Abstract
Phagocytic removal of apoptotic cells occurs efficiently in vivo such that even in tissues with significant apoptosis, very few apoptotic cells are detectable1. This is thought to be due to the release of find-me signals by apoptotic cells that recruit motile phagocytes such as monocytes, macrophages, and dendritic cells, leading to the prompt clearance of the dying cells2. However, the identity and in vivo relevance of such find-me signals are not well understood. Here, through several lines of evidence, we identify extracellular nucleotides as a critical apoptotic cell find-me signal. We demonstrate the caspase-dependent release of ATP and UTP (in equimolar quantities) during the early stages of apoptosis by primary thymocytes and cell lines. Purified nucleotides at these concentrations were sufficient to induce monocyte recruitment comparable to apoptotic cell supernatants. Enzymatic removal of ATP and UTP (by apyrase or ectopic CD39 expression) abrogated the ability of apoptotic cell supernatants to recruit monocytes in vitro and in vivo. We then identified the ATP/UTP receptor P2Y2 as a critical sensor of nucleotides released by apoptotic cells using RNAi depletion studies in monocytes, and macrophages from P2Y2-null mice3. The in vivo relevance of nucleotides in apoptotic cell clearance was revealed by two approaches. First, in a murine air-pouch model, apoptotic cell supernatants induced a three-fold greater recruitment of monocytes and macrophages compared to supernatants from healthy cells; this recruitment was abolished by depletion of nucleotides and significantly decreased in P2Y2−/− mice. Second, clearance of apoptotic thymocytes was significantly impaired by either depletion of nucleotides or interference with P2Y receptor function (by pharmacological inhibition, or in P2Y2−/− mice). These results identify nucleotides as a critical find-me cue released by apoptotic cells to promote P2Y2-dependent phagocyte recruitment, and provide strong evidence for a clear relationship between a find-me signal and efficient corpse clearance in vivo.
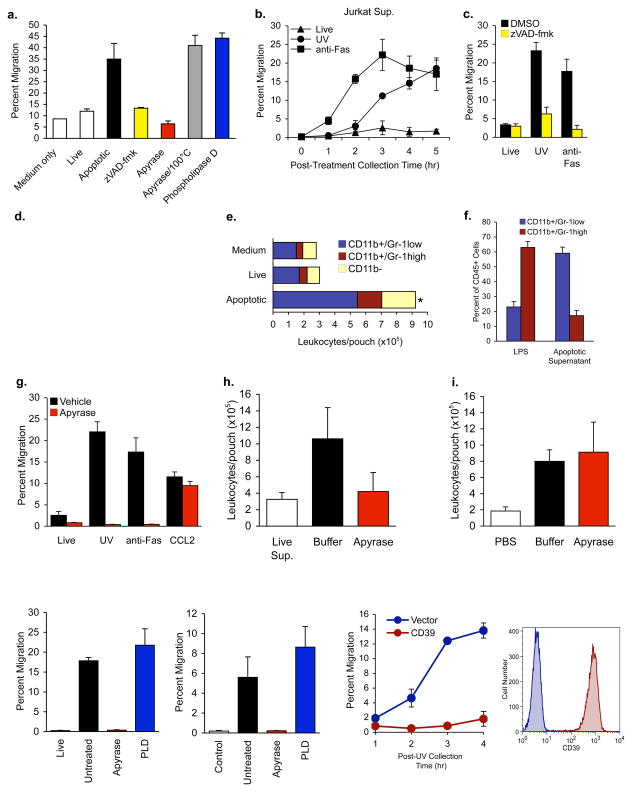
Most developing thymocytes (95%) undergo apoptosis; yet in steady-state only 1–2% are detectable as apoptotic4,5. It is hypothesized that dying thymocytes secrete soluble factors that attract resident phagocytes to promote prompt clearance2,6. To determine if apoptotic thymocytes release such factors, cell-free supernatants after apoptosis induction (by anti-Fas/CD95 crosslinking) were assessed for their ability to attract THP-1 monocytesor primary human monocytes in a transwell migration assay (Figure 1a and Supplemental Figure S2). Apoptotic supernatants caused a 3-fold increase in monocyte migration compared to supernatants of live thymocytes. Such release of chemotactic factors was also seen with Jurkat cells (a mature T cell line) induced to undergo Fas- or ultraviolet (UV)-mediated apoptosis (Figure 1b). There was no detectable increase in membrane permeability or leakage of cytoplasmic markers when the supernatants were collected (Supplemental Figure S1a-e and Figure 2g). Moreover, supernatants from cells pretreated with the caspase inhibitor zVAD-fmk prior to the induction of apoptosis failed to induce monocyte migration (Figure 1a, c), suggesting caspase-dependent and regulated chemoattractant release. In a time-course, chemotactic factor release correlated with the onset and progression of apoptosis (assessed by annexin V exposure and caspase 3/7 activation, Figure 1b, Supplemental Figure S1b,c). Lastly, the chemotactic factor(s) was soluble and heat-stable, since high-speed centrifugation or boiling of the supernatants did not affect the chemotactic potential (Supplemental Figure S3f).
Figure 1. Chemotactic factor released by apoptotic cells attracts monocytes in vitro and in vivo.
a, Migration of THP-1 monocytes through transwell(5 μm pore) to supernatants from control (‘live’) or Fas-induced apoptotic murine thymocytes, thymocytes pre-treated with caspase inhibitor (zVAD-fmk), apoptotic cell supernatants with apyrase, heat-inactivated apyrase or phospholipase D. The fraction of input monocytes that migrated to the lower chamber is shown. b, Attraction of monocytesby Jurkat T cell supernatants collected at the indicated times after apotosis induction via UV or anti-Fas for the indicated times. c, Monocyte attraction was inhibited by pre-treatment of Jurkat cells with zVAD-fmk prior to UV or anti-Fas treatment. d, Schematic for testing recruitment of leukocytes by apoptotic cell supernatants in the mouse air-pouch model. e, Recruitment to the air-pouch of macrophages and monocytes (CD11b+/Gr-1low) or neutrophils (CD11b+/Gr-1high) 24 hrs after injection of apoptotic cell supernatants. Eight mice per group, *p=0.02. f, Monocyte/macrophage and neutrophil populations recruited to the air-pouch 24 hrs after injection of LPS (1 μg) or apoptotic supernatants. Results are the average of six (LPS) and nine (apoptotic supernatant) mice. g, h, and i, Treatment of apoptotic cell supernatants with apyrase inhibits attraction of monocytes in vitro (g) or in the air-pouch model in vivo (h, i), but does not affect monocyte migration to the chemokine CCL2 (250 ng). Five (h) or three (i) mice per group, *p=0.005. j, k, Migration of monocytes to supernatants from apoptotic Jurkat or MCF-7/caspase-3 cells, supernatants being treated with apyrase or PLD. l, (right) CD39 surface expression on transfected Jurkat cells, and (left) monocyte migration to supernatants from CD39-overexpressing cells after UV treatment. Error bars indicate s.e.m.
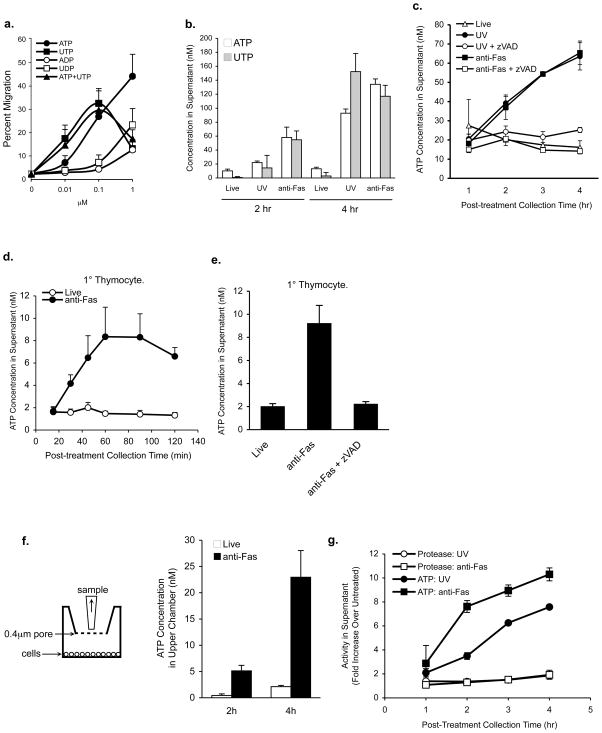
Figure 2. Regulated release of ATP and UTP as chemoattractants by apoptotic cells.
a, Migration of THP-1 monocytes to purified nucleotides at the indicated concentrations. b, Quantitation of ATP and UTP in supernatants of control and apoptotic Jurkat cells at 2 and 4hr after apoptosis induction. c, ATP level in supernatants of apoptotic Jurkat cells at indicated times and the inhibition by zVAD-fmk. d, ATP levels in supernatants of control or anti-Fas treated thymocytes for the indicated times. e, ATP levels in supernatants of thymocytes treated with zVAD-fmk before anti-Fas treatment. f, Left, Schematic of supernatant collection without disturbing the cells. Right, Quantitation of ATP that has diffused through the 0.4 μm filter into the medium from untreated (live) or anti-Fas treated Jurkat cells. g, Integrity of the cell membrane is retained when apoptotic cell supernatants are collected, as determined by ATP release but not leakage of cytoplasmic protease activity. Error bars indicate s.e.m.
We next tested whether find-me signal(s) in apoptotic cell supernatants could attract phagocytesin vivo. We used a murine dorsal air-pouch model (Figure 1d) where the supernatants from apoptotic or healthy cells were injected into sterile, subcutaneous air-pouches7. When cells in the air-pouch were recovered by lavage after 24 hr, apoptotic cell supernatants caused a 3-fold increase in the number of CD45+ leukocytes recruited compared to live cell supernatants or medium alone (Figure 1e, n=8, p=0.02). The total number of monocytes and macrophages (CD11b+/Gr-llow) in the lavage were increased about 3-fold compared to neutrophils (Gr-1high cells) (Figure 1e). By contrast, bacterial lipopolysaccharide (LPS) injection induced the recruitment of mostly Gr-1high neutrophils (Figure 1f). This is consistent with previous studies on preferential recruitment of monocyte/macrophages over inflammatory neutrophils to cells undergoing apoptosis8,9. F4/80+ macrophages recruited to the pouch could also engulf apoptotic Jurkat cells injected into the pouch (not shown). These data revealed the release of find-me signal(s) by apoptotic lymphocytes that attract monocytes in vitro and in vivo.
We then sought to determine the nature of the chemoattractant. Based on in vitro studies, the lipid lysophosphatidylcholine (LPC) was implicated as a find-me signal released by apoptotic MCF-7 cancer cells10. However, we did not observe monocyte migration to purified LPC over a range of concentrations (0.1–100 μM, data not shown); moreover, treatment of apoptotic cell supernatants with phospholipase D (PLD), an enzyme that promotes hydrolysis of LPC (see Supplemental Figure S3a,b), did not affect chemotactic activity of supernatants from apoptotic thymocytes, Jurkat cells or MCF-7 cells (Figure 1a, j, k). CX3CL1 (fractalkine) released by apoptotic Burkitt-lymphoma B cells can also act as a find-me signal11; however, THP-1 monocytes used here failed to show migration toward purified CX3CL1, and the anti-fractalkine depleting antibody did not block migration in our assays (data not shown). Thus, the find-me signal released by apoptotic primary thymocytes and Jurkat cells appeared to be distinct from those previously reported.
Subsequently, several lines of evidence suggested a role for extracellular nucleotides as a possible find-me signal. Treatment of apoptotic cell supernatants with recombinant apyrase, an enzyme that hydrolyzes nucleoside triphosphates and diphosphates to nucleoside monophosphates (e.g. ATP → ADP→ AMP), abolished the monocyte chemoattractant activity of apoptotic thymocytes, Jurkat and MCF-7 cells at all time points (Figure 1a, g, j, k, and Supplemental Figure S3e). Apyrase did not affect monocyte migration to chemokines CCL2 or CXCL12 (Figure 1g and data not shown). Importantly, treatment of apoptotic cell supernatants (but not CCL2) with apyrase prior to injection into the dorsal air-pouch also inhibited the attraction of leukocytes in vivo (Figure 1h, i). As another approach, we expressed in Jurkat cells the transmembrane protein CD39 (NTPDase-l), the primary mammalian ecto-apyrase responsible for NTP degradation by immune cells in vivo12 (see Supplemental Figure S4b); CD39 expression abrogated the chemoattractant activity in the supernatants of apoptotic Jurkat (Figure 1l). Neither apyrase treatment nor CD39 overexpression impaired the induction of apoptosis (Supplemental Figure S4a and Figure 4g). Heat-inactivation of the recombinant apyrase (prior to addition to apoptotic cell supernatants) abolished its effect (Figure 1a and Supplemental Figure S3d), suggesting a need for intact enzymatic activity. Thus, induction of apoptosis led to accumulation of extracellular nucleotides, capable of monocyte chemoattraction in vitro and in vivo.
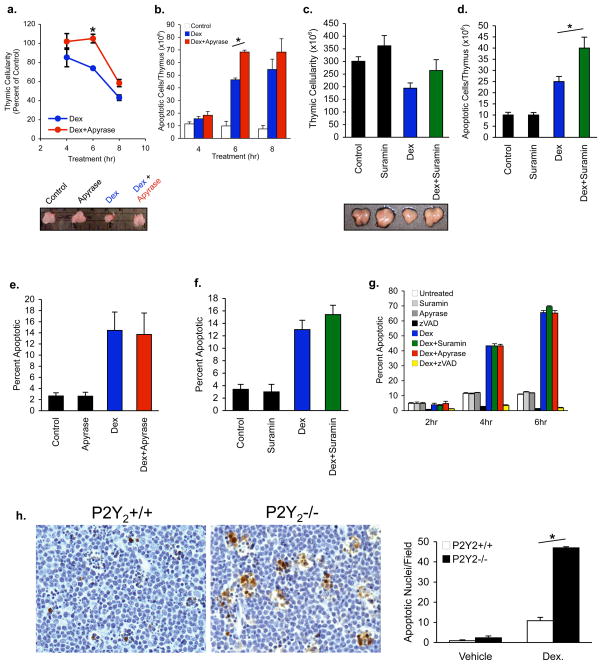
Figure 4. Interference with the nucleotide find-me signal or its sensing impairs clearance of apoptotic cells in the thymus.
a, b, C57BL/6 mice (4–6 wk) were injected i.p. with 250μg dexamethasone (Dex) for the indicated times, with or without apyrase, and the cellularity (a) or number of apoptotic cells per thymus (b) was determined (annexin V positive/propidium iodide-negative populations). Data in a were normalized to untreated animals within the same experimental group (4, 6 or 8hr). Representative thymus from each group is shown below. Data shown are representative of 2–4 experiments per time point using at least three mice/group, *p=0.03. c, d, Same as in a, b, except mice were injected with 6mg of suramin 1hr prior to Dex injection (6hr). Representative thymus from each group is shown below c. Four mice per group, *p=0.03. e, f, Effect of apyrase or suramin on percentage of apoptotic cells in vivo in the thymi of Dex treated mice (from a-d above). g, Apyrase (0.05U/mL) and suramin (100μM) do not affect Dex-induced thymocyte apoptosis in vitro. zVAD was included as a control. Percentage of apoptotic cells is shown. h, Left, Paraffin sections from thymi of wild-type mice and P2Y2−/− mice treated with Dex for 6hr (Apostain, brown. Hematoxylin, blue). Right, mean number of Apostain positive nuclei per field from 10–16 random fields per section per mouse, *p=0.001. Error bars indicate s.e.m.
Among the four naturally occurring extracellular nucleotides (ATP, ADP, UTP and UDP) ATP and UTP induced strong chemotactic activity in THP-1 (Figure 2a); in contrast, ADP and UDP exhibited partial activity at the highest concentrations tested (Figure 2a), but lower than NTPs. The migration was also stimulated by non-hydrolyzable ATPγS, but not adenosine (Supplemental Figure S5a), suggesting attraction primarily toward triphosphate nucleotides. When ATP and UTP levels in apoptotic cell supernatants were directly quantified (see methods), higher ATP and UTP could be detected as early as 2 hr after apoptosis induction, with further increase by 4 hours (Figure 2b). The concentration of ATP and UTP at the time point when the apoptotic supernatants induced maximal monocyte migration correlated well with the concentration at which pure ATP and UTP caused maximal migration (~100 nM) (Figure 2a, b and Figure 1b). Adding pure ATP and UTP to the upper chamber of the transwell, to disrupt the gradient, blocked the migration of monocytes to the lower chamber containing apoptotic cell supernatants (Supplemental Figure S6 a-c). Although ATP can promote chemokinesis/random migration of neutrophils13, addition of pure ATP or UTP only to the upper chamber did not induce migration of THP-1 cells to the lower chamber, indicating that the migration induced is not chemokinesis, rather chemotaxis (Supplemental Figure S6d). Furthermore, the nucleotide release from apoptotic cells is caspase-dependent, occurs after different types of apoptosis induction (DNA damage, receptor-mediated, and steroid-induced), occurred in primary thymocytes, Jurkat and epithelial cells undergoing apoptosis, and this ATP release correlates well with the induction of apoptosis (Figure 2c-e and Supplemental Figures S1b-d and S8). The release of ATP during apoptosis was not due to leakage of cellular contents or mechanical stress during handling of cells14 (Figure 2f, g). Collectively, these data strongly suggested a role for ATP and UTP as find-me signals important for phagocyte chemoattraction by apoptotic cells.
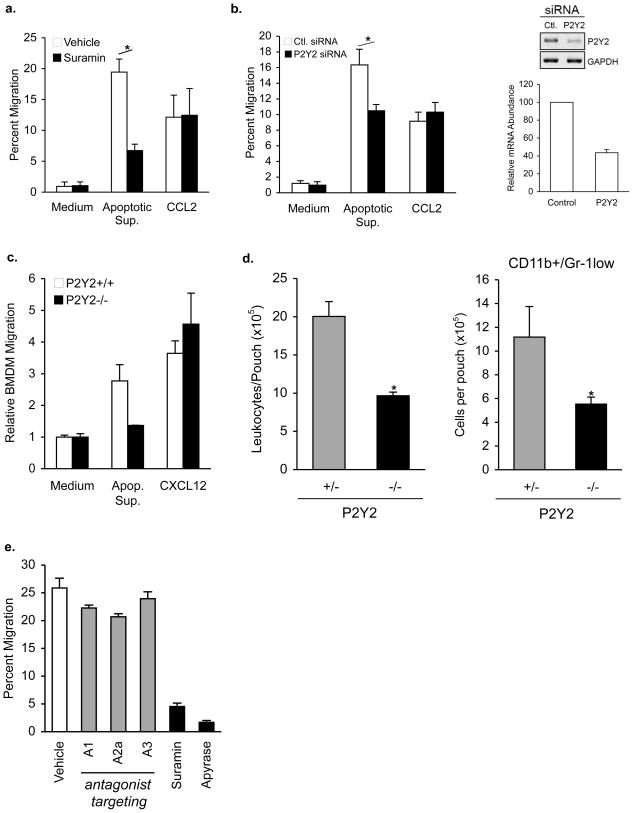
We then addressed how the phagocytes may ‘sense’ the extracellular nucleotides as a find-me cue. Leukocyte migration toward nucleotides has been shown to be dependent on members of the P2Y family of G protein-coupled receptors15,16. We tested the role of P2Y receptors on monocytes and macrophages in migration toward apoptotic cell supernatants. First, pretreatment of THP-l cells with suramin, a nonselective inhibitor of P2 family members, showed dose-dependent inhibition of migration to apoptotic supernatants, but not CCL2 (Figure 3a and Supplemental Figure S5c). After evaluating several P2Y family members through known P2Y antagonists to inhibit migration toward apoptotic cell supernatants, we focused on P2Y2 based on its known affinities for ATP and UTP (since both are released by apoptotic cells), and P2Y2 expression on monocytes and macrophages16. Although P2Y4 also fit the profile for ATP and UTP binding, THP-1 cells express no detectable P2Y4, yet strongly express P2Y2 mRNA (ref. 17 and data not shown). siRNA-mediated knockdown of P2Y2 in THP-1 monocytes led to a 60% decrease in P2Y2mRNA, and also partially inhibited migration to apoptotic cell supernatants (10.5% ± 0.8 versus 16.3% ± 2.0 for control siRNA, n=6, p=0.003), but did not affect migration to CCL2 or CXCL12 (Figure 3b and data not shown). Bone marrow derived macrophages (BMDM) from P2Y2−/− mice3 showed impaired migration to apoptotic supernatants, but their migration to CXCL12 was intact (Figure 3c). Importantly, when apoptotic cell supernatants were injected into the air-pouch of P2Y2-deficientmic e, there was a strong reduction in the recruitment of monocytes and macrophages to the pouch, indicating in vivo relevance of this receptor in sensing the find-me signal (Figure 3d). Although P2Y6 was shown to play a role in UDP-dependent leukocyte migration15,18 and phagocytosis by microglial cells19, neither RNAi toward P2Y6, nor the P2Y6 antagonist MRS2578 showed specific inhibition of migration (Supplemental Figure S5b and data not shown) Finally, addition of antagonists to the adenosine receptors A1, A2aand A3 13,20, or the A2 receptor agonistCGS21860 did not significantly affect the migration of monocytes to the apoptotic cell supernatants (Figure 3e and Supplemental Figure S5d). Moreover, adenosine itself did not induce migration of THP-1 cells and addition of exogenous adenosine did not affect migration to apoptotic cell supernatants (Supplemental Figure S5a, d). Together, these results identify the P2Y2 receptor on monocytes and macrophages as a critical sensor of the find-me signal released by apoptotic cells.
Figure 3. P2Y2 receptor on monocytes and macrophages as a sensor of ATP/UTP released by apoptotic cells.
a, Effect of pretreatment of monocytes with P2Y receptor antagonist suramin (100 μM) on migration to apoptotic cell supernatants or the chemokine CCL2 (50 ng/mL). n=3, *p=0.003. b, Migration of THP-1 transfected with siRNA specific for P2Y2 receptoror control siRNA. n=6, *p=0.03. Right, qPCR and agarose gel electrophoresis (inset, inverted image) analysis of P2Y2 receptor mRNA levels in siRNA transfected THP -1 cells. c, BMDM from P2Y2+/+ or P2Y2−/− mice were assessed for transwell migration to apoptotic Jurkat supernatant or CXCL12 (50ng/mL). d, Recruitment of CD45+ cells (left) and CD11b+/Gr-1low monocytes and macrophages (right) to the air-pouch of mice with the indicated P2Y2genotypes 24hr after injection of apoptotic Jurkat supernatants. Five (P2Y2+/+) and seven (P2Y2−/−) mice per group, *p≤0.03. e, THP-1 cells pretreated with antagonists targeting adenosine receptors A1, A2a and A3, apyrase or suramin prior to migration assay. Error bars indicate s.e.m.
To further test the importance of nucleotides as a find-me signal in an in vivo model of apoptosis, we used intraperitoneal (i.p.) injection of dexamethasone (Dex), where a large fraction of immature thymocytes undergo relatively synchronous apoptosis and phagocytic clearance5,21. Dex injection induced thymic apoptosis, with decline in thymus size and cellularity within 4 hrs, and by 8 hrs was less than half that of control-treated mice (Figure 4a). Treatment of thymocytes with Dex in vitro also induces apoptosis in a large fraction of the cells by 4 hrs (>40%) and 6 hrs (>60%) (Figure 4g). We asked whether apyrase-mediated destruction of nucleotides in vivo could influence Dex-mediated thymocyte apoptosis and clearance. Injection of apyrase significantly reversed the decline in thymus size and cellularity due to Dex treatment (especially at 6 hrs) (Figure 4a). This was not due to an effect of apyrase on the apoptotic process itself, as the fraction of cells undergoing apoptosis due to Dex was unchanged upon apyrase treatment in vivo or in vitro (Figure 4e,g). Interestingly, the total number of apoptotic cells remaining at 6 hr and 8 hr were increased in mice treated with apyrase+Dex, compared to Dex alone (Figure 4b). Since apyrase had no effect on the phagocytic capacity of macrophages (Supplemental Figure S9), apyrase-mediated destruction of the nucleotide find-me signal appeared to affect phagocyte recruitment, and in turn delay clearance.
In a complementary set of studies, we tested how disrupting the‘ sensing’ of the find-me signal affects apoptotic cell clearance. Injection of the P2Y inhibitor suramin prior to Dex reversed the diminution of the thymic cellularity and organ size seen with Dex alone (Figure 4c, d). The total number of apoptotic thymocytes also increased in suramin+Dex conditions (Figure 4d). As was the case with apyrase, suramin itself did not alter the induction of thymocyte apoptosis (Figure 4f, g). When we assessed the presence of apoptotic cells in the native thymic architecture (by immunohistochemistry using anti-ssDNA antibody), there were increased numbers of uncleared apoptotic cells in Dex+suramin treated conditions compared to Dex alone (Supplemental Figure S10). We also assessed whether genetic disruption of the putative find-me signal receptor P2Y2would affect apoptotic cell clearance in vivo. Following Dex injection, the number of apoptotic thymocytes in the thymi of P2Y2−/− mice was significantly increased compared to control mice (Figure 4h). Taken together, the disruption of a find-me signal circuit at the level of nucleotides or the sensing receptor significantly impairs the clearance of apoptotic thymocytes, without an apparent effect on the induction of apoptosis or engulfment.
The data presented in this report provide new insights on particular aspects of the programmed cell death process. First, this work identifies ATP and UTP as critical and non-redundant find-me signal released by apoptotic cells. To our knowledge, this is the first documentation of regulated and caspase-dependent release of nucleotides from apoptotic cells, with a functional secondary consequence. Since nucleotide release is seen in primary cells and cell lines (after different types of apoptotic stimuli), nucleotides may be a broadly used find-me signal. However, these data do not rule out other chemotactic factors that work alone or together with nucleotides. Second, these data establish a clear relationship between a find-me signal and efficient apoptotic cell clearance in vivo; the disruption of the ‘find-me signal’ circuit at the level of ATP/UTP or the receptors (P2Y) impaired the clearance of apoptotic thymocytes. Although we focused here on motile monocytes/macrophages, genetic studies in C. elegans, where healthy cells engulf the dying neighbors, have revealed a link between apoptosis and engulfment 22,23. How nucleotides might regulate engulfment by neighboring non-professional phagocytes6,24 remains to be determined.
Extracellular nucleotides at higher concentrations are considered pro-inflammatory25 (>1 μM, e.g. necrotic cells, Supplemental Figure S7), but nucleotides can also induce an anti-inflammatory response26. Besides serving as a find-me signal, whether nucleotides participate in anti-inflammatory signaling during engulfment remains to be determined. Bournazou et al recently showed that lactoferrin released by apoptotic cells inhibit neutrophil migration27. How nucleotides and lactoferrin concurrently promote monocyte migration while inhibiting neutrophil migration remains to be seen. Since failure to promptly clear dying cells can lead to autoimmunity and chronic inflammatory diseases9, phagocyte chemoattraction to apoptotic cells via nucleotides may have implications for human disease states related to failed clearance.
METHODS SUMMARY
Supernatant preparation
Jurkat cells (E6-1) at 2×106/mL in RPMI/5% heat-inactivated FBS/10 mM HEPES were treated with 250 ng/mL anti-Fas (CH11) or 100 mJ UVC. Freshly isolated thymocytes from 4–5 wk C57BL/6 mice were treated at 5×106/mL with crosslinked anti-Fas (5 μg/mL Jo2, 2 μg/mL protein G) in RPMI/1%BSA/10 mM HEPES. Supernatants were collected by two successive centrifugations at 2000g/4 min/4°C. zVAD-fmk (50 μM) pretreatment and treatment of supernatants by apyrase (0.025 U/mL) and phospholipase D (0.5 U/mL) was carried out 5 min at RT. MCF-7 supernatants were prepared as previously described10.
Migration Assays
THP-1 cells at 2×106/mL were placed on a 5 μm pore Transwell® with chemoattractant for 1 hr. Percent migration was determined by flow cytometry using AccuCount beads. For BMDM migration, 5×104 d7 BMDM cells were placed on a 5 μm pore Transwell®, incubated 2 hrs with Jurkat supernatants in RPMI/1%BSA/HEPESin lower chamber, and migration determined by Diff-Quick staining and microscopy.
In vivo experiments
Air-pouch experiments were performed as described7 using 8–12 wk C57BL/6 mice injected with 1 mL of 0.2 μm filtered supernatants. After 24 hrs, the pouch was lavaged and cells counted and analyzed by flow cytometry. For thymic clearance studies, 4–5 wk C57BL/6 mice were injected intraperitoneal with 250 μg of Dex, 5 U apyrase at midpoint of Dex treatment, and 6 mg of suramin 1 hr prior to Dex injection. Thymocytes were stained with annexin V and propidium iodide and beads added for quantitation by flow cytometry. For in situ apoptotic cell detection, thymi from female P2Y2−/− or age-matched C57BL/6 mice were stained by Apostain as described28.
Nucleotide measurement
ATP was measured by luciferase reaction as described29. UTP was quantified by the UDP-glucose pyrophosphorylase-based reaction30. Measurements were conducted on supernatants prepared from cells cultured in RPMI, 1% BSA, 10 mM HEPES.
Methods
Reagents
Purified nucleotides, adenosine, dexamethasone, etoposide, protein G, LPS and suramin were obtained from Sigma-Aldrich. Annexin V and other flow cytometry reagents were obtained from eBioscience. Other reagents were obtained as follows: recombinant apyrase (New England Biolabs), zVAD-fmk (Alexis Biochemicals), phospholipase D and ATPγS (EMD), anti-Fas (Jo2 anti-mouse clone, BD; CH11 anti-human clone, Millipore), siRNA (Dharmacon), purified lipids (Avanti Polar Lipids), chemokines (R & D). Adenosine receptor reagents were the kind gift of Dr. Joel Linden at the University of Virginia; the antagonists used (20μM each) were: 8-cyclopentyl-1,3-dipropylxanthine [CPX] (A1), ZM241 (A2a), MRS1191 (A3). Experiments were carried out comparing the stock solution of apyrase to the same solution dialyzed into PBS. Although the activity of the enzyme toward pure ATP in vitro was slightly decreased after dialysis of the enzyme (see Supplemental Figure S3c), the two preparations performed the same in all migration experiments we examined.
Air-pouch and thymic apoptotic cell clearance studies
All animal studies were carried out in accordance with the University of Virginia Animal Care and Use Committee guidelines and housed in a specific pathogen free facility. For air-pouch experiments, female C57BL/6 mice were used (Charles River Laboratories), except those conducted with P2Y2-deficient mice which included males and females (results were similar). P2Y2-deficient mice were obtained from Taconic3 with permission from Dr. Beverly Koller (UNC, Chapel Hill). Air -pouch experiments were performed as previously described7 using mice aged 8–12 weeks. Briefly, 5mL of 0.2μm filtered air was injected subcutaneously in the dorsal region. After 3 days, the same pouches were injected with 3mL of 0.2μm filtered air to maintain the pouch. Four days later the pouches were injected with 1mL of 0.2μm filtered supernatants from control or UV-treated Jurkat cells or with LPS (1μg) or CCL2 (250 ng). After 24 hrs, cells from the air-pouch were collected by lavage with 2mL HBSS/1% FBS twice. Harvested cells were resuspended in equal volumes and cell counting performed by hemacytometer and/or by flow cytometry. For analyses of specific populations in the air-pouch, cells were treated with anti-CD16/32 for 15 min on ice to block Fc receptor binding, followed by addition of the indicated fluorescently-labeled antibodies for 30 min on ice. After washing, the cells were analyzed on the FACS Canto (Becton Dickinson) instrument.
For thymic clearance studies, 4–5 week old mice were injected i.p. with 250 μg of dexamethasone in 300 μL PBS. Apyrase-treated mice were injected with 5 units of recombinant apyrase at the midpoint of the Dex treatment. Suramin-treated mice were injected intraperitoneally with 6 mg of suramin in 300 μL of PBS 1 hr prior to Dex injection. In all cases, control groups were treated with equivalent volumes of vehicle controls. After treatment, the thymus was collected and dissociated over a 70 μm mesh filter in cold HBSS/2% FBS and diluted 100-fold into 1× final annexin V binding buffer. Cells were incubated with annexin V and propidium iodide for 10 min at room temperature. AccuCount beads (Spherotech) were then added and the samples analyzed by flow cytometry (FACS Canto, BD) in duplicate.
Migration assays
Transwell migration assays were performed by applying 100μL of THP-1 cells at 2×106/mL to the upper chamber in the same culture medium as the chemoattractant in lower chamber (500μL) of a 24-well plate with 5 μm pore size transwells (Corning) for 1hr at 37°C, 5% CO2. The percentage of migrated cells was determined by FACS using 5.1μm AccuCount beads or by Cell Titer Glo (Promega) and calculated as the percentage of input cells. Bone marrow-derived macrophages (BMDM) were prepared as previously described7. Day 7 BMDM cells (5×104 were added to the upper well of a 5μm pore size transwell and incubated at 37°C, 5%CO2 for 2hr. The number of migrated cells on the underside of the membrane was determined by Diff-Quick staining and counting 5 random fields under 10× magnification per duplicate insert. For migration of primary human monocytes, PBMC from healthy donors were allowed to adhere on plastic dishes overnight at 37°C, 5%CO2. The next day, non-adherent cells were removed and adherent cells (monocyte enriched fraction) were collected resuspended in RPMI/1%BSA/HEPES and applied to the upper chamber of a 3μm pore size transwell at 5×104 cells/well. Cells that migrated to the lower chamber were collected, counted and stained for CD14. The data presented reflect the relative level of CD14+ monocytes migrated to the lower chamber based on post-migration CD14 staining.
siRNA and stable transfections
THP-1 cells were transfected with 1 μg of siRNA using the BTX Square Pulse T820 electroporator and used 72hr after transfection. The pA-Puro-hCD39 plasmid was generated by PCR cloning of the human CD39 cDNA (Open Biosystems) into the pA-Puro vector. For generation of the stable line, cells were electroporated with 10μg of linearized plasmid (pA-Puro-hCD39) and selected in puromycin for one week prior to clonal expansion and screening for cell surface expression of CD39.
Nucleotide Measurement
ATP was measured by the luciferin/lucifease assay via a LB953 AutoLumat luminometer (Berthold), as previously described29. UTP concentrations were quantified by the UDP-glucose pyrophosphorylase-based reaction, as described30. Briefly, 100μl samples were incubated in the presence of 0.5 U/mL UDPglucose pyrophosphorylase, 0.5U/mL inorganic pyrophosphatase, 1.6mM CaCl2, 2mM MgCl2, 25mM hydroxyethylpiperazine ethanesulfonic acid (HEPES) (pH 7.4), and ~100,000 cpm 1μM [14C]glucose-1P. Incubations lasted 1h at 30°C. Reactions were terminated by heating the samples at 95°C for 2 min. Conversion of [14C]glucose-1P to [14C]UTP was determined by HPLC. All nucleotide measurements were conducted on supernatants prepared from cells cultured in RPMI, 1% BSA, 10mM HEPES. For measurement of ATP without disturbing the cells, Jurkat cells were induced to undergo apoptosis in a 24 well plate and a 0.4μm pore transwell filter with 200μL medium was submerged in the well. The amount of ATP that diffused through the transwell was determined my acquiring samples from within the transwell at the indicated times.
Caspase activation and protease release
Caspase activation and protease release assays were performed using the Caspase-Glo and CytoTox-Glo (Promega) reagents according to manufacturer’s instructions.
Real-time PCR (qPCR)
cDNA was synthesized from 50 ng of RNA (RNeasy, Qiagen) using Superscript III (Invitrogen). qPCR was performed on the ABI Prism 7000 instrument using TaqMan probes (Applied Biosystems). Values shown are normalized to 18S levels.
Immunohistochemistry
Detection of apoptotic cells by Apostain (Alexis Biochemical) was carried out as previously described28.
Statistical analyses
Data are presented as the mean ± standard error of the mean (s.e.m.) unless otherwise noted. Statistical significance for individual data points was determined by the Student’s two-tailed t test. A p value less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank members of the Ravichandran lab and Dr. Kenneth Rock for helpful suggestions. We are grateful to Dr. Beverly Koller for use of the P2Y2 mice. We also thank Dr. Joel Linden for the adenosine receptor reagents, Dr. Ignacio Juncadella for lung epithelial cells, Dr. Kirsten Lauber and Dr. Sebastian Wesselborg for providing MCF-7/caspase-3 cells, and Robert Tacke for assistance with primary monocyte experiments. This work was supported by Public Health Service grants from the National Institutes of Health (to K.S.R. and N.L.), the American Cancer Society (M.R.E.) and the University of Virginia Farrow Fellowship (M.R.E.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing interests.
Author Contributions M.R.E. designed, performed and analyzed most of the experiments in this study with input from K.S.R. F.B.C. performed ATP quantitation experiments. P.T.C. helped with in vivo thymic apoptosis experiments. E.R.L. carried out HPLC analysis of supernatants. S.F.W. generated the CD39 expression plasmid and stable Jurkat cell lines. D.P. conducted phagocytosis experiments. A.K. and N.L. carried out the MS analysis and provided critical support in establishing the air-pouch model system. R.I.W. and J.J.L. carried out immunohistochemical detection of apoptotic cells in the thymus. M.O. and P.S. assisted with the BMDM generation and macrophage chemotaxis experiments. T.K.H. provided critical intellectual input in the preparation of the manuscript. K.S.R. provided overall coordination with respect to conception, design and supervision of the study. K.S.R. and M.R.E. wrote the manuscript with comments from co-authors.
References
- 1.Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–250. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell. 2004;14:277–287. doi: 10.1016/s1097-2765(04)00237-0. [DOI] [PubMed] [Google Scholar]
- 3.Homolya L, Watt WC, Lazarowski ER, Koller BH, Boucher RC. Nucleotide-regulated calcium signaling in lung fibroblasts and epithelial cells from normal and P2Y(2) receptor (−/−) mice. J Biol Chem. 1999;274:26454–26460. doi: 10.1074/jbc.274.37.26454. [DOI] [PubMed] [Google Scholar]
- 4.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 5.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–103. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 6.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 7.Kadl A, Galkina E, Leitinger N. Induction of CCR2-dependent macrophage accumulation by oxidized phospholipids in the air-pouch model of inflammation. Arthritis Rheum. 2009;60:1362–1371. doi: 10.1002/art.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huynh ML, Fadok VA, Henson PM. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J Clin Invest. 2002;109:41–50. doi: 10.1172/JCI11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savill J, Dransfield I, Gregory C, Haslett C. A blast from the past: clearance of apoptotic cells regulates immune responses. Nat Rev Immunol. 2002;2:965–975. doi: 10.1038/nri957. [DOI] [PubMed] [Google Scholar]
- 10.Lauber K, et al. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell. 2003;113:717–730. doi: 10.1016/s0092-8674(03)00422-7. [DOI] [PubMed] [Google Scholar]
- 11.Truman LA, et al. CX3CL1/fractalkine is released from apoptotic lymphocytes to stimulate macrophage chemotaxis. Blood. 2008;112:5026–5036. doi: 10.1182/blood-2008-06-162404. [DOI] [PubMed] [Google Scholar]
- 12.Mizumoto N, et al. CD39 is the dominant Langerhans cell-associated ecto-NTPDase: modulatory roles in inflammation and immune responsiveness. Nat Med. 2002;8:358–365. doi: 10.1038/nm0402-358. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, et al. ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science. 2006;314:1792–1795. doi: 10.1126/science.1132559. [DOI] [PubMed] [Google Scholar]
- 14.Lazarowski ER, Homolya L, Boucher RC, Harden TK. Direct demonstration of mechanically induced release of cellular UTP and its implication for uridine nucleotide receptor activation. J Biol Chem. 1997;272:24348–24354. doi: 10.1074/jbc.272.39.24348. [DOI] [PubMed] [Google Scholar]
- 15.Myrtek D, Idzko M. Chemotactic activity of extracellular nucleotideson human immune cells. Purinergic Signal. 2007;3:5–11. doi: 10.1007/s11302-006-9032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnstock G, Knight GE. Cellular distribution and functions of P2 receptor subtypes in different systems. Int Rev Cytol. 2004;240:31–304. doi: 10.1016/S0074-7696(04)40002-3. [DOI] [PubMed] [Google Scholar]
- 17.Moore DJ, et al. Expression pattern of human P2Y receptor subtypes: a quantitative reverse transcription-polymerase chain reaction study. Biochim Biophys Acta. 2001;1521:107–119. doi: 10.1016/s0167-4781(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 18.Idzko M, et al. Characterization of the biological activities of uridine diphosphate in human dendritic cells: Influence on chemotaxis and CXCL8 release. J Cell Physiol. 2004;201:286–293. doi: 10.1002/jcp.20070. [DOI] [PubMed] [Google Scholar]
- 19.Koizumi S, et al. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature. 2007;446:1091–1095. doi: 10.1038/nature05704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasko G, Linden J, Cronstein B, Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawane K, et al. Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat Immunol. 2003;4:138–144. doi: 10.1038/ni881. [DOI] [PubMed] [Google Scholar]
- 22.Hoeppner DJ, Hengartner MO, Schnabel R. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature. 2001;412:202–206. doi: 10.1038/35084103. [DOI] [PubMed] [Google Scholar]
- 23.Reddien PW, Cameron S, Horvitz HR. Phagocytosis promotes programmed cell death in C. elegans. Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- 24.Monks J, Smith-Steinhart C, Kruk ER, Fadok VA, Henson PM. Epithelial cells remove apoptotic epithelial cells during post-lactation involution of the mouse mammary gland. Biol Reprod. 2008;78:586–594. doi: 10.1095/biolreprod.107.065045. [DOI] [PubMed] [Google Scholar]
- 25.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.la Sala A, et al. Alerting and tuning the immune response by extracellular nucleotides. J Leukoc Biol. 2003;73:339–343. doi: 10.1189/jlb.0802418. [DOI] [PubMed] [Google Scholar]
- 27.Bournazou I, et al. Apoptotic human cells inhibit migration of granulocytes via release of lactoferrin. J Clin Invest. 2009;119:20–32. doi: 10.1172/JCI36226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lysiak JJ, Turner SD, Turner TT. Molecular pathway of germ cell apoptosis following ischemia/reperfusion of the rat testis. Biol Reprod. 2000;63:1465–1472. doi: 10.1095/biolreprod63.5.1465. [DOI] [PubMed] [Google Scholar]
- 29.Lazarowski ER, Boucher RC, Harden TK. Constitutive release of ATP and evidence for major contribution of ecto-nucleotide pyrophosphatase and nucleoside diphosphokinase to extracellular nucleotide concentrations. J Biol Chem. 2000;275:31061–31068. doi: 10.1074/jbc.M003255200. [DOI] [PubMed] [Google Scholar]
- 30.Lazarowski ER, Harden TK. Quantitation of extracellular UTP using a sensitive enzymatic assay. Br J Pharmacol. 1999;127:1272–1278. doi: 10.1038/sj.bjp.0702654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.