Abstract
The structural underpinnings of enzyme substrate specificity are investigated in a pair of copper amine oxidases (CAOs) from Hansenula polymorpha (HPAO-1 and HPAO-2). The X-ray crystal structure (to 2.0 Å resolution) and steady state kinetic data of the second copper amine oxidase (HPAO-2) are presented for comparison to HPAO-1. Despite 34 % sequence identity and superimposable active site residues implicated in catalysis, the enzymes vary considerably in their substrate entry channel. The previously studied CAO, HPAO-1, has a narrow substrate channel. In contrast HPAO-2 has a wide funnel-shaped substrate channel, which also contains a side-chamber. In addition, there are a number of amino acid changes within the channels of HPAO-2 and HPAO-1 that may sterically impact the ability of substrates to form covalent Schiff base catalytic intermediates and to initiate chemistry. These differences can partially explain the greatly different substrate specificities as characterized by kcat/Km value differences: in HPAO-1, the kcat/Km for methylamine is 330-fold greater than for benzylamine, whereas in HPAO-2 it is benzylamine that is the better substrate by 750-fold. In HPAO-2 an inflated Dkcat/Km(methylamine) in relation to Dkcat/Km(benzylamine) indicates that proton abstraction has been impeded more than substrate release. In HPAO-1, Dkcat/Km(S) changes little with the slow substrate, and indicates a similar increase in the energy barriers that control both substrate binding and subsequent catalysis. In neither case is kcat/Km for the second substrate, O2, significantly altered. These results reinforce the modular nature of the active sites of CAOs and show that multiple factors contribute to substrate specificity and catalytic efficiency. In HPAO-1, the enzyme with the smaller substrate binding pocket, both initial substrate binding and proton loss are affected by an increase in substrate size, while in HPAO-2, the enzyme with the larger substrate binding pocket, the rate of proton loss is differentially affected when a phenyl substituent in substrate is reduced to the size of a methyl group.
Copper amine oxidases (CAOs)1 are virtually ubiquitous in aerobic organisms, and catalyze the oxidative deamination of primary amines in the following overall reaction:
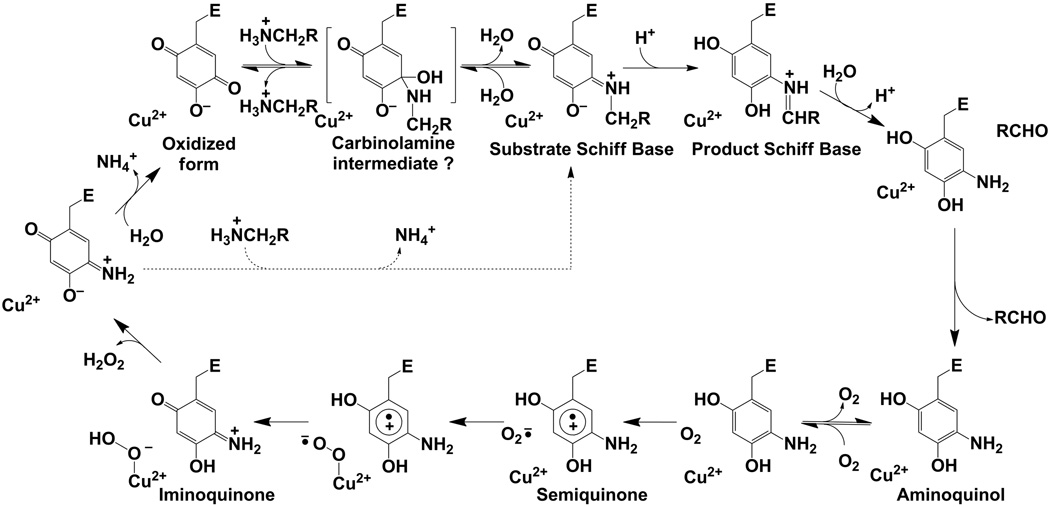
While usually sharing only 20–40 % amino acid sequence identity, features of the active sites are almost identical across CAOs from widely varying species. These include the protein-derived enzymatic cofactor, 2,4,5-trihydroxyphenylalanine quinone (TPQ), an active site metal, copper, and its three histidine ligands (1, 2). The CAO mechanism is ping-pong, and proceeds via Schiff base chemistry creating a covalent adduct of the amine substrate with the TPQ (Scheme 7). A conserved Asp residue acts as the catalytic base. The release of aldehyde product at the end of the enzymatic reductive half-reaction leaves the cofactor as a two-electron reduced aminoquinol containing a substrate-derived nitrogen. In the oxidative half-reaction the reduced enzyme is converted to a one-electron reduced N-semiquinone and then an oxidized iminoquinone through the reduction of molecular oxygen to hydrogen peroxide. This is followed by hydrolysis of the iminoquinone to regenerate the resting TPQ quinone and release ammonia.
Scheme 7.
Proposed reaction mechanism for HPAO-1, indicating intermediates along the pathway. Brackets indicate the probable, but unconfirmed, existence of a carbinolamine intermediate. The dashed arrow indicates that substrate can react directly with the iminoquinone to form the substrate Schiff base when substrate concentrations are high.
Despite these similarities, CAOs from different organisms preferentially react with primary amine structures that range from small aliphatic amines to peptides. The CAO from Escherichia coli, for example, prefers aromatic monoamines (3), while the previously characterized CAO from Hansenula polymorpha (HPAO-1) is most active against small aliphatic amines (4). The structural basis for this difference in substrate specificity has remained elusive.
Copper amine oxidases are not unique in their variable substrate specificity. Among other classes of enzymes, e.g., aminotransferases (5) and flavin-containing monoamine oxidases (MAOs) (6), the basis of isoenzyme specificity has been explored. Protein conformational flexibility has been suggested as a source of substrate specificity in aspartate aminotransferase and aromatic amino acid aminotransferase (7), whereas differences in static hydrophobic and aromatic π-π interactions in the substrate binding pocket have been implicated in MAO-A (8). It is rare for paralogous enzymes to demonstrate a strong yet inverted discrimination between two different substrates. Thus, most investigations have focused on understanding the relative rates of preferred over poor substrates in a single enzyme, rather than between isoenzymes.
The first described CAO from the methylotrophic yeast H. polymorpha (HPAO-1) was heterologously expressed in Saccharomyces cerevisiae and purified in 1994 (4) and subsequently studied by X-ray crystallography (9). Although a peroxisomal protein in its native host organism, it was found in the cytosol of S. cerevisiae during recombinant expression (10). Characterized as a “methylamine oxidase,” HPAO-1 demonstrated much greater activity against methylamine than benzylamine. However, CAO extraction from H. polymorpha grown on benzylamine-enriched media had indicated the presence of a second CAO with a distinct catalytic activity towards benzylamine (4). In the case of mammals, multiple CAO isoenzymes have also been established, annotated as aoc-1, aoc-2 and aoc-3 in the human genome (11). Comparison of the enzymatic activities of the human kidney diamine oxidase (12) to human vascular adhesion protein-1 (13) indicates broad, though largely non-overlapping substrate specificities. Although these two human enzymes have also been characterized structurally (14–16), their broad substrate specificities make it difficult to rationalize the features that control substrate discrimination.
In this work, we report detailed kinetic and crystallographic characterization of a second copper amine oxidase from H. polymorpha (HPAO-2) for comparison with HPAO-1. In particular, kcat/Km data indicate a marked reversal of substrate selectivity between the two isoenzymes when their activities are interrogated with a small aliphatic vs. an aromatic amine. Using the X-ray crystal structure of HPAO-2 reported herein to compare to that of HPAO-1, we can begin to decipher the structural origins of this divergent substrate specificity. Residue changes between HPAO-1 and HPAO-2 that likely play roles in determining substrate specificity have been identified close to the active site and are discussed in the context of the available kinetic parameters and corresponding isotope effects.
MATERIALS AND METHODS
Cloning
Due to the unavailability of the H. polymorpha genome at the start of the investigation, the hpao-2 gene (1967 base pairs) was cloned out of the H. polymorpha genome by “genome walking,” using iterations of primers to progressively amplify the entire gene. Initially, mass spectrometry of trypsin-digested protein isolated from cell extracts of H. polymorpha grown on benzylamine-enriched growth media that demonstrated CAO activity, yielded peptide sequences distinct from that of the original HPAO (HPAO-1). Degenerated primers were designed based on the new peptide sequences and subsequent PCR resulted in a 600 bp gene fragment of hpao-2. The 5' end and 3' end sequences of the fragment were then used for upstream and downstream genome walking to yield 1967 base pairs of full length hpao-2 sequence. The identity of the full-length gene was verified by comparison to the now available H. polymorpha genome (Rhein Biotech, personal communication). The hpao-2 gene was inserted into the S. cerevisiae expression vector, pYES2 (Invitrogen), under inducible control by the GAL1 promoter, via cloned KpnI and XbaI restriction enzyme cut sites present in the multiple cloning site. The full DNA-derived protein sequence and its comparison to HPAO-1 are given in Figure S1 of Supporting Information.
Expression
Invitrogen’s INVSC-1 S. cerevisiae cells were transformed with selection for uracil-independent growth, conferred by the ura3 gene on pYES2. Importantly, S. cerevisiae contains no CAO genes, and thus no background CAO expression. Starter cultures were grown in URA− minimal media containing 6.7 % yeast nitrogen base without amino acids, 0.77 g/L complete supplemental mixture minus uracil (MP Biomedicals) with 2 % raffinose for carbon source at 30°C and 225 rpm in a New Brunswick Scientific Innova 4300 temperature controlled platform shaker. At an OD600 of ~ 5, cells were diluted to an OD600 of 0.4 into URA− minimal media with 30 µM CuSO4, 1 % raffinose and 2 % galactose for induction of expression. Induced cultures were grown in 1.5 L volumes in 4 L flasks. After 24 hours induction cells were pelleted by centrifugation in a Sorvall RC 5C Plus centrifuge at 5,000 rpm in an SLA-3000 fixed angle rotor for 5 min at 4 °C. Typically, 9 L of cell culture yields 60–70 g of cell pellet.
Purification
Protein purification followed the previously developed purification strategy for HPAO-1 with some modifications (4). Cells were lysed with glass beads (425–600 µm, Sigma G8772) in the presence of four protease inhibitors: 0.5 mM phenylmethylsulfonyl fluoride, 5 mM 1,10-phenanthroline, 1 µM pepstatin A, and 5 µM E-64 (Sigma). Approximately 150 ml glass beads were combined with a ~ 250 ml suspension of cell pellet and cold 10 mM potassium phosphate, pH 7.2 in a bead beater. Cells were lysed by five cycles of grind for 5 min, rest for 2 min, with changes in the ice and water surrounding the bead-beating chamber between cycles. The cell extract was then separated from cellular debris by centrifugation in a Sorvall RC 5C Plus centrifuge in ~ 40 ml aliquots at 12,000 rpm in an SS-34 fixed angle rotor at 4°C for 1 hr.
The lysate was loaded onto a ~ 200 ml (5 cm diameter, ~ 20 cm height) Q Sepharose column, pre-equilibrated with 5 mM potassium phosphate, pH 7.2. This column was washed with 1 L of 5 mM potassium phosphate, pH 7.2. Protein was eluted as follows: 300 ml total volume of a linear gradient of 5 to 100 mM potassium phosphate, pH 7.2, followed by 200 ml of 100 mM potassium phosphate, pH 7.2, then 300 ml total volume of a linear gradient of 100 to 400 mM potassium phosphate, pH 7.2, and finally 300 ml of 400 mM potassium phosphate, pH 7.2. 10 ml fractions were collected at 1 min/fraction (elution by gravity). The column was washed with 1 L of 400 mM potassium phosphate + 1 M NaCl between preparations and then re-equilibrated with ~ 1 L of 5 mM potassium phosphate, pH 7.2.
Fractions 28–58 were pooled at 4°C and dialyzed (Pierce SnakeSkin Pleated Dialysis Tubing 10,000 MWCO #68100) into 4 L of 50 mM potassium phosphate, pH 7.2 overnight at 4°C. Dialyzed pooled fractions were then centrifuged at 12,000 rpm (1 hr, 4°C) to separate out precipitated protein. Cleared pooled fractions were concentrated to approximately 6 ml using Amicon concentrators with polyethersulfone, 50,000 NMWL ultrafiltration membranes.
2 ml aliquots of the concentrated pool were centrifuged at 13,000 rpm for 5 min, 4°C, in a benchtop microcentrifuge to clear any precipitation prior to loading onto an S-300 column. This was run overnight, by elution with 50 mM potassium phosphate, pH 7.2. Fractions were collected every 10 min at a flow rate of 0.2 ml/min (for ~ 2 ml/fraction).
Pooled fractions 46–58 were concentrated using an Amicon concentrator with polyethersulfone, 50,000 NMWL ultrafiltration membranes. After reaching ~ 2–3 ml final volume, 50–100 µl aliquots of purified protein were made, snap frozen in liquid nitrogen and placed at −20°C for short-term storage or −80°C for long-term storage. Concentrated purified HPAO-2 samples were peach in color.
Protein Characterization
Total protein concentration was measured by Bradford assay (Bio-Rad). Typical total protein yields varied from 150 to 300 mg per 9 L cell culture (at a 90 % purity, as judged by SDS-PAGE).
The presence of a quinone cofactor in HPAO-2 was tested by both quinone stain and phenylhydrazine assay. Reaction with phenylhydrazine resulted in the formation of a peak at 448 nm, as previously observed in HPAO-1 (4). The very rapid formation of the product phenylhydrazone at room temperature in 100 mM potassium phosphate, pH 7.2, necessitated the use of a Hi-Tech Scientific stopped flow instrument to estimate a rate constant. The concentration of TPQ was further quantified by reaction with phenylhydrazine HCl. The phenylhydrazine assay used a 5-fold molar excess of a freshly prepared solution of phenylhydrazine HCl (in 100 mM potassium phosphate, pH 7.2) to HPAO-2 protein concentration, as determined by Bradford assay. The change in absorbance at 448 nm was measured on a Hewlett-Packard 8452A diode-array spectrophotometer. An extinction coefficient previously determined for HPAO-1 (40,500 M−1cm−1) and a subunit molecular weight of 72,000 were used to calculate the percentage of TPQ present in protein (17). The presence of TPQ cofactor was also observed by redox stain, specifically via reaction of the protein-bound quinone with nitroblue tetrazolium (NBT) (17). After separation of HPAO-2 from contaminants by SDS-PAGE, protein was electroblotted onto nitrocellulose membrane and washed in the dark with 0.24 mM NBT in 2 M sodium glycinate (pH 10).
Purified HPAO-2 was N-terminal sequenced at the Stanford PAN facility (http://cmgm.stanford.edu/pan/) by Edman degradation.
Kinetic Measurements
Methylamine, [1,1,1,2H3] methylamine, and benzylamine hydrochlorides were purchased from Sigma and used without further purification. [1,1-2H2] benzylamine hydrochloride was prepared as previously described (18). Purity of synthesized [1,1-2H2] benzylamine hydrochloride and commercial [1,1,1,2H3] methylamine were verified by NMR with no evidence for contamination by protium substrate found. In all cases with protio- and deuterio- substrates, NMR spectra also indicated no significant level of chemical contaminant.
Observation of benzyaldehyde formation from benzylamine (19) served as a preliminary enzyme activity assay during the purification of HPAO-2 and in subsequent kinetic studies of benzylamine. Benzyaldehyde absorbs at 250 nm (ε = 12,800 M−1 cm−1), and its production is easily followed on a Hewlett-Packard 8452A diode-array spectrophotometer. Enzyme activity varying methylamine was monitored by oxygen consumption with a Clark oxygen electrode. Unless otherwise noted, all kinetic studies were conducted at 25 °C, in 100 mM potassium phosphate, pH 7.2, controlled for 300 mM ionic strength by the addition of KCl (4). In HPAO-2 assays, the range of concentration for amine substrates was: 0.5–20 mM for methylamine and 1–200 µM for benzylamine. In HPAO-1, 0.05–1.5 mM methylamine and 0.2–5 mM benzylamine were used for steady state kinetic rate parameter determination. Buffers used for the pH studies were: 100 mM potassium phosphate, pH 6.2–8.2, 25 mM sodium pyrophosphate, pH 8.6, and 0.1 M potassium carbonate, pH 9.2–9.5 (Sigma). Ionic strength was maintained at 300 mM with KCl for potassium-based buffers, and NaCl for sodium-based buffer. These were carried out at 1–150 µM benzylamine at 25°C. Enzyme activity varying oxygen, at saturating amine concentrations, for determination of kcat/Km(O2) values was monitored by oxygen consumption with a Clark oxygen electrode. The kinetic parameters, kcat and kcat/Km, were derived from data fit to the Michaelis-Menton equation by nonlinear regression using Kaleidagraph (Abelbeck Software).
Crystallization
HPAO-2 in 50 mM potassium phosphate, pH 7.2, was buffer exchanged into 20 mM HEPES, pH 7.0, and concentrated to 23 mg/ml for crystallization. HPAO-2 crystals were grown by sitting drop vapor diffusion using a 1:1 volume ratio (6 µL total) of purified HPAO-2 and mother liquor solution (0.50–0.75 M potassium sodium tartrate tetrahydrate in 0.10 M phosphate, pH 6.0 ȓ7.5) at 20 °C. Crystals grew as clusters and physical manipulation was necessary to separate individual crystals. Crystals were soaked in 25 % high purity glycerol (Hampton Research) mixed with mother liquor taken directly from the crystallization well for 5 min prior to flash-freezing in liquid nitrogen.
Structure Determination and Refinement
X-ray diffraction data were collected from a single crystal at 100 K using an undulator source (wavelength of 0.979 Å) and an ADSC Quantum 315r detector at the Advanced Photon Source, Argonne National Laboratory (Beamline 19-ID, SBC-CAT). Data were processed using HKL2000 and SCALEPACK (20). Molecular replacement was carried out using PHASER and MOLREP from the CCP4 suite (21) with a polyalanine truncated search model based on a previously deposited HPAO-1 model (PDB ID: 2oov) (22).
An initial model was built into the experimental electron density and manual adjustments were carried out in Coot (23). Model refinement was performed using Refmac5 with 5 % of data excluded from refinement to enable calculation of a free R-factor (24). ARPWaters was used to place water molecules into the model at peaks greater than 3.0σ in the 2Fo - Fc map (25). Cycles of model building and refinement were performed until Fo - Fc difference peaks did not make structural sense, and appeared to be within the noise level of the electron density map.
RESULTS
Characterization of HPAO-2 and Comparison to HPAO-1
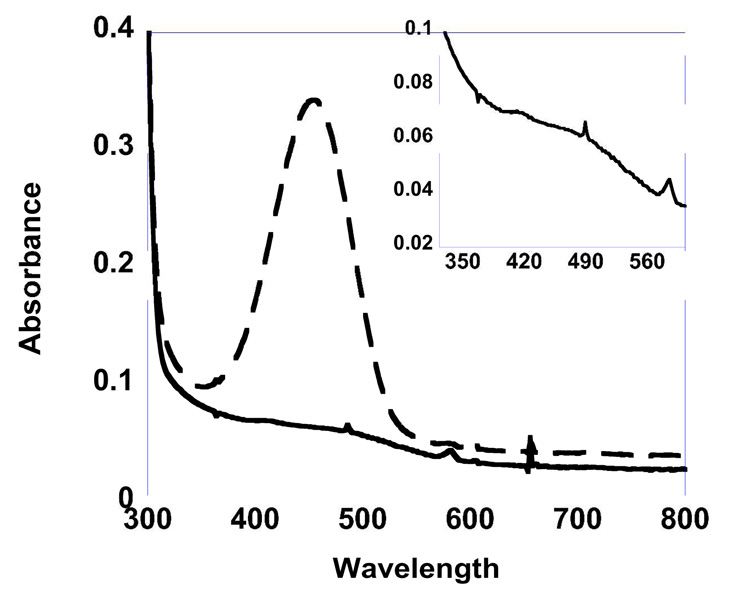
Cell extracts at 24 hrs of induction contained a band at ca. 72 kDa that is absent in the non-transformed control yeast cell extracts. The size of the protein corresponds to the estimated size of HPAO-2 calculated from the primary sequence. After purification, the resulting protein constituted > 90 % of total protein, as observed by SDS-PAGE (Supporting Information Figure S2). The net effect of the purification protocol was to increase the relative amounts of HPAO-2 to the contaminant bands. Some of the contaminant bands also stained on an NBT blot, indicating probable breakdown of HPAO-2 (data not shown). HPAO-1 migrates slightly faster by SDS-PAGE, suggesting an overall smaller primary structure; however, the DNA-derived primary protein sequence of HPAO-1 predicts a slightly larger protein than HPAO-2. Post-translational modifications may account for this difference as HPAO-2 has an additional predicted glycosylation site. Concentrated purified HPAO-2 was a vibrant peach color, characterized by a broad shoulder in the UV/Vis around 480 nm (Figure 1). N-terminal sequencing of the protein matched the expected N-terminus from the HPAO-2 DNA sequence, except for the absence of the initial Met (Supporting Information Figure S3). The N-terminus of HPAO-1 is longer by 20 amino acids and contains a peroxisomal targeting sequence (Supporting Information Figure S3).
Figure 1.
UV-visible spectra of 20 µM HPAO-2 before (bold line) and after (dashed line) 5 min incubation with 100 µM phenylhydrazine HCl in 100 mM potassium phosphate buffer, pH 7.2 at 25°C. Inset shows 20 µM HPAO-2 before incubation with pheynylhydrazine HCl blown up to show a shoulder at 480 nm, as observed in other TPQ containing enzymes.
The presence of a quinone cofactor was tested by both quinone stain and phenylhydrazine assay. Nitroblue tetrazolium staining resulted in a purple band similar to that observed with HPAO-1 (data not shown). Reaction with phenylhydrazine resulted in the rapid formation of a peak in the region seen previously with other CAOs (~450 nm) corresponding to the phenylhydrazone (Figure 1). The rate of phenylhydrazone formation in HPAO-2 (87 ± 3 s−1) is over 104 × faster than that of HPAO-1 (7.8 × 10−3 ± 7.0 × 10−4 s−1). Typical yields of phenylhydrazone correspond to ~ 50 % of subunit concentration.
Copper content of purified protein was measured by elemental analysis. Initial analyses indicated 2 moles of copper per monomer. However, after overnight dialysis against 1 mM EDTA, followed by dialysis against 0.1 M potassium phosphate buffer, pH 7.2, the expected one copper per monomer was observed. The presence of the loosely bound second copper did not affect enzyme turnover rates, as kinetic measurements performed with the dialyzed enzyme yielded the same rates as those with non-dialyzed enzyme samples (Supporting Information Table S1). Thus, there appears to be an adventitious metal binding site that plays no role in catalysis.
Kinetic Properties
The near-unity Dkcat values observed for HPAO-2 with both substrates indicate that proton abstraction is not significantly rate-limiting with either substrate (Table 1). The lower limit for the rate constant reflecting the proton abstraction step is thus defined by the respective kcat values. Conversely, HPAO-1 has an elevated Dkcat for the slower substrate benzylamine (Dkcat(benzylamine) is 5.9, whereas Dkcat(methylamine) is 1.7), pointing to a higher relative energy barrier for proton loss from the poor substrate relative to other steps following substrate binding. The overall turnover rate (kcat) for benzylamine is also down 94-fold relative to methylamine. Stopped flow kinetic studies of HPAO-1 have confirmed the oxidative half-reaction is the major determinant of rate during turnover with methylamine (26).
Table 1.
Steady State Kinetic Parameters for HPAO-1 and HPAO-2 at 25 °C, 100 mM Potassium Phosphate, pH 7.2, Ionic Strength Maintained at 300 mM with KCl.
| kcat (s−1) | kcat/Km(S)a M−1s−1 | kcat/Km(O2)b M−1s−1 | Dkcat | Dkcat/Km(S) | |
|---|---|---|---|---|---|
| HPAO-2 | |||||
| Benzylamine | 8.1 ± 0.2 | 9 ± 1 × 105 | 4.7 ± 0.6 × 105 | 1.00 ± 0.04 | 2.0 ± 0.3 |
| Methylamine | 2.18 ± 0.06 | 1.2 ± 0.1 × 103 | 1.6 ± 0.3 × 105 | 1.36 ± 0.08 | 18.5 ± 0.1 |
| HPAO-1 | |||||
| Benzylamine | 6.6 ± 0.3 × 10−2 | 9 ± 1 × 101 | 8.1 ± 2.1 × 104 | 5.9 ± 0.7 | 3 ± 1 |
| Methylaminec | 6.2 ± 0.2 | 3.0 ± 0.6 × 104 | 4 ± 1 × 105 | 1.7 ± 0.1 | 4.3 ± 0.2 |
Amine substrate.
Measured at saturating amine substrate and varying oxygen. This led to some variation in ionic strength: HPAO-2, benzylamine, µ = 250 mM and methylamine, µ = 300 mM; HPAO-1, benzylamine, µ = 257 mM and methylamine, µ = 250 mM.
Data from ref 26; experimental conditions identical to those herein, except at pH 7.0 instead of pH 7.2.
Overall, the changes in kcat between the different amine substrates are smaller than the changes in kcat/Km(S) (Table 1). The second order rate constant reflects turnover under low substrate conditions and varies by 750-fold between the two substrates for HPAO-2. The change in kcat/Km(S) between substrates observed with HPAO-1 is similar, but somewhat smaller (330-fold). As such, HPAO-2 shows a clear preference for bulkier aromatic amines and HPAO-1 shows a preference for short aliphatic amines. These differences in kcat/Km(S) originate from substrate-dependent rate differences in one or more steps that occur between binding of substrate to enzyme up to and including the first irreversible step (Scheme 7).
In contrast, kcat/Km(O2) changes little between the two substrates and two isoenzymes (Table 1). This suggests that steps from oxygen binding through the irreversible oxygen reduction step vary little between the two substrates, as expected for the ping-pong mechanism proposed for HPAO-1 (Scheme 7). The small reduction in kcat/Km(O2) for methylamine turnover (as compared to benzylamine) in HPAO-2 could suggest oxygen binding to an enzyme form that has retained formaldehyde and is unreactive to O2 until product is released. Bound aldehyde product has been shown to significantly impede the oxidative half-reaction in E. coli CAO crystals, where the restraints imposed by the lattice lead to very slow product release (27).
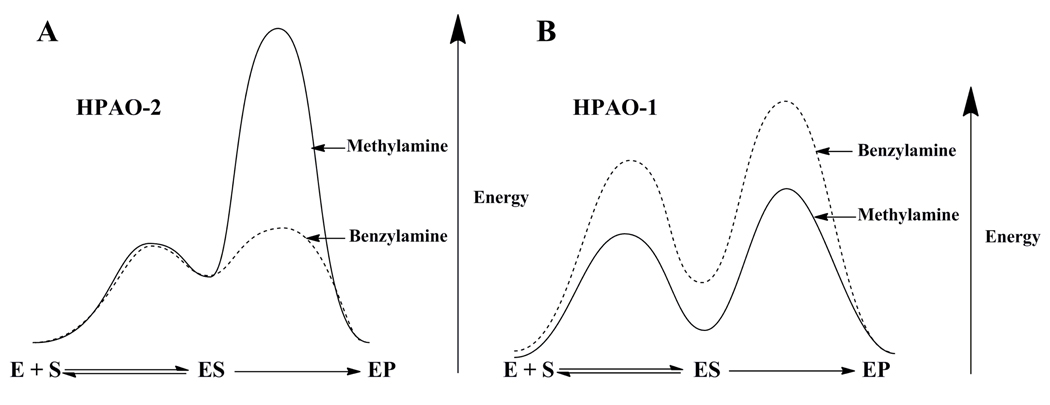
Comparison of isotope effects on kcat/Km(S) on the two isoenzymes with both substrates demonstrate that the extent of rate limitation by proton abstraction differ on steps in the reductive half-reaction. Isotope effects on kcat/Km(S) with HPAO-2 indicate that proton abstraction is clearly more ratelimiting for methylamine than benzylamine, as Dkcat/Km(S) is 2.0 for benzylamine and 18.5 for methylamine (Table 1). The increase in contribution of the proton abstraction step to kcat/Km is likely due to an increase in the free energy barrier for proton abstraction, rather than a decrease in the barrier for methylamine binding (Scheme 8A), as this substrate is smaller than the preferred substrate benzylamine, and thus should have no trouble accessing the active site. In the case of HPAO-1, the similar Dkcat/Km values (benzylamine 3.0 vs. methylamine 4.3, Table 1) indicate that proton abstraction is similarly rate-limiting with respect to substrate binding for both substrates, despite the large reduction in rate when using benzylamine as substrate. This implies that both substrate binding/release and proton abstraction are affected to a similar extent (Scheme 8B).
Scheme 8.
Proposed free energy diagrams that represent kcat/Km(S). A. HPAO-2. B. HPAO-1.
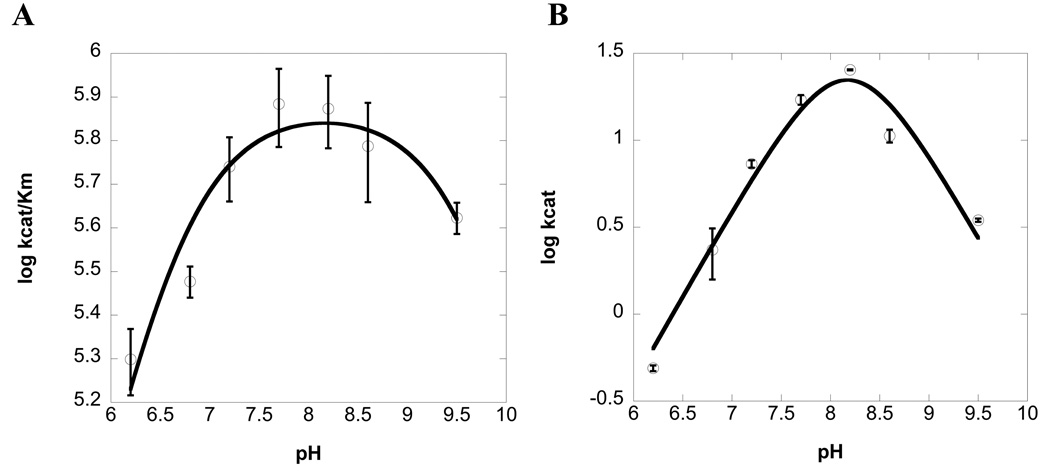
The impact of pH on catalysis has also been examined for HPAO-2 (benzylamine as substrate) for comparison to HPAO-1 (methylamine as substrate (28)). As summarized in Figure 2, the pH optima for both kcat/Km(S) and kcat are ca. 8–8.2 and similar to HPAO-1 (28). The fact that these pH optima are elevated above pH 7 is consistent with the fact that HPAO-1 is found in the peroxisome with a pH of 8.2 in S. cerevisiae (29). The intracellular location of HPAO-2 is unknown, however the absence of an extended N-terminus in relation to HPAO-1 suggests the site of function may be cytosolic (Supporting Information Figure S3).
Figure 2.
HPAO-2 pH profiles of (a) kcat/Km(benzylamine) (pKa1 = 6.6, pKa2 = 9.7), and (b) kcat(pKa1 = 6.0, pKa2 = 7.6, pKa3 = 8.9).
Crystal Structure of HPAO-2 in comparison to HPAO-1: Overall fold and active site
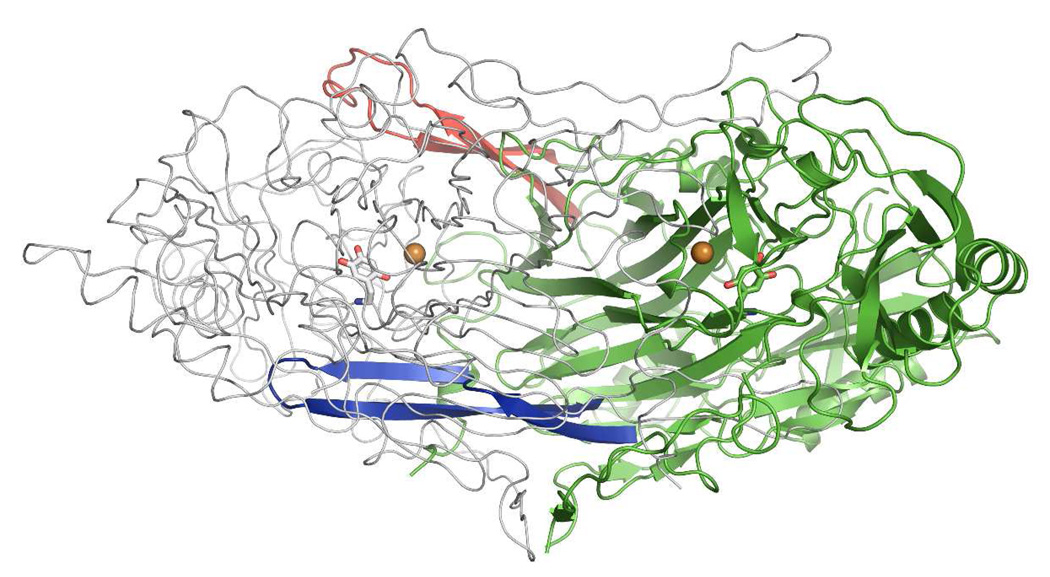
Table 2 contains the data collection, processing and refinement statistics for HPAO-2 to a resolution of 2.0 Å. Final R−-work and R-free values are 14.5 % and 19.1 %, respectively (for quality of electron density see Supporting Information Figure S4). Three polypeptide chains, or 1.5 physiological HPAO-2 dimers, were seen in the crystallographic asymmetric unit (ASU) in space group C2. The overall structure of the HPAO-2 dimer is identical in fold to HPAO-1 and other CAOs (Figure 3, Supporting Information Figure S5) (2, 9, 14–16, 30–33). Superimposition of the HPAO-2 and HPAO-1 homodimer yields an rmsd for main chain atoms of 0.99 Å (Supporting Information Figure S6). HPAO-2 consists of three domains arranged along its primary sequence (D2-D4). The amine oxidase from E. coli contains an additional protruding “stalk” domain (D1) at the N-terminus that is not present in the other CAO crystal structures (2). Two small α/β domains (D2 and D3) are formed by the N-terminal portion of HPAO-2, while the larger catalytic domain (D4) is composed of the C-terminal portion of the chain. As seen in HPAO-1, the catalytic domain (D4) of HPAO-2 contains a complex antiparallel β-sandwich fold (9). Two β-hairpin arms protrude from each monomer, and form part of the intricate interaction within the HPAO-2 homodimer, (Figure 3). Some CAOs from different sources, such as E. coli, pea seedling, and human kidney, bind a calcium or manganese ion at an additional site on the periphery of the protein (2, 12, 16, 31). Like HPAO-1, no evidence for this second metal binding site was observed in the HPAO-2 crystal structure (9). Unusually, an adventitious second copper ion is present in HPAO-2 samples following initial protein purification. Even though the protein used in crystallization had not been dialyzed against EDTA, which removes this loosely bound copper, there was no evidence in the electron density to indicate where this site may be located within the protein. HPAO-2 contains 8 cysteine residues per monomer, but only two form a disulfide bond (Cys 321 and Cys 347). In HPAO-1, the equivalent two cysteine residues (12 Cys in total) also form the single disulfide bond of HPAO-1 (Cys 338 and Cys 364) (9).
Table 2.
X-ray Crystallographic Data Collection, Processing and Refinement Statistics for HPAO-2.
| Data Collection and Processing Statistics | |
|---|---|
| Detector type | ADSC Quantum 315r |
| Beam-line and source | 19-ID SBC-CAT, Advanced Photon Source |
| Temperature (K) | 100 |
| Space group | C2 |
| Unit Cell (Å) | 288.5 × 91.1 × 151.1 |
| β=117.2° | |
| Number of molecules in the unit cell, Z | 6 |
| Wavelength (Å) | 0.9785 |
| Resolution (Å)a | 50.0–2.0 (2.07–2.00) |
| Unique reflections | 210,228 |
| Completeness (%)a | 90.6 (69.3) |
| Rmergeab | 0.089 (0.319) |
| I/σIa | 13.4 (3.0) |
| Redundancya | 3.5 (3.1) |
| Crystallographic Refinement and Model Statistics | |
| Resolution range (Å)a | 37.4–2.00 (2.06–2.01) |
| No. of reflections in working seta | 200,161 (10,684) |
| No. of reflections in test seta | 10,514 (539) |
| R-work (%)a,c | 14.5 (21.9) |
| R-free (%)a,d | 19.1 (27.3) |
| No. of non-hydrogen atoms | |
| No. of amino acid residues | 1,899 |
| No. of protein atoms | 15,321 |
| No. of solvent molecules | 3,199 |
| No. of Cu2+ ions | 3 |
| No. of other atoms | 91 |
| Rmsd from ideal geometry | |
| Bond lengths (Å) | 0.026 |
| Bond angles (°) | 2.0 |
| Ramachadran plot | |
| Energetically favored regions (%) | 89.2 |
| Additionally allowed regions (%) | 10.4 |
| Generously allowed regions (%) | 0.2 |
| Disallowed regions (%) | 0.2 |
| Average B-factor (Å2) | |
| Main Chain | 24.4 |
| Side Chain | 26.3 |
| Ligands | 43.8 |
| Solvent Atoms | 44.2 |
Numbers in parentheses refer to the highest resolution shell.
Rmerge = ∑hkl∑i|Ihkl,i-〈Ihkl〉|/∑hkl∑iIhkl,i, where I is the observed intensity and 〈I〉 is the average intensity for multiple measurements.
R-work = Σ||Fo| - |Fc||/ Σ|Fo|, where |Fo| = observed structure factor amplitude and |Fc| = calculated structure factor amplitude for 95% of the data used in refinement.
R-free, R-factor based on 5% of the data excluded from refinement.
Figure 3.
Overall view of the HPAO-2 dimer highlighting the two β-hairpin arms. One monomer is displayed in green ribbon, except for the two β-hairpin arms that are colored in blue (corresponding to residues 348 ȓ 376) and red (corresponding to residues 454 ȓ 481). The second monomer is displayed in grey Cα trace. The TPQs are drawn in stick colored by atom type, and copper atoms are displayed as gold spheres. Figure produced using PyMOL (http://www.pymol.org/).
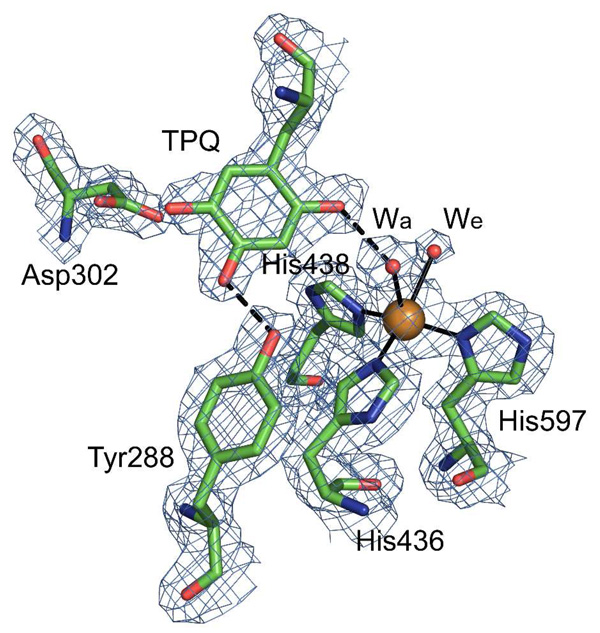
The HPAO-2 active site is deeply buried within the protein interior. The two active sites within the functional HPAO-2 homodimer each contain a TPQ cofactor (42.2 Å distance between Cα of each TPQ cofactor) and a cupric ion (34.7 Å distance between coppers in the dimer) coordinated by three conserved histidine ligands in a distorted square pyramidal geometry (Figure 4). Two well-ordered water molecules are ligated to the copper ion: one sits in an axial position (Wa) bridging the O2 of TPQ and the copper, and the other in an equatorial position (We) nearly planar with the Cu2+ and nitrogen atoms of its three histidine ligands (Figure 4). This latter solvent position is variably occupied in different CAO crystal structures, and when modeled has a higher average B-factor than the axial water. We was clearly observed in all three subunits within the crystallographic ASU of the HPAO-2 electron density, whereas it was only modeled in two of the six subunits in the HPAO-1 structure ASU, and thus, this water appears to be more labile in HPAO-1 than HPAO-2 (22). The electron density for TPQ clearly indicates that > 90 % of the active sites in the crystal contain fully formed TPQ (Figure 4), even though treatment with phenylhydrazine of the protein preparation used in crystallization indicated only ~ 50 % TPQ was present. However, the observation of only 50% TPQ could arise from half-of-sites reactivity, which is known to occur in other CAOs, and as indicated in the crystal structure the true TPQ content could actually be close to 100% (34–36). The TPQ cofactor adopts a single orientation with its O2 atom closest to the copper center and O5 atom closest to the conserved catalytic base, Asp302. It is thus poised for substrate attack at C5 to form the substrate Schiff base intermediate, and represents a conformation competent for catalysis termed “copper off”. There is no evidence of the “copper on” conformation in which O4 of TPQ is directly ligated to the copper displacing the axial water ligand (Wa). This conformation is considered not competent for catalysis as the C5 is inaccessible to amine substrate but is often present in CAO crystal structures, including HPAO-1 where it forms ~30 % of the TPQ conformers (22).
Figure 4.
Active site of HPAO-2 with 2Fo – Fc electron density. Residues are colored by atom type (carbon, green). Hydrogen bonds are indicated by dashed lines. Ligand-to-metal interactions are indicated by solid lines. The copper ion is shown as a gold sphere. Water molecules are depicted as red spheres (Wa indicates the axial water ligand to the copper; We the equatorial water ligand). 2Fo – Fc electron density contoured at 1.5σ is shown as a blue mesh. Figure produced using PyMOL (http://www.pymol.org/).
Crystal Structure of HPAO-2 in comparison to HPAO-1: Substrate amine entry channel
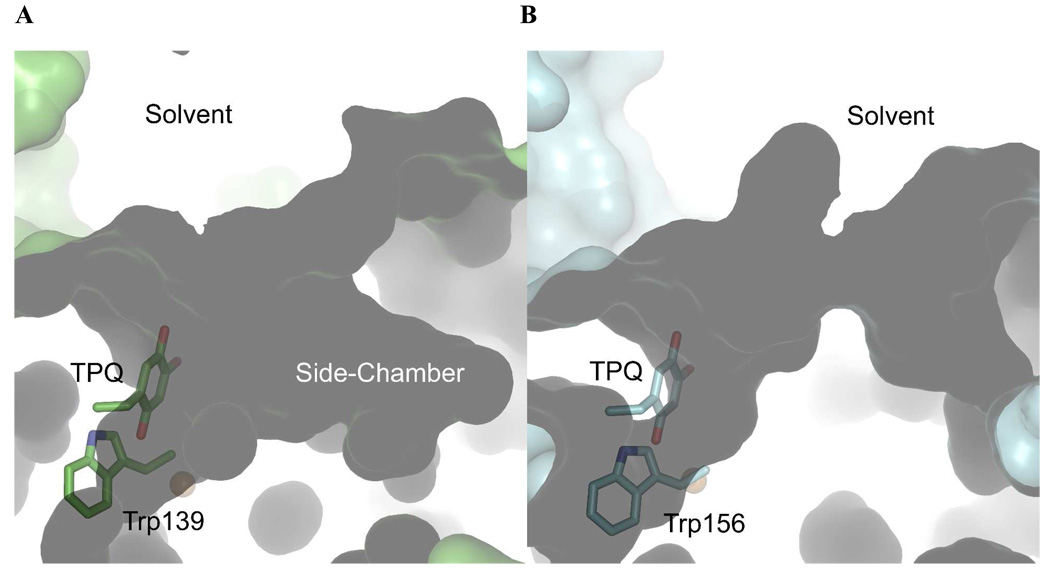
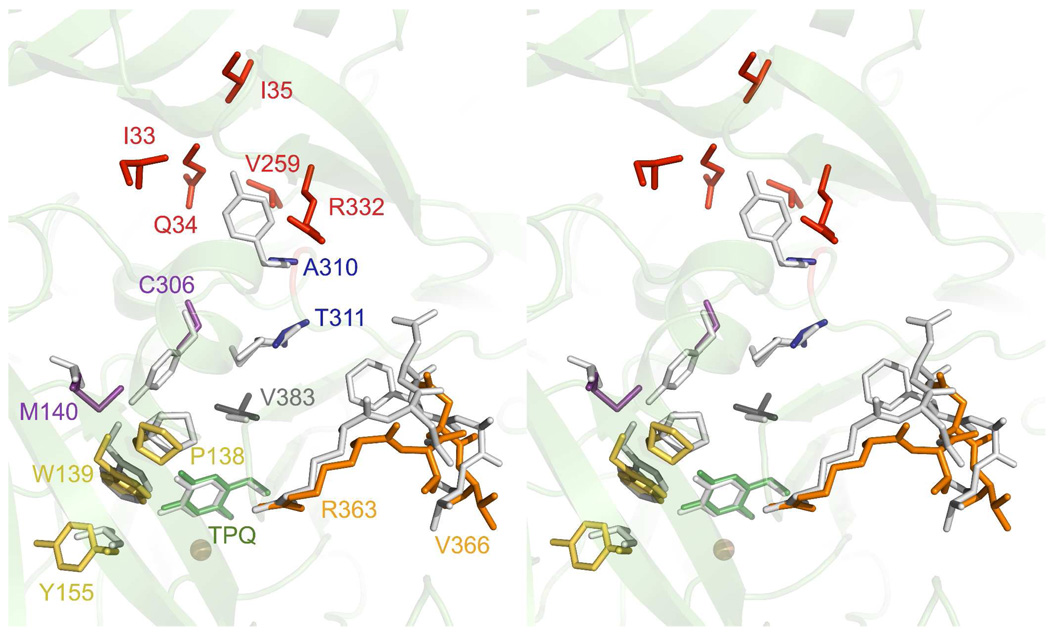
Although the residues important for chemistry are structurally identical between HPAO-2 and HPAO-1, the substrate channels differ considerably. HPAO-2 has a broad, funnel-shaped substrate entry channel compared to HPAO-1 (Figure 5). The widening of the channel mouth of HPAO-2 is primarily due to a two-residue deletion in the loop of the β-hairpin arm from the other monomer that reaches across close to the active site (blue in Figure 3). This arm contains a conserved His (359 and 376 in HPAO-2 and −1 respectively) that forms a hydrogen bond to the conserved acidic residue following TPQ in the protein sequence. In HPAO-1, the loop is defined by Arg380-Asp-Asn-Phe-Ala-Thr385, whereas in HPAO-2, the Phe-Ala are deleted to give a four-residue β-hairpin turn consisting of Arg363-Thr-Asn-Val366 (orange in Figure 6). Interestingly, HPAO-2 also has a side-chamber just below the mouth of the main channel, which is large enough to accommodate and retain a bulkier aromatic substrate like benzylamine or the outgoing product of its reaction, benzaldehyde (Figure 5, Supporting Information Figure S7). The chamber is accessed by a mouth that widens out to a diameter of 5.1 Å, and is 5.4 Å deep. The chamber has three small pockets at its deepest point that present backbone polar groups (Ile33, Gln34, and Ile35) that could hydrogen bond to a substrate amine group (red in Figure 6, Supporting Information Figure S7A). One side of the chamber is hydrophobic, which is formed by the alkyl chain of Arg332, Val259 and Ala310, and would enable a favorable interaction with an aromatic moiety (red except Ala310 which is blue in Figure 6, Supporting Information Figure S7B). Interestingly, the opposite wall contains a high density of oxygen atoms, which gives a slight negative electrostatic potential to this side of the chamber.
Figure 5.
Surface representation of substrate entry channels in (A) HPAO-2, and (B) HPAO-1. TPQ and the “gating” Trp are drawn in stick colored by atom type ((A) carbon, green; (B) carbon, blue), and copper as a gold sphere. Figure produced using PyMOL (http://www.pymol.org/).
Figure 6.
Stereo overlay between key residues in HPAO-2 and HPAO-1 that define the substrate channel. Only HPAO-2 residues are numbered, and the side-chains are colored as specified in the main text. The structurally corresponding residues in HPAO-1 are drawn as white sticks, except in the case of the red HPAO-2 side-chains that define the side-chamber, which is absent in HPAO-1. The fold of HPAO-2 is depicted in green ribbon. Figure produced using PyMOL (http://www.pymol.org/).
Between the side-chamber and the active site there are two key sequentially adjacent residue side-chain changes that widen the HPAO-2 channel from that of HPAO-1, these being Ala310-Thr311 in HPAO-2 compared to Tyr327-Met328 in HPAO-1 (Table 3, blue in Figure 6).
Table 3.
Residue Changes Between HPAO-2 and HPAO-1 that Likely Impact Accommodation of Substrate and Catalytic Intermediate Structures.
| HPAO-2 | HPAO-1 |
|---|---|
| Met140 | Thr157 |
| Tyr155 | Leu174 |
| Cys306 | Tyr323 |
| Ala310 | Tyr327 |
| Thr311 | Met328 |
| Val383 | Ala402 |
At the base of the substrate entry channel, where substrate undergoes chemistry to form product, there is a conserved Trp in HPAO-2 and −1 (Trp139 and Trp156 respectively, Figure 5 and yellow in Figure 6) that in other CAOs is generally a Tyr or Phe (2, 9, 14, 15, 30–33). This residue has been referred to as a “gate” to the active site (33). It has two major conformers; one blocking access to TPQ (normally observed in resting state CAO crystal structures, although the side-chain can also be disordered), and the other rotated out of the way (observed in Schiff base intermediates generated in an active site base mutant, and suicide inhibitor complexes that mimic Schiff base intermediates) (15, 36–38). Interestingly, the “gate” hypothesis does not appear to pertain to mammalian CAOs. The corresponding tyrosine residue in structures of CAO from bovine plasma, human diamine oxidase, and human vascular adhesion protein-1 are in an intermediate position halfway between the “open” and “closed” conformations (14–16, 32). Due to HPAO-2 and −1 having the larger Trp amino acid at this position, the “gate” is always open in these structures, and has B-factors that are equivalent to those of surrounding residues indicating a lower mobility than in other CAOs. However, the angle of the Trp side-chain is different between the two enzymes, and the main-chain is shifted such that the channel in HPAO-1 is more constricted than in HPAO-2 (Figure 5). This appears to primarily result from the substitution of Leu174 in HPAO-1 to Tyr155 in HPAO-2 (Table 3, yellow in Figure 6). This residue does not directly form part of the channel wall, but sits behind the 5-membered ring of Trp pushing it into the channel in HPAO-1. The conserved Pro residue that is N-terminal to the Trp in both structures rigidifies the main-chain, reducing the possibility for compensating flexibility in the channel (yellow in Figure 6). On the other side of the channel the main-chain is also moved inwards in HPAO-1 constricting the channel compared to HPAO-2. This is again a “knock-on” effect due to a two-residue insertion into the tip of the β-hairpin that reaches across from the other monomer of the homodimer, and was discussed above (blue in Figure 3, orange in Figure 6). HPAO-1 has a two-residue insertion Phe383-Ala384 compared to HPAO-2, and it is the side-chain of Phe383 that pushes the main-chain containing Ala402 (Val383 in HPAO-2) into the substrate channel (Table 3, colored dark grey in Figure 6). Although Val383 is a larger side-chain than Ala402, the displacement of the main-chain places the end of these two residue side-chains at the same place, but the effect N-terminal to these residues is an overall displacement constricting the HPAO-1 channel. The coincident positioning of the end of the side-chains Val383 in HPAO-2 and Ala402 in HPAO-1 appears to have an important role, as TPQ is sequentially close to this position (HPAO-2, 386; HPAO-1, 405) and the structures become coincident immediately C-terminal to Val383/Ala402.
Finally, there are two side-chain changes between HPAO-2 and −1 within the substrate entry channel that would interact with bulky R groups during the reductive half-reaction; in HPAO-2 Met140 and Cys306 have replaced Thr157 and Tyr323, respectively, in HPAO-1 (Table 3, magenta in Figure 6).
DISCUSSION
HPAO-2 is the second copper amine oxidase to be isolated and characterized from H. polymorpha. The marked change in substrate specificity is largely manifested in kcat/Km, which is 750-fold greater for benzylamine than methylamine. Complementarily, the kcat/Km values for the previously studied CAO from the same organism, HPAO-1, are opposite in specificity. Hence, the isolation and characterization of two copper amine oxidases from the same organism with 67 % sequence homology and 34 % sequence identity, but inverted substrate specificities, begins to address the nature of substrate specificity in these kinetically diverse enzymes.
The substrate entry and binding pockets of the two enzymes are clearly very structurally different from comparison of the X-ray crystal structures (Figure 5). The wider mouth of the entry channel of HPAO-2 would be more likely to capture and funnel the larger benzylamine from bulk solvent into the heart of the enzyme. In HPAO-1 the channel is almost as narrow at its entrance as it is close to the active site. This would restrict the orientations in which bulky aromatic substrates could enter the channel, which is likely to reduce their probability of capture.
In the amine entry channel in HPAO-2 there is a defined side-chamber (Figure 5A). This chamber could easily accommodate benzylamine, and the three pockets at the deepest part of the chamber have polar groups that could hydrogen bond with the amine group of substrate (Supporting Information Figure S7). This could be considered a holding area or “anteroom” that increases the local concentration of substrate near the active site. It is also possible that binding of substrate into this side-chamber away from the main portion of the entry channel could facilitate movement of product out of the enzyme. Interestingly, there has been another “anteroom” identified in CAOs that is proposed to have a similar function, but in this case for substrate O2 (22, 30, 32). This anteroom lies on the opposite side of the copper from the amine entry channel and is structurally conserved among CAOs. In contrast, the amine anteroom appears to be a particular feature of HPAO-2, and is unlikely to play a significant role in the kinetics of the enzyme.
The large changes in kcat/Km between the substrates of both enzymes suggest that substrate binding at the site of chemistry plays a role in determining substrate preference. However, selection against the smaller substrate methylamine for HPAO-2 cannot depend on sterics alone, since the substrate-binding pocket could accommodate both substrates. During catalysis the formation of tetrahedral intermediates are postulated through nucleophilic attack on C5 of cofactor, as in the initial attack of amine to form the substrate Schiff base (Scheme 7). TPQ in its active “copper off” conformation sits in a wedge-shaped environment in which the C2-C3 side of the ring forms tight packing interactions with the protein, while the C5-C6 side of the ring does not. This additional space next to C5 is proposed to be important for the accommodation of tetrahedral intermediates during nucleophilic attack (39). The C5 environment is controlled primarily by two invariant residues, the Asp catalytic base and the Asn residue (385 in HPAO-2 and 404 in HPAO-1) N-terminal to TPQ. The head-group of the Asn packs against one face of the TPQ ring, meaning that amine attack can only occur from the other face of the ring that lies by the catalytic base (Supporting Information Figure S8). The importance of this residue in orienting the TPQ cofactor has been demonstrated. Mutation of Asn404 to Ala in HPAO-1 leads to the accumulation of product Schiff base upon reaction with methylamine (40). However, for bulkier substrates, such as benzylamine, the orientation of amine attack and the accommodation of subsequent Schiff base intermediates are controlled by the next sphere of residues that line the entry channel just outside the catalytic active site. A key pair of residues are juxtaposed spatially with the large/small residue pattern being reversed in the two enzymes, Met140 and Cys306 in HPAO-2, are replaced by Thr157 and Tyr323 respectively in HPAO-1 (Table 3, magenta in Figure 6, Supporting Information Figure S9). This has the effect of moving the position of a small pocket such that bulkier R groups can access the HPAO-2 active site. Benzylamine, as a substrate, is unusual in that its product Schiff base is fully conjugated and thus planar, and the shape of the HPAO-2 substrate channel by the active site is more complementary to this species than that of HPAO-1 (Supporting Information Figure S9).
In HPAO-2, the effect of substrate deuteration on kcat/Km is of particular value in understanding the reduced reactivity with methylamine, specifically the large Dkcat/Km for methylamine as compared to the more optimal substrate, benzylamine. The magnitude of the methylamine Dkcat/Km (18.5 ± 0.1) is very close to the intrinsic isotope effect for CAOs (41), implying that proton abstraction has become rate-limiting for the poor substrate. The origin of the reduced kcat/Km(S) value is thus attributed in part to an impaired proton abstraction step (Scheme 8A). Considering the shape complementarity for benzylamine in the entry channel of HPAO-2, a preferential decrease in the rate of proton abstraction is likely due to difficulty in achieving a precise alignment between the methylamine substrate Schiff base and the active site base. It was also possible that the faster benzylamine rate could reflect an increased conjugation in its resulting carbanion/product Schiff complex. However, a comparison of kcat/Km(S) for the aromatic phenylethylamine oxidation (2.6 × 105 M−1s−1) at the non-conjugated C-1 position to the benzylamine value (Table 1) indicates a similar or elevated value.
Although the HPAO-1 and −2 structures overlay exactly with regard to the positions of the catalytic Asp base, “copper off” TPQ, and the ordered water molecules between them, TPQ appears to have a lower mobility in HPAO-2 compared to HPAO-1. This is supported by the fact that in the 3 subunits of the crystallographic ASU of HPAO-2 there is no evidence for the “copper on” conformer of TPQ, whereas in 4 out of the 6 subunits in the HPAO-1 structure, there is a substantial proportion of “copper on”. The side-chain B-factors of the cofactor and surrounding residues, normalized to the overall B-factor of each structure, also support that TPQ is less mobile in HPAO-2 than HPAO-1. This feature may exacerbate the ability of HPAO-2 to orient the reactive carbon of the methylamine Schiff base with the catalytic Asp. We note that there is some isotope effect on kcat/Km for benzylamine with HPAO-2, indicating that proton abstraction contributes to kcat/Km for this substrate as well as for methylamine. It is, therefore, very likely that additional factors contribute to the 103-fold rate reduction with methylamine oxidation by HPAO-2.
In the case of HPAO-1, similar Dkcat/Km values are observed for both substrates, while the second order rate constant is reduced ca. 330-fold for benzylamine. The most likely explanation is that both proton abstraction and substrate binding/release are slowed when the enzyme is challenged with the larger substrate (Scheme 8B). The increase in rate limitation by proton abstraction for benzylamine is also reflected in Dkcat, which is elevated relative to methylamine (Dkcat(benzylamine) is 5.9), whereas, Dkcat(methylamine) is 1.7). In previous studies of HPAO-1 with methylamine, three partially rate-determining steps have been documented that include aldehyde release, first electron transfer to oxygen and hydrogen peroxide and/or ammonium release (26). Loss of a proton from C-1 of the substrate Schiff base must contribute significantly to the decreased rate of turnover with benzylamine, since all steps after oxygen binding are common to both substrates, and therefore cannot account for the difference in turnover rates (Scheme 7).
Significantly, the differences in kcat/Km(S) are not mirrored in kcat/Km(O2). The oxygen binding and reactivity sites, copper and copper ligands on the two enzymes are structurally similar. All the residues that define the O2 anteroom close to the copper are different between HPAO-1 and −2, but the nature of the area is still hydrophobic (Supporting Information Table S2). This is true in other CAOs, where the residues are not conserved but the hydrophobic nature of the pocket is maintained (22, 32, 42, 43). Hence, the kinetic and structural data suggest a modular structure for both HPAOs, in which changes at the amine substrate site are not propagated to the pocket where oxygen undergoes reaction. Furthermore, the similar kcat/Km(O2) values for both isoenzymes, regardless of substrate, support the ping-pong mechanism proposed for HPAO-1 (Scheme 7). The stark contrast between the substrate-dependent changes in kcat/Km(S) and the substrate-independent kcat/Km(O2) highlights the independence of the oxygen binding and reduction sites from the structural determinants affecting amine kinetics.
CONCLUSIONS
In this study, we have investigated the origins of the substrate specificity in the copper amine oxidases designated HPAO-1 and HPAO-2. From the newly solved crystal structure of HPAO-2 in comparison with that of HPAO-1, we can begin to see how the narrow binding channel of HPAO-1 may predispose that CAO isoenzyme to reaction with smaller aliphatic amines via changes in substrate binding energies. However, selection against the smaller aliphatic substrate in the wide funnel-like substrate binding channel of HPAO-2 cannot be due to sterics alone. A significant increase in Dkcat/Km values for the poorer methylamine substrate relative to the preferred substrate indicates that proton abstraction has become rate-determining with this substrate. An impairment of the positioning of the methylamine substrate Schiff base in HPAO-2 affords the most likely explanation for the increase in Dkcat/Km(S), while the reduction in the absolute magnitude of kcat/Km(S) likely arises from numerous factors. Conversely, in HPAO-1 both binding of substrate and release of product, as well as the proton abstraction step, appear to be hindered under catalysis with the bulkier aromatic amine. The present combination of kinetic and structural characterization reveals the multiplicity of factors that enters into the differential discrimination between large vs. small substrates at the active sites of a pair of paralogous enzymes catalyzing identical chemical reactions.
Supplementary Material
Supporting Informational material is available free of charge via the Internet at http://pubs.acs.org and contains the following:
Tables
Table S1: Comparison of kcat values in EDTA-dialyzed vs. non-dialyzed HPAO-2 samples.
Table S2: Residue changes that define the O2 anteroom.
Figures
Figure S1: HPAO-1 and HPAO-2 sequence alignment.
Figure S2: 10% SDS PAGE gel of HPAO-2.
Figure S3: Comparison of the N-terminal sequence of HPAO-2 and HPAO-1.
Figure S4: Stereo graphic of the 2Fo – Fc electron density quality in HPAO-2.
Figure S5: Overall fold of the HPAO-2 dimer.
Figure S6: Ribbon diagrams showing an overlay of HPAO-1 and HPAO-2.
Figure S7: Graphics of the side-chamber located in the HPAO-2 entry channel.
Figure S8: TPQ environment showing that only one face of the ring can support nucleophilic attack.
Figure S9: Model of the product Schiff base of benzylamine with TPQ.
ACKNOWLEDGMENTS
Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute, and we thank Can Ergenekan for his support. X-ray data were collected at the Kahlert Structural Biology Laboratory (KSBL) at The University of Minnesota and beam-line 19-ID-D, SBC-CAT at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL. The work was performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U. S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02–06CH11357. We thank Ed Hoeffner for KSBL support, and Steve Ginell and the staff at Sector 19, APS for their support. We would also like to thank Peder Cedervall and Brandon Goblirsch for help generating figures and Rhein Biotech for the access to the H. polymorpha Genome Data Base.
Footnotes
This work was supported by NIH grants, GM39296 to J.P.K., GM66569 to C.M.W. and Chemistry-Biology Interface Training Grant GM-008700 to B.J.J, and Minnesota Medical Foundation grant 3714-9221-06, Office of the Dean of the Graduate School of the University of Minnesota grant 21087, and a Minnesota Partnership for Biotechnology and Medical Genomics grant SPAP-05-0013-P-FY06 to C.M.W.
Co-ordinates and structure factors have been deposited in the Protein Data Bank as entry 3loy.
Abbreviations: CAO, copper amine oxidase; HPAO, Hansenula polymorpha copper amine oxidase; TPQ, 2,4,5-trihydroxyphenylalanine quinone; MAO, flavin-containing monoamine oxidase; NBT, nitroblue tetrazolium; ASU, asymmetric unit.
REFERENCES
- 1.Mure M, Mills SA, Klinman JP. Catalytic mechanism of the topa quinone containing copper amine oxidases. Biochemistry. 2002;41:9269–9278. doi: 10.1021/bi020246b. [DOI] [PubMed] [Google Scholar]
- 2.Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, Corner AS, Phillips SE, McPherson MJ, Knowles PF. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 Å resolution. Structure. 1995;3:1171–1184. doi: 10.1016/s0969-2126(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 3.Roh JH, Suzuki H, Azakami H, Yamashita M, Murooka Y, Kumagai H. Purification, Characterization, and Crystallization of Monoamine-Oxidase from Escherichia-Coli K-12. Bioscience Biotechnology and Biochemistry. 1994;58:1652–1656. doi: 10.1271/bbb.58.1652. [DOI] [PubMed] [Google Scholar]
- 4.Cai DY, Klinman JP. Copper amine oxidase - Heterologous expression, purification, and characterization of an active enzyme in Saccharomyces cerevisiae. Biochemistry. 1994;33:7647–7653. doi: 10.1021/bi00190a019. [DOI] [PubMed] [Google Scholar]
- 5.Cronin CN, Kirsch JF. Role of arginine-292 in the substrate specificity of aspartate aminotransferase as examined by site-directed mutagenesis. Biochemistry. 1988;27:4572–4579. doi: 10.1021/bi00412a052. [DOI] [PubMed] [Google Scholar]
- 6.Youdim MB, Edmondson D, Tipton KF. The therapeutic potential of monoamine oxidase inhibitors. Nature Reviews. 2006;7:295–309. doi: 10.1038/nrn1883. [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi S, Nobe Y, Yasuoka J, Wakamiya T, Kusumoto S, Kuramitsu S. Enzyme flexibility: a new concept in recognition of hydrophobic substrates. J. Biochemistry. 1997;122:55–63. doi: 10.1093/oxfordjournals.jbchem.a021740. [DOI] [PubMed] [Google Scholar]
- 8.Tsugeno Y, Ito A. A key amino acid responsible for substrate selectivity of monoamine oxidase A and B. J Biol Chem. 1997;272:14033–14036. doi: 10.1074/jbc.272.22.14033. [DOI] [PubMed] [Google Scholar]
- 9.Li RB, Klinman JP, Mathews FS. Copper amine oxidase from Hansenula polymorpha: the crystal structure determined at 2.4 angstrom resolution reveals the active conformation. Structure. 1998;6:293–307. doi: 10.1016/s0969-2126(98)00033-1. [DOI] [PubMed] [Google Scholar]
- 10.De Hoop MJ, Valkema R, Kienhuis CB, Hoyer MA, Ab G. The peroxisomal import signal of amine oxidase from the yeast Hansenula polymorpha is not universal. Yeast. 1992;8:243–252. doi: 10.1002/yea.320080402. [DOI] [PubMed] [Google Scholar]
- 11.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Elmore BO, Bollinger JA, Dooley DM. Human kidney diamine oxidase: heterologous expression, purification, and characterization. J Biol Inorg Chem. 2002;7:565–579. doi: 10.1007/s00775-001-0331-1. [DOI] [PubMed] [Google Scholar]
- 13.Marti L, Abella A, De La Cruz X, Garcia-Vicente S, Unzeta M, Carpene C, Palacin M, Testar X, Orozco M, Zorzano A. Exploring the binding mode of semicarbazide-sensitive amine oxidase/VAP-1: identification of novel substrates with insulin-like activity. J Med Chem. 2004;47:4865–4874. doi: 10.1021/jm0499211. [DOI] [PubMed] [Google Scholar]
- 14.Airenne TT, Nymalm Y, Kidron H, Smith DJ, Pihlavisto M, Salmi M, Jalkanen S, Johnson MS, Salminen TA. Crystal structure of the human vascular adhesion protein-1: unique structural features with functional implications. Protein Science. 2005;14:1964–1974. doi: 10.1110/ps.051438105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jakobsson E, Nilsson J, Ogg D, Kleywegt GJ. Structure of human semicarbazide-sensitive amine oxidase/vascular adhesion protein-1. Acta Crystallogr D Biol Crystallogr. 2005;61:1550–1562. doi: 10.1107/S0907444905028805. [DOI] [PubMed] [Google Scholar]
- 16.McGrath AP, Hilmer KM, Collyer CA, Shepard EM, Elmore BO, Brown DE, Dooley DM, Guss JM. Structure and inhibition of human diamine oxidase. Biochemistry. 2009;48:9810–9822. doi: 10.1021/bi9014192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DuBois JL, Klinman JP. Methods for characterizing TPQ-containing proteins. Quinones and Quinone Enzymes. 2004;Pt A 276:17–31. doi: 10.1016/S0076-6879(04)78002-7. [DOI] [PubMed] [Google Scholar]
- 18.Neumann R, Hevey R, Abeles RH. The action of plasma amine oxidase on beta-haloamines. Evidence for proton abstraction in the oxidative reaction. J Biol Chem. 1975;250:6362–6367. [PubMed] [Google Scholar]
- 19.Hartmann C, Klinman JP. Structure-function studies of substrate oxidation by bovine serum amine oxidase: relationship to cofactor structure and mechanism. Biochemistry. 1991;30:4605–4611. doi: 10.1021/bi00232a035. [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Johnson BJ, Cohen J, Welford RW, Pearson AR, Schulten K, Klinman JP, Wilmot CM. Exploring molecular oxygen pathways in Hansenula polymorpha copper-containing amine oxidase. J Biol Chem. 2007;282:17767–17776. doi: 10.1074/jbc.M701308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature Protocols. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi K, Klinman JP. Relationship of stopped flow to steady state parameters in the dimeric copper amine oxidase from Hansenula polymorpha and the role of zinc in inhibiting activity at alternate copper-containing subunits. Biochemistry. 2006;45:4683–4694. doi: 10.1021/bi0521893. [DOI] [PubMed] [Google Scholar]
- 27.Wilmot CM, Hajdu J, McPherson MJ, Knowles PF, Phillips SE. Visualization of dioxygen bound to copper during enzyme catalysis. Science. 1999;286:1724–1728. doi: 10.1126/science.286.5445.1724. [DOI] [PubMed] [Google Scholar]
- 28.Hevel JM, Mills SA, Klinman JP. Mutation of a strictly conserved, active-site residue alters substrate specificity and cofactor biogenesis in a copper amine oxidase. Biochemistry. 1999;38:3683–3693. doi: 10.1021/bi982199m. [DOI] [PubMed] [Google Scholar]
- 29.van Roermund CW, de Jong MLIJ, van Marle J, Dansen TB, Wanders RJ, Waterham HR. The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J Cell Sci. 2004;117:4231–4237. doi: 10.1242/jcs.01305. [DOI] [PubMed] [Google Scholar]
- 30.Duff AP, Cohen AE, Ellis PJ, Kuchar JA, Langley DB, Shepard EM, Dooley DM, Freeman HC, Guss JM. The crystal structure of Pichia pastoris lysyl oxidase. Biochemistry. 2003;42:15148–15157. doi: 10.1021/bi035338v. [DOI] [PubMed] [Google Scholar]
- 31.Kumar V, Dooley DM, Freeman HC, Guss JM, Harvey I, McGuirl MA, Wilce MC, Zubak VM. Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 Å resolution. Structure. 1996;4:943–955. doi: 10.1016/s0969-2126(96)00101-3. [DOI] [PubMed] [Google Scholar]
- 32.Lunelli M, Di Paolo ML, Biadene M, Calderone V, Battistutta R, Scarpa M, Rigo A, Zanotti G. Crystal structure of amine oxidase from bovine serum. J Mol Biol. 2005;346:991–1004. doi: 10.1016/j.jmb.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 33.Wilce MCJ, Dooley DM, Freeman HC, Guss JM, Matsunami H, McIntire WS, Ruggiero CE, Tanizawa K, Yamaguchi H. Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: Implications for the biogenesis of topaquinone. Biochemistry. 1997;36:16116–16133. doi: 10.1021/bi971797i. [DOI] [PubMed] [Google Scholar]
- 34.De Biase D, Agostinelli E, De Matteis G, Mondovi B, Morpurgo L. Half-of-the-sites reactivity of bovine serum amine oxidase. Reactivity and chemical identity of the second site. Eur J Biochem. 1996;237:93–99. doi: 10.1111/j.1432-1033.1996.0093n.x. [DOI] [PubMed] [Google Scholar]
- 35.Frebort I, Toyama H, Matsushita K, Adachi O. Half-site reactivity with p-nitrophenylhydrazine and subunit separation of the dimeric copper-containing amine oxidase from Aspergillus niger. Biochemistry and molecular biology international. 1995;36:1207–1216. [PubMed] [Google Scholar]
- 36.Wilmot CM, Murray JM, Alton G, Parsons MR, Convery MA, Blakeley V, Corner AS, Palcic MM, Knowles PF, McPherson MJ, Phillips SE. Catalytic mechanism of the quinoenzyme amine oxidase from Escherichia coli: exploring the reductive half-reaction. Biochemistry. 1997;36:1608–1620. doi: 10.1021/bi962205j. [DOI] [PubMed] [Google Scholar]
- 37.Chiu YC, Okajima T, Murakawa T, Uchida M, Taki M, Hirota S, Kim M, Yamaguchi H, Kawano Y, Kamiya N, Kuroda S, Hayashi H, Yamamoto Y, Tanizawa K. Kinetic and structural studies on the catalytic role of the aspartic acid residue conserved in copper amine oxidase. Biochemistry. 2006;45:4105–4120. doi: 10.1021/bi052464l. [DOI] [PubMed] [Google Scholar]
- 38.Wilmot CM, Saysell CG, Blessington A, Conn DA, Kurtis CR, McPherson MJ, Knowles PF, Phillips SE. Medical implications from the crystal structure of a copper-containing amine oxidase complexed with the antidepressant drug tranylcypromine. FEBS Lett. 2004;576:301–305. doi: 10.1016/j.febslet.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 39.Murray JM, Saysell CG, Wilmot CM, Tambyrajah WS, Jaeger J, Knowles PF, Phillips SEV, McPherson MJ. The Active Site Base Controls Cofactor Reactivity in Escherichia coli Amine Oxidase: X-ray Crystallographic Studies with Mutational Variants. Biochemistry. 1999;38:8217–8227. doi: 10.1021/bi9900469. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz B, Green EL, Sanders-Loehr J, Klinman JP. Relationship between Conserved Consensus Site Residues and the Productive Conformation for the TPQ Cofactor in a Copper-Containing Amine Oxidase from Yeast. Biochemistry. 1998;37:16591–16600. doi: 10.1021/bi981541s. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann C, Klinman JP. Structure-function studies of substrate oxidation by bovine serum amine oxidase: relationship to cofactor structure and mechanism. Biochemistry. 1991;30:4605–4611. doi: 10.1021/bi00232a035. [DOI] [PubMed] [Google Scholar]
- 42.Duff AP, Trambaiolo DM, Cohen AE, Ellis PJ, Juda GA, Shepard EM, Langley DB, Dooley DM, Freeman HC, Guss JM. Using xenon as a probe for dioxygen-binding sites in copper amine oxidases. J Mol Biol. 2004;344:599–607. doi: 10.1016/j.jmb.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 43.Pirrat P, Smith MA, Pearson AR, McPherson MJ, Phillips SEV. Structure of a xenon derivative of Escherichia coli copper amine oxidase: confirmation of the proposed oxygen-entry pathway. Acta Crystallogr F Struc Biol & Crystallization Comm. 2008;64:1105–1109. doi: 10.1107/S1744309108036373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Informational material is available free of charge via the Internet at http://pubs.acs.org and contains the following:
Tables
Table S1: Comparison of kcat values in EDTA-dialyzed vs. non-dialyzed HPAO-2 samples.
Table S2: Residue changes that define the O2 anteroom.
Figures
Figure S1: HPAO-1 and HPAO-2 sequence alignment.
Figure S2: 10% SDS PAGE gel of HPAO-2.
Figure S3: Comparison of the N-terminal sequence of HPAO-2 and HPAO-1.
Figure S4: Stereo graphic of the 2Fo – Fc electron density quality in HPAO-2.
Figure S5: Overall fold of the HPAO-2 dimer.
Figure S6: Ribbon diagrams showing an overlay of HPAO-1 and HPAO-2.
Figure S7: Graphics of the side-chamber located in the HPAO-2 entry channel.
Figure S8: TPQ environment showing that only one face of the ring can support nucleophilic attack.
Figure S9: Model of the product Schiff base of benzylamine with TPQ.