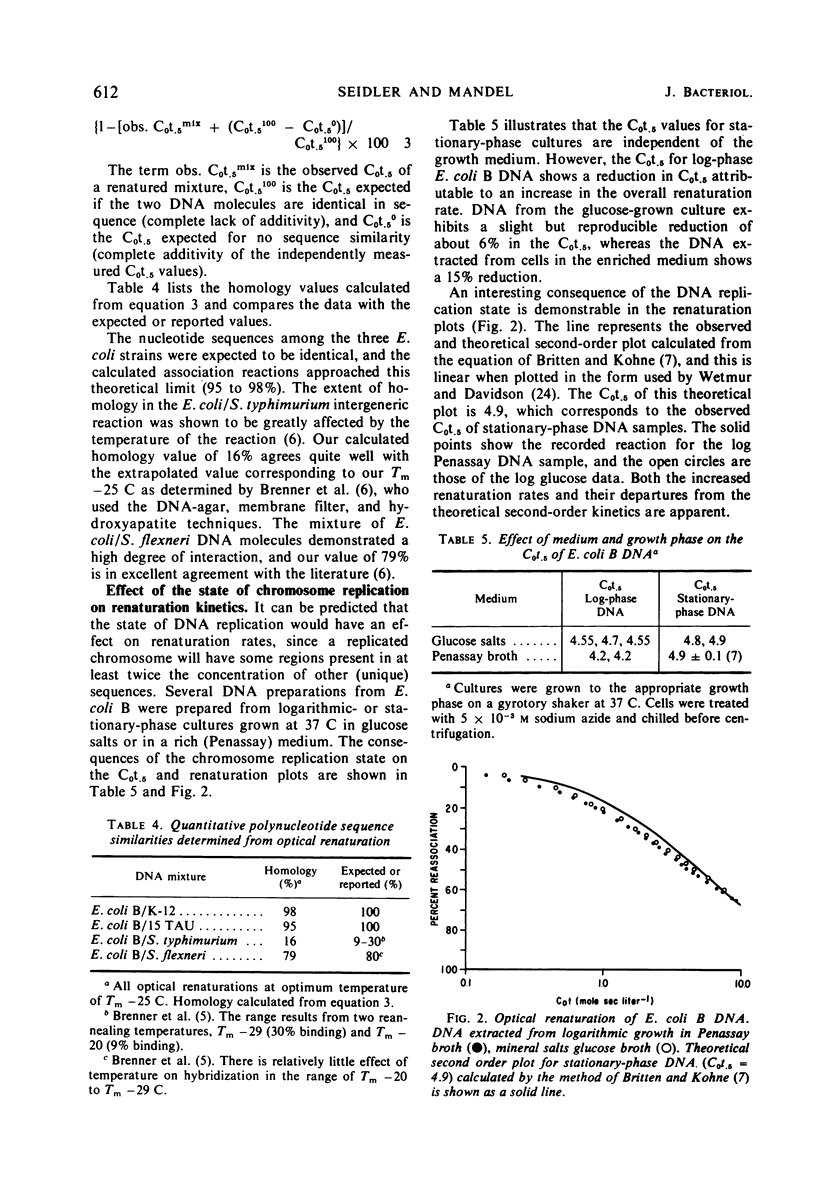
The base composition of a deoxyribonucleic acid (DNA) sample affects its intrinsic rate of renaturation. In agreement with the information of Wetmur and Davidson, it was established that high guanosine plus cytosine (GC) DNA renatures faster than expected from analytical measurement of its molecular weight. A calculated correction factor of 1.8% of the observed C0t.5 is required for every mole per cent GC difference from 51% GC. The correction factor is now established in the range of 32 to 65% GC. Renaturation of DNA mixtures prepared from pairs of organisms has been studied. When no similarity existed between the two organisms, the observed C0t.5 of the mixture was the sum of the independently determined C0t.5 values. Lack of additivity was correlated with similarities in polynucleotide sequence of the reassociating DNA molecules. A quantitative relationship was formulated to relate C0t.5 values of renatured DNA mixtures to per cent binding (“homology”). Finally, it was demonstrated that DNA prepared from log-phase cells renatures faster than stationary-phase DNA and also departs from theoretical second-order kinetics.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bak A. L., Black F. T., Christiansen C., Freundt E. A. Genome size of mycoplasmal DNA. Nature. 1969 Dec 20;224(5225):1209–1210. doi: 10.1038/2241209a0. [DOI] [PubMed] [Google Scholar]
- Ballard R. W., Palleroni N. J., Doudoroff M., Stanier R. Y., Mandel M. Taxonomy of the aerobic pseudomonads: Pseudomonas cepacia, P. marginata, P. alliicola and P. caryophylli. J Gen Microbiol. 1970 Feb;60(2):199–214. doi: 10.1099/00221287-60-2-199. [DOI] [PubMed] [Google Scholar]
- Bicknell J. N., Douglas H. C. Nucleic acid homologies among species of Saccharomyces. J Bacteriol. 1970 Feb;101(2):505–512. doi: 10.1128/jb.101.2.505-512.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode H. R., Morowitz H. J. Size and structure of the Mycoplasma hominis H39 chromosome. J Mol Biol. 1967 Jan 28;23(2):191–199. doi: 10.1016/s0022-2836(67)80026-3. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Johnson K. E., Citarella R. V., Falkow S. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol. 1969 May;98(2):637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Hirschman S. Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J Mol Biol. 1965 Sep;13(2):407–427. doi: 10.1016/s0022-2836(65)80106-1. [DOI] [PubMed] [Google Scholar]
- Gillis M., De Ley J., De Cleene M. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970 Jan;12(1):143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Helmstetter C. E. DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J Mol Biol. 1968 Feb 14;31(3):507–518. doi: 10.1016/0022-2836(68)90424-5. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- Johnson J. L., Ordal E. J. Deoxyribonucleic acid homology in bacterial taxonomy: effect of incubation temperature on reaction specificity. J Bacteriol. 1968 Mar;95(3):893–900. doi: 10.1128/jb.95.3.893-900.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T. Estimate of the genome size of various microorganisms. J Bacteriol. 1969 Jun;98(3):1400–1401. doi: 10.1128/jb.98.3.1400-1401.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Mandel M. New approaches to bacterial taxonomy: perspective and prospects. Annu Rev Microbiol. 1969;23:239–274. doi: 10.1146/annurev.mi.23.100169.001323. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Base sequence homology and renaturation studies of the deoxyribonucleic acid of extremely halophilic bacteria. J Bacteriol. 1969 Jul;99(1):255–262. doi: 10.1128/jb.99.1.255-262.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowitz H. J., Bode H. R., Kirk R. G. The nucleic acids of mycoplasma. Ann N Y Acad Sci. 1967 Jul 28;143(1):110–114. doi: 10.1111/j.1749-6632.1967.tb27650.x. [DOI] [PubMed] [Google Scholar]
- Rogul M., Brendle J. J., Haapala D. K., Alexander A. D. Nucleic acid similarities among Pseudomonas pseudomallei, Pseudomonas multivorans, and Actinobacillus mallei. J Bacteriol. 1970 Mar;101(3):827–835. doi: 10.1128/jb.101.3.827-835.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler R. J., Starr M. P. Isolation and characterization of host-independent Bdellovibrios. J Bacteriol. 1969 Nov;100(2):769–785. doi: 10.1128/jb.100.2.769-785.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- YOSHIKAWA H., O'SULLIVAN A., SUEOKA N. SEQUENTIAL REPLICATION OF THE BACILLUS SUBTILIS CHROMOSOME. 3. REGULATION OF INITIATION. Proc Natl Acad Sci U S A. 1964 Oct;52:973–980. doi: 10.1073/pnas.52.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]



