Abstract
Background
The overall impact of influenza virus infection in immunocompromised patients is largely unknown. Antigenic drift and genetic variations during prolonged influenza infection have been demonstrated. In this report we describe a multi-drug resistant H3N2 influenza virus isolated from an immunocompromised patient after 5 days of therapy.
Methods
Multiple nasal washes were collected from an infected patient and viral isolates characterized. Sensitivity to antiviral agents was evaluated. Fitness and transmissibility was assessed in ferrets and tissue culture.
Results
An in-frame 4 amino acid deletion emerged in the neuraminidase gene of an H3N2 virus after 5 days of oseltamivir therapy. No other changes in the NA or HA genes were noted. Drug sensitivity assays revealed resistance to oseltamivir (>10-fold increase in IC50) and reduction of sensitivity to zanamivir (3 to 7-fold increase in IC50/EC50). No change in fitness or transmissibility was observed.
Conclusions
An in-frame NA gene deletion was rapidly selected for in an immunocompromised patient, resulting in decreased sensitivity of the isolate to available neuraminidase inhibitors without a change in fitness or transmissibility. This carries implications for our understanding of the emergence of antiviral resistance and treatment of patients with influenza A infection, especially those who are immunocompromised.
Keywords: Influenza, Neuraminidase inhibitors, Antiviral resistance, Immunocompromised host, Respiratory viruses
Introduction
Despite progress in vaccine and antiviral development, influenza virus infection remains a major public health problem, as reflected by more than 36,000 annual deaths in the United States [1]. In pandemic years, increased numbers of infections and deaths have been noted [2–4]. The current novel H1N1 pandemic highlights the unpredictability of how new strains of influenza A virus can emerge and rapidly spread across the world [5]. These pandemic and novel viruses continue to pose health risks to individuals and a significant economic and public health challenge to healthcare providers and governments worldwide.
Antiviral agents are now commonly used to mitigate the impact of influenza infection. Many governments stockpile these agents, and in recent years their use by healthcare providers to treat influenza has increased. Due to recent widespread resistance of seasonal A/H3N2 viruses to the adamantanes since 2005 [6, 7], neuraminidase inhibitors (NAI) have increasingly become the drug of choice [8]. Oseltamivir and the inhaled agent zanamivir have been shown to be clinically effective when given early in the course of disease, decreasing both duration of viral shedding and complications [9]. Until recently, resistance to these agents has been limited [10, 11], but during the 2008–2009 influenza season nearly 100% of the circulating A/H1N1 viruses in many countries were found to contain the H275Y mutation (N1 numbering) conferring resistance to oseltamivir [12–14]. The recent A/H3N2 and the new pandemic A/H1N1 viruses, although resistant to the adamantanes, have retained sensitivity to NAI, but a small but increasing number of cases have also been reported in which oseltamivir resistance has been observed [8, 11, 15]. Resistance to zanamivir has not been widely reported, but this drug has not been in widespread use, and has limited usefulness in severely ill patients due to the nature of its administration and possible adverse effects [16, 17].
As a consequence of the emergence of drug-resistant influenza viruses, the Centers for Disease Control and the Infectious Disease Society of America recently adopted new guidelines for the treatment of influenza, recommending the use of zanamivir or combination therapy of oseltamivir with an adamantane to treat seasonal influenza empirically [8, 18]. NAI use will likely continue to increase during the current pandemic [19–21] and future influenza seasons, making it extremely important to investigate how antiviral resistance emerges.
Severe disease due to influenza infection occurs most often in individuals with underlying illnesses, and the current H1N1 pandemic continues to follow this trend [22, 23]. Immunocompromised individuals are one such high risk group [24–27]. The impact of influenza on this growing population is still largely unknown. Prolonged illness and viral shedding, simultaneous infection with two subtypes, and viral genetic variation and antigenic drift during a single prolonged illness has been demonstrated in immunocompromised individuals [28–31].
Recently a novel 4 amino acid deletion in the NA gene at positions 245–248 in an A/H3N2 virus conferring resistance to oseltamivir was identified [32]. This deletion was found in the presence of other known NA resistance mutations in a virus isolated from a severely immunocompromised 3 year old who had a prolonged infection of nearly a year, and had received a total of 107 days of neuraminidase inhibitor therapy. This patient received three months of continuous oseltamivir prior to the time of isolation of this mutant virus [31, 32]. The authors hypothesized that the NA deletion may have been detrimental to viral fitness, but this was not fully evaluated in their study.
We describe the rapid emergence of an identical deletion at positions 245–248 in an adult immunocompromised patient infected with an influenza A/H3N2 virus (A/Bethesda/NIH12-0/2008). This is only the second reported instance of this deletion, and it is the first time it has been reported in the absence of other NAI resistance mutations. Additionally, the deletion appeared after only 5 days of oseltamivir therapy. We evaluated the effect of this deletion on viral replication and transmissibility and observed no reduction in replicative fitness or in contact transmissibility in ferrets. Finally, we observed that this mutation was retained after transmission between mammalian hosts.
Methods
Clinical Case
A 43 year old male with mantle cell lymphoma six months post allogeneic stem cell transplant was admitted for rectal pain and a fluctuant mass. The patient had a history of CMV reactivation and the development of agranulocytosis. He received a dose of rituximab five days prior to admission.
On admission the patient complained of dry cough and a mild sore throat. The next day he underwent incision and drainage of a perirectal abscess and was treated empirically with IV piperacillin-tazobactam. Abscess cultures grew an Enterococcus species, a swab specimen grew HSV, and appropriate therapy was administered.
Three days after admission he developed fever to 38.5°C, coryza, fatigue, cough, and nasal discharge. Physical examination revealed no evidence of retained or secondary infection at the incision site. Imaging studies were unremarkable.
Laboratory diagnostic evaluation was notable for severe neutropenia (absolute neutrophil count 120 cells/μl). A nasal wash was performed and a rapid influenza A test was positive. On the same day, the patient was started on oseltamivir phosphate by mouth 75mg twice daily.
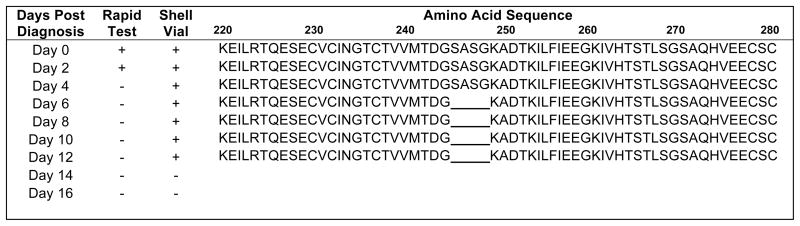
The patient was enrolled in a clinical study of influenza (NIH protocol: “Influenza in Normal and Immunocompromised Hosts”), and a skilled respiratory therapist performed nasal washes every other day during his infection. The patient manifested fever for a total of 4 days and upper respiratory symptoms for two weeks. He remained on oseltamivir phosphate for a total of 14 days. Two days after initial diagnosis another nasal wash revealed a positive rapid test and culture. After that time all rapid tests were negative, but the patient continued to have positive influenza cultures for a total of 12 days (Figure 1).
Figure 1.
Viral Shedding, diagnostics, and partial neuraminidase (NA) amino acid sequence. Nasal washes were collected from the patient every other day for 16 days. Nasal wash and shell vial cultures were performed on each sample (results shown). Nucleic acid sequencing of the NA gene was performed on all isolated viruses. Amino acid sequence shown here from position 220 to 280. A 4 amino acid deletion was noted as of day 6 at position 245–248. This deletion was present in all isolates collected after day 6 as shown.
Sequence analysis
Reverse-transcription polymerase chain reaction (RT-PCR) and sequencing was performed on the primary specimens and viral isolates as previously described [33]. Influenza virus hemagglutinin (HA) and neuraminidase (NA) sequences determined in this study have been deposited in GenBank (GenBank accession no. GU294117-GU294118). Analysis of the initial nasal wash specimen included sequencing of 50 PCR products that were cloned using the Topo TA cloning kit (Invitrogen, Carlsbad, CA).
NA activity and antiviral susceptibility
NA activity and antiviral susceptibility was measured using 50 μl of 100 μM methyl-umbelliferyl-N-acetyl neuraminic acid (MU-NANA, Sigma, St. Louis, MO) substrate as described previously [34]. An amount of virus that resulted in a signal of approximately 1×105 relative fluorescence units (RFU) was added to serial dilutions of oseltamivir and zanamivir (GlaxoSmithKline, Research Triangle Park, NC) in a 96-well plate format. A/Wisconsin/67/2005 (H3N2) was used as control. Total protein of each preparation was determined by bicinchoninic acid (BCA) assay following the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The 50% inhibitory concentration (IC50) of each drug was determined by regression analysis (Prism, GraphPad Software Inc., La Jolla, CA).
Virus replication and antiviral susceptibility
A high throughput cell-based assay was used to determine the 50% effective concentration (EC50) as described [35]. Assays were performed to test sensitivity to amantadine (Sigma), oseltamivir, and zanamivir. A/Wisconsin/67/2005 (H3N2) was used as control.
Plaque reduction assay
MDCK cells were inoculated with 100 plaque forming units (pfu) of virus per well. Ten-fold dilutions of 0.0001μM to 100μM oseltamivir tartrate solution (F. Hoffman-La Roche Ltd., Basel Switzerland) or zanamivir solution (GlaxoSmithKline, Brentford, UK) was added to each well of a six well plate, with one well containing no drug. Plaques were counted after 48 hours. Plaque reduction assays performed in triplicate.
Viral replication assay
MDCK cells were infected with 0.01 multiplicity of infection (MOI) of each viral isolate. Plates were incubated at 37°C for 1 hour. Cells were washed, 3ml DMEM containing 1μg/ml TPCK-Trypsin added, and incubated. Every twelve hours, 500μl of supernatant was collected over a 72 hour time period. Virus titers of each supernatant were determined in triplicate using standard plaque assay technique [36] and reported as mean pfu/ml.
Ferret contact transmission
Sixteen seronegative 4 month old male ferrets were housed in pairs in separate cages with individual airflow. Two pairs (four ferrets) were inoculated intranasally with 106 pfu of influenza virus isolated from the patient on initial sampling, and two pairs were inoculated with 106 pfu of the virus isolated from the patient 8 days after diagnosis. After 48 hours, two uninoculated ferrets were placed in each cage with inoculated ferrets. Daily weights, temperatures, and nasal washes were performed. Nasal wash titers were determined by standard plaque assay technique, and real-time RT-PCR was performed as previously described [37]. Amplification and sequencing of the NA gene was performed on each viral isolate recovered to determine if the NA gene sequence remained stable after transmission and infection. Animal experiments were performed following NIH Institutional Animal Care and Use Committee approved protocols and guidelines.
Results
Identification and sequencing of viral isolates
Viruses isolated from patient nasal washes collected on day 0 (time of initial diagnosis), 2, 4, 6, 8, 10, and 12 were characterized. NA genes were amplified and direct sequencing revealed that on days 0, 2, and 4 the NA gene was similar to the wild-type sequence of NA genes found in other A/Wisconsin/67/2005-like H3N2 viruses circulating during the 2007–2008 season. The virus isolated on day 6 and all subsequent isolates had a 12 nucleotide/4 amino acid in-frame deletion in the NA gene, corresponding to amino acid positions 245–248 (Figure 1). No other coding changes in the NA gene were noted, and no other known NAI resistance mutations were observed. The HA gene of all viral isolates were also sequenced and showed no coding changes.
To determine whether this deletion was present at the time of diagnosis, a 600 bp region spanning the area of deletion of the NA gene was amplified from the nasal wash specimen collected on day 0. Sequencing of 50 clones did not reveal any deletions at amino acid position 245–248.
Antiviral sensitivity
The IC50 of NAI was determined using an NA activity assay. The IC50 of oseltamivir for the virus containing the deletion isolated from the patient on day 8 (16.37nM, 95% CI 12.72–21.07) showed a significant > 90-fold increase from the day 0 wild-type virus (0.18nM, 95% CI 0.18–0.19). A 3-fold increase in the IC50 of zanamivir was also noted between the wild-type day 0 virus and the day 8 virus with an increase from 4.0 (95% CI 3.60–4.44) to 12.70 (95% CI 12.08–13.37) respectively (Table 1). In addition to reduced sensitivity to antivirals, NA enzymatic activity associated with the day 8 virus was less than the day 0 isolate. Even though the day 0 isolate had a lower level of NA activity/μg than A/Wisconsin/67/2005, another circulating H3N2 virus, it retained sensitivity to oseltamivir and showed very modest decrease in sensitivity to zanamivir.
Table 1.
NA Activity and Antiviral Sensitivity
| NA Activity (μU/μg total protein) | |||
| A/Wisconsin/67/2005 | Day 0 | Day 8 (deletion in NA of AA 245–248) | |
| 85.3 | 6.0 | 1.4 | |
| NA-inhibition Assay IC50 (95% CI) | |||
| A/Wisconsin/67/2005 | Day 0 | Day 8 (deletion in NA of AA 245–248) | |
| Oseltamivir (nM) | 0.29 (0.27–0.32) | 0.18 (0.18–0.19) | 16.37 (12.72–21.07) |
| Zanamivir (nM) | 3.07 (2.80–3.36) | 4.00 (3.60 – 4.44) | 12.70 (12.08–13.37) |
| High Throughput Assay EC50 (95% CI) | |||
| A/Wisconsin/67/2005 | Day 0 | Day 8 (deletion in NA of AA 245–248) | |
| Aman tadine (μM) | >200 | >200 | >200 |
| Oseltamivir (nM) | 0.37 (0.34–0.42) | 0.21 (0.19–0.23) | 442.60 (378.00–518.20) |
| Zanamivir (nM) | 6.16 (5.79–6.57) | 18.85 (17.26–20.58) | 131.40 (113.10–152.80) |
| Plaque Reduction Assay IC50 (95% CI) | |||
| Day 0 | Day 8 (deletion in NA of AA 245–248) | ||
| Oseltamivir (nM) | 0.38 (0.21–0.73) | 49.70 (33.28–74.22) | |
| Zanamivir (nM) | 15.58 (3.696 to 65.65) | 224.50 (119.30 to 422.40) | |
A high throughput tissue culture assay demonstrated that both day 0 and day 8 virus isolates were resistant to amantadine, with a 50% effective concentration (EC50) of >200μM. The EC50 of oseltamivir of the wild-type virus isolated from day 0 was >2000-fold lower than the EC50 of the deletion containing virus isolated on day 8. The EC50 of zanamivir was also increased 7-fold in the virus containing the deletion from the wild-type day 0 virus, and 21-fold higher than the control virus (Table 1).
Standard plaque reduction assays showed similar results to the high throughput assay demonstrating a significantly higher IC50 to oseltamivir in the virus containing the deletion with a >49-fold increase, and an increase of 14-fold in zanamivir IC50 (Table 1).
The greater resistance demonstrated in these replication-dependent assays is likely to reflect not only the amplification of virus required for read-out but also reduced amount of NA in each virion. Comparison of the NA activity per μg total protein in each virus preparation showed that the day 8 isolate had approximately 3 times less NA than the day 0 isolate (1.4 μU and 6.0 μU per μg respectively). Interestingly, these isolates have substantially less NA activity than A/Wisconsin/67/2005 (85.3 μUnits/μg) that was used as a control.
Viral fitness in cell culture
The virus isolated on day 8 containing the NA deletion showed no significant difference in viral replicative fitness in MDCK cells compared to the day 0 wild-type isolate. Both viruses reached peak titers (105 pfu/ml) at 36–48 hours (Figure 2).
Figure 2.
Viral replication kinetics in MDCK cells at 37°C. Cellswereinfected with 0.01 MOI of virus. Supernatants from infected MDCK cells were collected at 12 hour intervals and viral titer determined by plaque assay (see Methods). Cultures and measurements were performed in triplicate. Error bars represent SEM.
Ferret infection and contact transmission
All ferrets intranasally inoculated with either wild-type and NA mutant virus developed similar mild illness associated with significant nasal shedding of influenza A virus. Virus was detectable 48 hours after inoculation by culture and RT-PCR from the nasal washes of all 4 ferrets in each group. No significant difference in detectable levels of virus was noted between the ferrets inoculated with the virus isolated on day 0 or the day 8 virus containing the NA deletion (Figure 3).
Figure 3.
Ferret inoculation and transmission. All four ferrets inoculated with each virus were shedding detectable virus 48 hours after inoculation. In both groups, three of four (75%) of ferrets placed in contact with the inoculated ferrets developed active infection with detectable viral shedding by 48 hours after contact. No significant difference in mean viral titer was seen between those ferrets infected with the wild-type or deletion containing virus. Plaque assays to determine viral titer on each nasal wash samples were repeated three separate times, and points represent mean viral titer of all four ferrets in each group. Error bars represent SEM.
Three of four (75%) ferrets in each group that were not inoculated but placed in the cages with the inoculated ferrets were found to have detectable levels of virus in nasal wash fluid 48 hours after contact. No significant difference in the viral titer was observed between the ferrets placed in contact with day 0 or day 8 virus inoculated ferrets (Figure 3).
Discussion
We describe the isolation and characterization of an influenza A/H3N2 virus isolated containing a four amino acid deletion in the NA 5 days after initiation of oseltamivir therapy. This deletion was recently reported in another immunocompromised patient; however, in the previous instance, the patient had an unusually long duration of illness (nearly 9 months), with the resistant strain isolated only after 3 months of continuous oseltamivir therapy. That patient had received multiple antiviral drugs, and the deletion occurred in the presence of other NA mutations previously associated with NAI resistance, including E119V and I222V [31]. In that report, it was hypothesized that the deletion may have led to some decrease in viral fitness since it appeared only after months of continuous antiviral selective pressure. A reverse genetics-produced virus containing the deletion was studied in cell culture, but no conclusive evidence of this deletion’s effect on viral fitness was obtained [32].
In the present report, the NA deletion appeared rapidly in an immunocompromised adult who demonstrated two weeks of viral shedding and illness. A much shorter duration of NAI selective pressure was present in this instance before the appearance of the resistance mutation; we note that the recommended course of oseltamivir for influenza treatment is 5 days, but is often given longer to immunocompromised patients. Thus, this report demonstrates that NAI resistance can occur, apparently in response to antiviral pressure, within the standard duration of administration. Although we cannot rule out the possibility that this deletion existed in a small subpopulation of the virus when the host became infected, the absence of this deletion in the 50 NA clones sequenced from the original clinical specimen suggests that if it was present, it was only a minor component. Given the available evidence, it appears likely that the deletion was rapidly selected for due to treatment with oseltamivir, since by day 6 of treatment the deletion-containing virus was the dominant viral genotype. Unlike the previously reported instance of this deletion, this isolate did not bear other mutations commonly associated with NAI resistance.
No firm definition of a resistant IC50 for neuraminidase inhibitors exists, but a change in IC50 of 10-fold or greater between a single virus pre- and post-treatment is commonly considered the hallmark of resistance [38]. As was shown in the previous characterization of a virus with this deletion [32], we found that the current isolate had a greater than 10-fold increase in IC50 for oseltamivir. These data, along with the apparent lack of clinical efficacy of the NAI in this case, indicate that this deletion causes clinically significant resistance to oseltamivir. Also of interest is the reduction of sensitivity to zanamivir, primarily in cell culture and to a lesser degree in NA activity assays. This change may or may not correlate with clinically significant resistance; further studies are needed.
Characterization of the viral isolate containing this naturally occurring deletion revealed no differences in replicative fitness or transmissibility when compared to the deletion-free isolate. No effect on the virus’s ability to grow in cell culture was seen, as judged by their similar peak titers and growth curves. More compelling than the cell culture findings, we also observed similar duration and magnitude of shedding from ferrets infected with both isolates. Thus in standard models of influenza replication and transmission, our data suggest that this deletion had no detrimental effect on viral fitness.
While the factors underlying influenza evolution and selection are incompletely understood, transmissibility is likely a very important factor in determining whether a multi-drug resistant virus could successfully establish itself as a dominant strain in humans. The recent, unexpected emergence of an NAI-resistant seasonal A/H1N1 virus containing the NA H275Y mutation, with no apparent detriment to its transmissibility, is ample evidence that NAI resistance and evolutionary fitness need not be mutually exclusive.
The independent emergence of this NAI resistance-conferring deletion in two instances and the good viral fitness and efficient transmission of the isolate in a mammalian host raises the possibility of an H3N2 virus containing this 4 amino acid deletion becoming more common as did H275Y containing H1N1 isolates. With clinically significant resistance to the adamantanes and oseltamivir, and decreased sensitivity to zanamivir, this is a disconcerting possibility. That both occasions involved immunocompromised patients with prolonged illness due to influenza suggests that careful attention to diagnosis, treatment, and prevention of spread is warranted. Additionally, the value of rigorous surveillance in identifying and tracking the emergence of similar mutations cannot be underestimated. Such efforts could obviously be helpful in containing the emergence of resistant viruses, but moreover, they would also inform our understanding of the evolution of influenza and help us to assess whether the immunocompromised are an important source for the emergence of antiviral resistance.
Conclusion
Widespread emergence of antiviral resistance and identification of cases such as the one identified here suggest that resistance to neuraminidase inhibitors and the adamantanes is likely to increase as influenza viruses continue to evolve. The rapid emergence of oseltamivir resistance in an already amantadine resistant A/H3N2 virus highlights the difficulties we face in treating influenza, especially those patients who are susceptible to prolonged infection such as immunocompromised individuals. The prompt emergence of this multi-drug resistant virus during clinical NAI therapy along with the lack of apparent detriment to viral fitness and transmissibility suggest that this resistance-conferring NA gene deletion as well as other resistance conferring mutations may not be isolated events and may become more widespread as usage of NAI increases, especially in the immunocompromised or chronically ill. Future studies to evaluate influenza treatment and prevention strategies for immunocompromised individuals, will need to include careful consideration of the rapid development of drug resistance during antiviral treatment.
Acknowledgments
The intramural program of the NIAID at the NIH and the FDA supported this work. Authors have no conflict of interests. The authors would like to acknowledge Dr. Jeffrey Cohen for his support of the clinical protocol. This work was presented in part at the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy, September 2009.
References
- 1.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289:179–86. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 2.Russell CA, Jones TC, Barr IG, et al. The global circulation of seasonal influenza A (H3N2) viruses. Science. 2008;320:340–6. doi: 10.1126/science.1154137. [DOI] [PubMed] [Google Scholar]
- 3.Stohr K. Influenza--WHO cares. Lancet Infect Dis. 2002;2:517. doi: 10.1016/s1473-3099(02)00366-3. [DOI] [PubMed] [Google Scholar]
- 4.Morens DM, Taubenberger JK, Fauci AS. The persistent legacy of the 1918 influenza virus. N Engl J Med. 2009;361:225–9. doi: 10.1056/NEJMp0904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. Update: novel influenza A (H1N1) virus infections - worldwide, May 6, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:453–8. [PubMed] [Google Scholar]
- 6.Bright RA, Medina MJ, Xu X, et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–81. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 7.Deyde VM, Xu X, Bright RA, et al. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis. 2007;196:249–57. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 8.Harper SA, Bradley JS, Englund JA, et al. Seasonal influenza in adults and children--diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:1003–32. doi: 10.1086/604670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jefferson TO, Demicheli V, Di Pietrantonj C, Jones M, Rivetti D. Neuraminidase inhibitors for preventing and treating influenza in healthy adults. Cochrane Database Syst Rev. 2006;3:CD001265. doi: 10.1002/14651858.CD001265.pub2. [DOI] [PubMed] [Google Scholar]
- 10.Monto AS, McKimm-Breschkin JL, Macken C, et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother. 2006;50:2395–402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. Monitoring of neuraminidase inhibitor resistance among clinical influenza virus isolates in Japan during the 2003–2006 influenza seasons. Wkly Epidemiol Rec. 2007;82:149–50. [PubMed] [Google Scholar]
- 12.WHO. Influenza A(H1N1) virus resistance to oseltamivir - 2008/2009 influenza season, northern hemisphere. Geneva: World Health organization; 2009. ed_ns.pdf HNw, ed. Vol. September 3, 2009. [Google Scholar]
- 13.Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360:953–6. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 14.Layne SP, Monto AS, Taubenberger JK. Pandemic influenza: an inconvenient mutation. Science. 2009;323:1560–1. doi: 10.1126/science.323.5921.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC. Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients - Seattle, Washington, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:893–6. [PubMed] [Google Scholar]
- 16.Williamson JC, Pegram PS. Respiratory distress associated with zanamivir. N Engl J Med. 2000;342:661–2. doi: 10.1056/NEJM200003023420914. [DOI] [PubMed] [Google Scholar]
- 17.GSK. Zanamivir Package Insert. Philadelphia: GSK; 2008. [Google Scholar]
- 18.Surveillance for the 2009 pandemic influenza A (H1N1) virus and seasonal influenza viruses - New Zealand, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:918–21. [PubMed] [Google Scholar]
- 19.Anderson RM. How well are we managing the influenza A/H1N1 pandemic in the UK? BMJ. 2009;339:b2897. doi: 10.1136/bmj.b2897. [DOI] [PubMed] [Google Scholar]
- 20.Hirst TM. A/H1N1 pandemic. Too soon for congratulations. BMJ. 2009;339:b3248. doi: 10.1136/bmj.b3248. [DOI] [PubMed] [Google Scholar]
- 21.Kmietowicz Z. England to launch special flu service next week to take pressure off primary care. BMJ. 2009;339:b2932. doi: 10.1136/bmj.b2932. [DOI] [PubMed] [Google Scholar]
- 22.CDC. Hospitalized patients with novel influenza A (H1N1) virus infection - California, April-May, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:536–41. [PubMed] [Google Scholar]
- 23.Vaillant L, La Ruche G, Tarantola A, Barboza P. Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro Surveill. 2009;14 doi: 10.2807/ese.14.33.19309-en. [DOI] [PubMed] [Google Scholar]
- 24.Malavaud S, Malavaud B, Sandres K, et al. Nosocomial outbreak of influenza virus A (H3N2) infection in a solid organ transplant department. Transplantation. 2001;72:535–7. doi: 10.1097/00007890-200108150-00032. [DOI] [PubMed] [Google Scholar]
- 25.Yousuf HM, Englund J, Couch R, et al. Influenza among hospitalized adults with leukemia. Clin Infect Dis. 1997;24:1095–9. doi: 10.1086/513648. [DOI] [PubMed] [Google Scholar]
- 26.Safrin S, Rush JD, Mills J. Influenza in patients with human immunodeficiency virus infection. Chest. 1990;98:33–7. doi: 10.1378/chest.98.1.33. [DOI] [PubMed] [Google Scholar]
- 27.Nichols WG, Guthrie KA, Corey L, Boeckh M. Influenza infections after hematopoietic stem cell transplantation: risk factors, mortality, and the effect of antiviral therapy. Clin Infect Dis. 2004;39:1300–6. doi: 10.1086/425004. [DOI] [PubMed] [Google Scholar]
- 28.McMinn P, Carrello A, Cole C, Baker D, Hampson A. Antigenic drift of influenza A (H3N2) virus in a persistently infected immunocompromised host is similar to that occurring in the community. Clin Infect Dis. 1999;29:456–8. doi: 10.1086/520243. [DOI] [PubMed] [Google Scholar]
- 29.Rocha E, Cox NJ, Black RA, Harmon MW, Harrison CJ, Kendal AP. Antigenic and genetic variation in influenza A (H1N1) virus isolates recovered from a persistently infected immunodeficient child. J Virol. 1991;65:2340–50. doi: 10.1128/jvi.65.5.2340-2350.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishikawa F, Sugiyama T. Direct isolation of H1N2 recombinant virus from a throat swab of a patient simultaneously infected with H1N1 and H3N2 influenza A viruses. J Clin Microbiol. 1983;18:425–7. doi: 10.1128/jcm.18.2.425-427.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baz M, Abed Y, McDonald J, Boivin G. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis. 2006;43:1555–61. doi: 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 32.Abed Y, Baz M, Boivin G. A novel neuraminidase deletion mutation conferring resistance to oseltamivir in clinical influenza A/H3N2 virus. J Infect Dis. 2009;199:180–3. doi: 10.1086/595736. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 34.Gubareva LV, Webster RG, Hayden FG. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob Agents Chemother. 2001;45:3403–8. doi: 10.1128/AAC.45.12.3403-3408.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eichelberger MC, Hassantoufighi A, Wu M, Li M. Neuraminidase activity provides a practical read-out for a high throughput influenza antiviral screening assay. Virol J. 2008;5:109. doi: 10.1186/1743-422X-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szretter KJ, Baa KJ. In: Influenza: Propagation, Quantification, and Storage. RC, editor. Hoboken, NJ: John Wiley and Sons; 2007. Current protocols in immunology. [Google Scholar]
- 37.Krafft AE, Russell KL, Hawksworth AW, et al. Evaluation of PCR testing of ethanol-fixed nasal swab specimens as an augmented surveillance strategy for influenza virus and adenovirus identification. J Clin Microbiol. 2005;43:1768–75. doi: 10.1128/JCM.43.4.1768-1775.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zambon M, Hayden FG. Position statement: global neuraminidase inhibitor susceptibility network. Antiviral Res. 2001;49:147–56. doi: 10.1016/s0166-3542(01)00124-3. [DOI] [PubMed] [Google Scholar]