Abstract
The efficient removal of apoptotic cells is critical for the physiological well-being of the organism1â4; defects in corpse removal have been linked to autoimmune disease4, 5. While several players regulating the early steps of corpse recognition and internalization have been characterized6, the molecules and mechanisms relevant to the subsequent processing of the internalized corpses are poorly understood. Here, we identify a novel pathway for the processing of internalized apoptotic cells in C. elegans and in mammals. First, we show that RAB-5 and RAB-7 are sequentially recruited to phagosomes containing apoptotic corpses as they mature within phagocytes, and that both proteins are required for efficient corpse clearance. We then used targeted genetic screens to identify players regulating the recruitment and/or retention of Rab5 and Rab7 to phagosomes. Seven members of the HOPS complex (a Rab7 activator/effector complex) were required for Rab7 localization or retention on phagosomes. In an effort to identify factors that regulate Rab5 recruitment, we undertook an unbiased reverse genetic screen and identified 61 genes potentially required for corpse removal. In-depth analysis of two candidate genes, vps-34 and dyn-1/dynamin, showed accumulation of internalized, but undegraded corpses within abnormal phagosomes that are defective in RAB-5 recruitment. Using a series of genetic and biochemical experiments in worms and mammalian cells, we ordered these proteins in a pathway, with DYN-1 functioning upstream of VPS-34, in the recruitment/retention of Rab5 to the nascent phagosome. Further, we identified a novel biochemical complex containing Vps34, dynamin and Rab5GDP, providing a mechanism for Rab5 recruitment to the nascent phagosome.
Removal of apoptotic cells (engulfment) is an essential process that occurs throughout life in multi-cellular animals as part of development, homeostasis, and wound healing1â4, 7, 8. Engulfment) can be broken down into a series of steps, comprising recognition, internalization, phagosome maturation and finally lysosomal degradation of the apoptotic cell by the phagocyte. In mammals, impaired clearance of apoptotic cell corpses can lead to exposure of autoantigens, resulting in onset of autoimmune diseases, such as systemic lupus erythematosus and chronic polyarthritis4, 9, 10. Modulation of the engulfment process is therefore a potential therapeutic target in these conditions. One of the fundamental challenges in understanding how defects in engulfment of apoptotic cells translates into diseased states is the identification of critical players involved in corpse removal and how these proteins orchestrate the different stages of engulfment.
The nematode C. elegans represents a powerful genetic tool for the study of programmed cell death11, 12. Large numbers of cells are induced to die during two periods in the life of a worm: during embryonic and larval morphogenesis and during germ cell development13. Genetic studies have identified two evolutionarily conserved signaling pathways involved in the recognition and internalization of apoptotic cells. The first pathway includes the adapter protein CED-2/CrkII14 functioning together with two other proteins, CED-12/ELMO and CED-5/Dock18015â18, which exhibit GEF activity towards CED-10/Rac119. In the second pathway, two transmembrane proteins, CED-1/LRP1/MEGF1020â22 and CED-7/ABCA1/ABCA723â25, may mediate recognition of the apoptotic cell and function upstream of the adapter protein CED-6/GULP21, 26; these proteins have also been suggested to signal to CED-10/Rac1 during engulfment27, 28. Activation of CED-10/Rac1 promotes rearrangement of the actin cytoskeleton, permitting pseudopod extension around the apoptotic prey and ultimately internalization of the cell corpse into a membrane bound phagosome29, 30.
Studies in mammalian cells and in model organisms have shed light on the early steps of engulfment, such as the recognition and internalization of dying cells, but the identity of proteins required for subsequent steps, such as the maturation of phagosomes containing apoptotic cells into acidic structures, remain unclear. Proteomic approaches using latex beads internalized by cultured cells have identified a large number of phagosome-associated proteins31. How these candidates coordinately function in phagosome maturation and processing remains to be determined. It is also unknown to what degree proteins identified in these screens would overlap with those containing apoptotic cells, which engage different signaling pathways for internalization and can produce different immunological outcomes32, 33.
Here, we use C. elegans as a model system to identify a novel pathway for the maturation of internalized apoptotic cell corpses. First, we show that the GTPases RAB-5 and RAB-7 are required for efficient corpse degradation following internalization, and that RAB-5 and RAB-7 are recruited to phagosomes containing apoptotic cells in a temporally distinct manner. Using a series of targeted and unbiased genetic screens in the nematode, we identify a number of genes functionally required at various stages during phagosome maturation and order them into a linear pathway. Finally, we identify a novel mode of signaling wherein dynamin-Vps34 complex is required for efficient Rab5 recruitment to the nascent phagosome and subsequent corpse processing.
Results
RAB-5 and RAB-7 are required for efficient phagosome maturation
The C. elegans adult hermaphrodite gonad is composed of two U-shaped tubes joined at a central uterus; germ cell nuclei sharing a common cytoplasm line the periphery of the tube, forming a large syncytium34. When germ cells die, they cellularize away from the common syncytium and condense, generating cell corpses that appear as ârefractileâ bodies by differential interference contrast (DIC) microscopy35. Apoptotic germ cells are rapidly recognized and internalized by the surrounding gonadal sheath cells that encase the germ line35; 131 somatic cells also die during embryonic development and larval morphogenesis, where they are engulfed by neighboring âbystanderâ cells36. Little is known regarding the ultimate fate of internalized corpses in C. elegans.
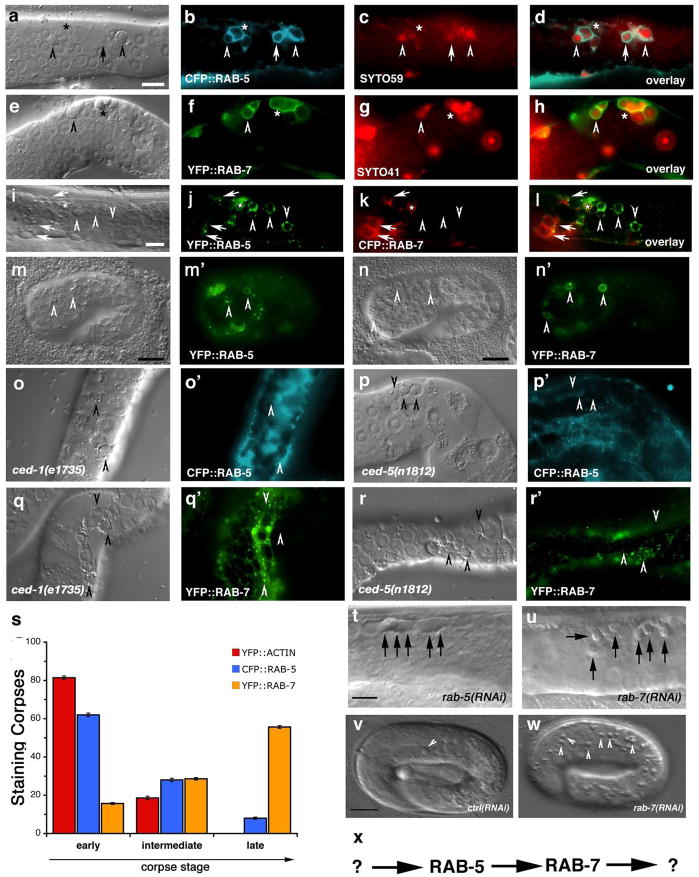
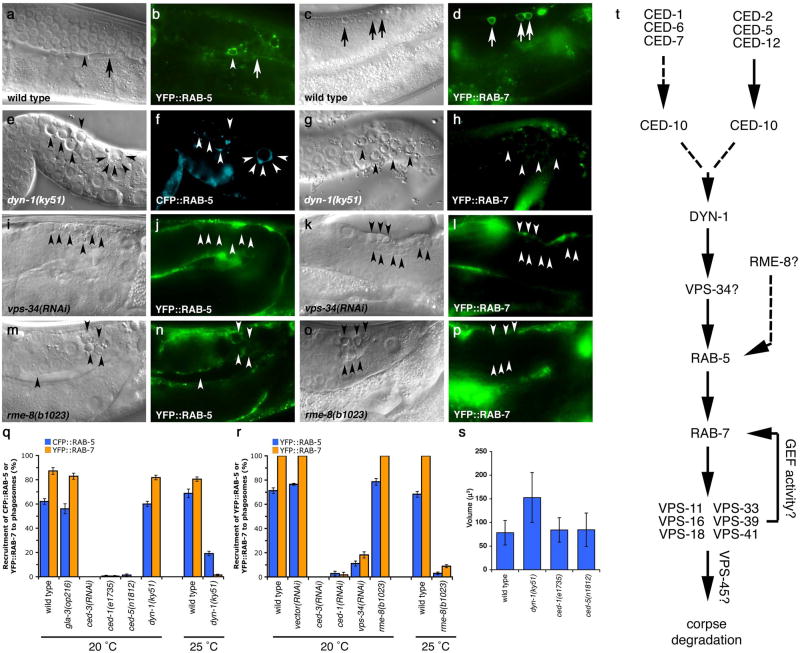
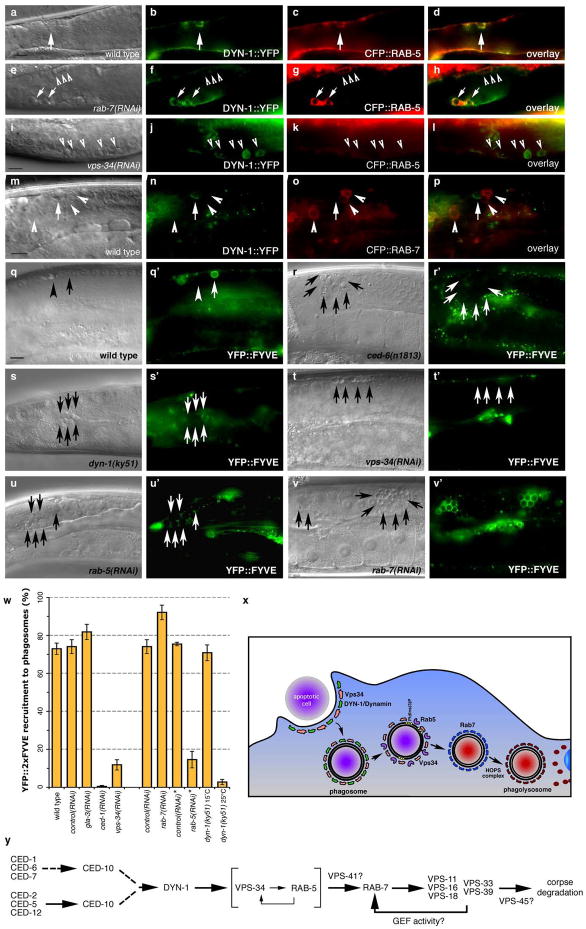
To better understand maturation of apoptotic cell-containing phagosomes, we first addressed the importance of the small GTPases Rab5 and Rab7 that have been shown to localize to phagosomes containing internalized bacteria37. We generated transgenic worms expressing CFP::RAB-5 or YFP::RAB-7 and assayed their localization around apoptotic cells in the adult hermaphrodite gonad; these markers largely localized to punctate structures likely representing early and late endosomes, respectively (Supplementary Figure S1). Apoptotic germ cells could be readily visualized inside RAB-5 or RAB-7 staining structures (Figure 1, aâh). RAB-5 preferentially localized around early, uncondensed cell corpses, when cells are being actively internalized29 (Supplementary Figure S2). CFP::RAB-5 did not extensively colocalize with YFP::actin around the apoptotic cell (data not shown), suggesting RAB-5 is recruited following corpse internalization and actin disassembly. Further, RAB-5-positive phagosomes stained weakly positive with SYTO dyes, which mark engulfed apoptotic cells in acidic lysosomal structures29, 38 (Figure 1, c, quantitated in s). In contrast, YFP::RAB-7 localized around late stage, highly-refractile apoptotic cell corpses (Figure 1f, s), most of which stained brightly with SYTO dyes (Figure 1, g). Multiple corpses were occasionally observed within RAB-5 or RAB-7 staining structures, suggesting that phagosomes may fuse following corpse recognition and internalization or that multiple corpses may be taken up within the same phagosome (Figure 1, b, f asterisks). The recruitment of RAB-5 and RAB-7 around apoptotic cells was severely compromised in worms deficient in ced-1 or ced-5, which represent members of two partially redundant pathways required for corpse phagocytosis, thus definitively placing RAB-5 and RAB-7 recruitment to the phagosome at a step downstream of corpse internalization (Figure 1, oâr, quantitated in Figure 2, g). Apoptotic cells generated during embryonic development also entered RAB-5 and RAB-7-positive phagosomes, suggesting that phagosome maturation utilize a similar machinery to process internalized corpses during both embryonic and germ cell development (Figure 1, mân).
Figure 1. RAB-5 and RAB-7 are required for efficient corpse clearance.
Bright field images represent DIC micrographs. Arrows and arrowheads indicate apoptotic germ cells or the indicated fluorescently tagged protein localized around apoptotic germ cells in dissected gonads. Chart shows number of halos normalized to corpse number; Error bars represent s.e.m. n>10 animals for each condition. SYTO41 or SYTO59 dye was used according to spectral requirements for co-localization with YFP or CFP. Size bar, 10 Îm.
(aâl) In wild-type worms, CFP::RAB-5 (aâd) and YFP::RAB-7 (eâh) highlight SYTO-positive, late stage internalized germ cell corpses (d, h, arrowheads) as well as corpses that stain weakly with SYTO dyes (c, g). Occasionally, more than one corpse can be seen as RAB-5 or RAB-7 positive structures (d, h, asterisks). YFP::RAB-5 (j, arrows) and CFP::RAB-7 (k, arrowheads) stain distinct sets of phagosomes (l), though the two makers are occasionally seen on the same phagosome (l, asterisk).
(mân) 1.5-fold embryos were assessed for localization of YFP::RAB-5 (m, mâ, arrowheads) and YFP::RAB-7 (n, nâ, arrowheads) around phagosomes containing apoptotic cells arising during developmental morphogenesis.
(oâr) Mutation of either ced-1 or ced-5 severely reduces recruitment of CFP::RAB-5 (oâ, pâ) and YFP::RAB-7 (qâ, râ) around apoptotic cell corpses (oâr, arrowheads).
(s) RAB-5 and RAB-7 stain discrete stages during corpse engulfment, with RAB-5 preferentially localizing around early corpses and RAB-7 localizing around late apoptotic cell corpses (quantitated in s). Corpse staging is described in Supplementary Figure S2.
(tâw) RNA interference against either rab-5 (t) or rab-7 (u), but not control RNAi (v and data not shown) resulted in increased numbers of undegraded refractile corpses in the adult hermaphrodite gonad or during developmental morphogenesis (w). Note: rab-5(RNAi) resulted in incompletely penetrant larval arrest; the animals scored (t) represent escapers that grew into adults. Corpse number is quantitated in Table 1.
(x) Schematic of the pathway for phagosome maturation containing apoptotic cells, based on the data above.
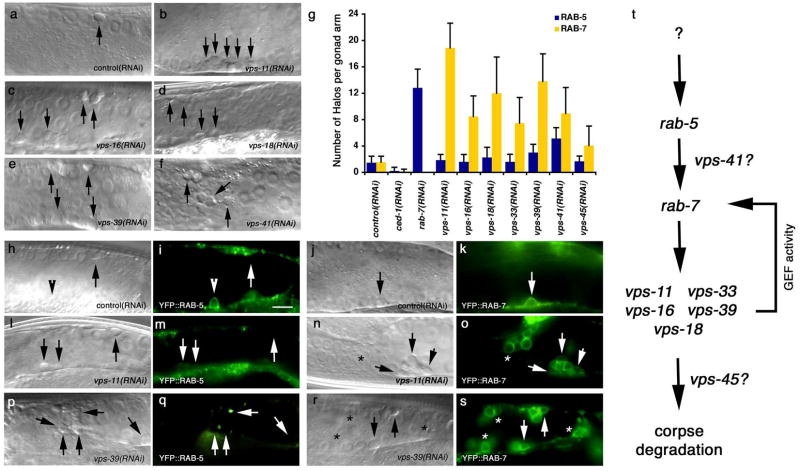
Figure 2. The HOPS complex functions downstream of RAB-7 during maturation of phagosomes containing apoptotic cells.
Bright field images represent DIC micrographs. Arrows and arrowheads indicate apoptotic germ cells or protein localized around apoptotic germ cells; asterisks represent RAB-7 staining phagosomes that contain non-refractile, partially degraded apoptotic cells. Error bars represent s.d. Size bar represents 10 Îm.
(aâf) RNA interference against indicated members of the HOPS complex resulted in increased numbers of undegraded, refractile cell corpses in the gonad.
(g) The increase in number of YFP::RAB-5 and YFP::RAB-7 positive phagosomes (as a marker for arrest in maturation of phagosomes containing corpses) was quantitated in animals treated with RNAi targeting ced-1, rab-7 or indicated members of the HOPS complex In rab-7(RNAi) animals, there is an accumulation of corpses in RAB-5(+) phagosomes, while the failure to see halos in ced-1 RNAi is due to defects in early stages of engulfment.
(hâs) The number of RAB-5-positive phagosomes in animals treated with vps-11 and vps-39 RNAi (m,q) were similar to control RNAi treated nematodes (i), while the number of RAB-7-positive phagosomes (o, s) were increased compared to control (k). Quantitated in (g).
(t) Schematic of the genetic pathway for phagosome maturation containing apoptotic cells, based on the data above.
Our above data suggests that recruitment of RAB-5 and RAB-7 may mark distinct steps during phagosome maturation. If this were the case, we would predict that RAB-5 and RAB-7 would co-localize only transiently on phagosomes, during âRab conversionâ39. When we made double transgenic worms expressing both YFP::RAB-5 and CFP::RAB-7, we found minimal co-localization between these two markers (Figure 1, iâl, arrows vs. arrowheads). Occasionally, we could find phagosomes that stained with both markers (Figure 1, iâl, asterisk), suggesting that RAB-5 and RAB-7 may transiently mark the same phagosome.
Even though RAB-5 and RAB-7 were recruited to phagosomes, their functional requirement during corpse removal was unknown. RNA interference (RNAi) directed against rab-5 or rab-7 resulted in accumulation of refractile corpses in the adult hermaphrodite gonad (Table 1, Figure 1, t, u) and also during embryogenesis [rab-7(RNAi), Figure 1w compared to ctrl(RNAi), Figure 1v]. Further, in rab-7(RNAi) worms, phagosomes containing apoptotic germ cells were arrested at the RAB-5-positive stage (Figure 2, g) suggesting that RAB-5 functions upstream of RAB-7 in a linear pathway for phagosome maturation (Figure 1, x).
Table 1.
A targeted screen identifies genes required for phagosome maturation
| Genotype | Corpse Number (DIC) | n |
|---|---|---|
| wild type | 2.3 Â 0.7 | 10 |
| wild type 25ÂC | 3.4 Â 1.4 | 15 |
| ced-1(e1735) | 18.2 Â 4.4 | 10 |
| ced-1(RNAi) | 11.2 Â 3.1 | 10 |
| rab-5(RNAi) | 10.1 Â 4.6 | 30 |
| rab-7(RNAi) | 25.4 Â 9.3 | 30 |
| vps-34(RNAi) | 8.4 Â 2.1 | 30 |
| nsf-1(RNAi) | 1.5 Â 0.8 | 15 |
|
Rab5 GEFs (Vps9-domain containing proteins) | ||
| tag-333(gk431) (RIN1) | 1.6 Â 0.7 | 20 |
| rme-6(b1014) (RabEx-5) | 2.1 Â 0.9 | 10 |
| rabx-5(RNAi) (RabEx-5) | 1.8 Â 0.9 | 30 |
| rabx-5(tm512) | 1.5 Â 0.8 | 20 |
| rme-6(b1014); rabx-5(RNAi) | 2.0 Â 0.9 | 10 |
| tag-333(gk431); rme-6(b1014) | 1.4 Â 1.0 | 20 |
| tag-333(gk431); rme-6(b1014); rabx-5(RNAi) | 1.5 Â 0.7 | 15 |
|
VPS/HOPS complex | ||
| vps-11(RNAi) | 10.8 Â 2.6 | 25 |
| vps-16(RNAi) | 13.7 Â 3.8 | 30 |
| vps-18(RNAi) | 12.7 Â 4.1 | 30 |
| vps-33(RNAi) | 6.1 Â 2.4 | 30 |
| vps-39(RNAi) | 11.8 Â 4.2 | 40 |
| vps-41(RNAi) | 13.1 Â 7.0 | 30 |
| vps-45(RNAi) | 11.4 Â 5.3 | 40 |
|
FYVE-domain containing proteins | ||
| aka-1(ok707) (SARA) | 1.4 Â 0.7 | 20 |
| rabs-5(ok1513) (Rabenosyn-5) (25ÂC) | 1.1 Â 0.5 | 20 |
| VT23B5.2(ok912) (BWF1) | 1.4 Â 0.9 | 20 |
| eea-1(ok1040) (EEA1) | 1.8 Â 0.9 | 30 |
| tag-77(gk206) (FGD6) | 1.5 Â 0.7 | 20 |
| mtm-6(ok330) (MTM6) | 1.3 Â 0.8 | 20 |
| exc-5(rh232) (FGD1) | 1.2 Â 1.1 | 20 |
| ZK632.12(RNAi) (Pafin2) | 2.1 Â 1.1 | 30 |
| hgrs-1(RNAi) (HRS) | 1.7 Â 1.5 | 20 |
| lst-2(RNAi) (LZ-FYVE) | 1.8 Â 1.1 | 30 |
| F22G12,4(RNAi) (Rabankyrin) | 2.3 Â 1.0 | 20 |
| mtm-3(RNAi) (MTM3) | 1.9 Â 0.9 | 30 |
| wdfy-2(RNAi) (WDFY2) | 2.4 Â 2.1 | 20 |
| ppk-3(RNAi) (PIP5K) | 2.3 Â 1.7 | 30 |
|
PX-domain containing proteins | ||
| Y116A8c.26(RNAi) (sorting nexin 13) | 7.4 Â 3.4 | 45 |
| F17H10.3 (RNAi) (sorting nexin 17) | 6.5 Â 2.8 | 39 |
| lst-4(RNAi) (sorting nexin 18) | 8.0 Â 2.7 | 30 |
| F55C5.7(RNAi) (ribosomal S6 kinase) | 2.5 Â 2.2 | 35 |
| F13E9.1(RNAi) (Nischarin) | 2.5 Â 1.3 | 15 |
| F25H2.2 (RNAi) (sorting nexin 27) | 2.3 Â 1.1 | 15 |
| tag-157(RNAi) (sorting nexin 12) | 2.7 Â 2.7 | 25 |
| Y59A8b.22\(RNAi) (sorting nexin 5) | 2.1 Â 1.0 | 15 |
| Y48E1b.14(RNAi) (sorting nexin 14) | 2.3 Â 1.6 | 15 |
| snx-1(RNAi) (sorting nexin 1) | 1.4 Â 0.8 | 15 |
| pld-1(RNAi) (phospholipase D) | 1.7 Â 1.0 | 15 |
| F39B1.1(RNAi) (PI3KC2a) | 1.5 Â 0.7 | 15 |
Number of refractile cell corpses were scored in the 12-hour adult hermaphrodite gonad. Whether a particular gene was analyzed through the use of existing mutant strains or through RNAi is indicated. Mammalian homologue is shown in parentheses next to gene name (when different from nematode name). rab-5 worms arrest during larval development; worms scored represent escapees. Data shown is average  s.d. Mutations used were backcrossed at least twice.
While refractile cell corpses in rab-5(RNAi) and rab-7(RNAi) most likely arose due to defects in phagosome maturation, increased numbers of refractile cell corpses could have also resulted from increased physiological cell death or decreased corpse internalization. When we used a YFP::actin transgenic strain to monitor corpse internalization (and indirectly programmed cell death) in the gonad, the numbers of cells undergoing internalization in rab-5 and rab-7 deficient worms showed no difference compared to wild type. Thus, the increased corpse number appears to be primarily due to defects in phagosome maturation (see Supplementary Figure S2). Taken together, these data suggest that RAB-5 and RAB-7 function downstream of corpse internalization, with RAB-5 functioning upstream of RAB-7 (Figure 1, x), and both GTPases being critical for proper clearance of apoptotic cells in vivo.
A targeted screen for players thought to regulate RAB-5 and RAB-7 reveals a linear pathway for phagosome maturation
We next used a candidate gene approach to test the importance during cell corpse clearance of genes known to regulate RAB-5 signaling during endocytosis. Mammalian and yeast Rab5 GTP exchange factors (GEFs), which promote cycling of GTPases from the GDP to GTP-bound conformation, all share a canonical Vps9 domain40. There are three Vps9-domain containing proteins in the nematode, rme-6, rabx-5, and tag-33341. RME-6 and RABX-5 are both homologues of mammalian RabEx-5 and are required for generation of RAB-5(+) endosomes in coelomocytes42 and in the somatic sheath cells (Supplementary Figure S1). Surprisingly, neither the single mutants rme-6, rabx-5 or tag-333 nor the triple mutant rme-6(b1014); tag-333(gk431); rabx-5(RNAi) worms showed any defects in corpse removal, suggesting that this class of proteins is not required for RAB-5 function during apoptotic cell clearance.
We then asked whether RAB-5 effectors, which bind activated, GTP-bound RAB-5, play a role in apoptotic cell clearance. Analysis of worms harboring mutations in the nematode homologues of the FYVE-domain containing RAB-5 effectors EEA1 (EEA-1) and Rabenosyn-5 (RABS-5) had no obvious defects in corpse degradation, although the same mutant strains have previously been shown to display defects during endocytosis in theworm43â45 (Table 1). This was somewhat surprising, as studies monitoring phagosome maturation in mammalian cells routinely use EEA1 protein as a marker for the early endosome. It was possible that eea-1(ok1040) represented a complex rearrangement rather than a well-defined deletion; however, we could not detect eea-1 mRNA by RT-PCR, suggesting that this is indeed a null allele.
EEA-1 and RABS-5 are hypothesized to function as a tethering complex during a âconcentratingâ stage of endocytosis, where multiple vesicles may fuse into RAB-5 positive structures39. To address the importance of fusion events during corpse clearance, we scored worms deficient in the ATPase NSF-1, the nematode homologue of N-ethylmaleimide-sensitive factor (NSF) and a key component of the vesicular fusion machinery on early endosomes46. Loss of NSF-1 does not result in increased corpse number (Table 1), suggesting that a concentrating step is dispensable for efficient corpse degradation; knockdown of nsf-1 mRNA was efficient under these conditions (86%, as determined from RT-PCR performed on dissected gonads) (Supplementary Figure S3). Additionally, a YFP::NSF-1 fusion protein did not localize around corpses undergoing engulfment, and nsf-1(RNAi) also greatly reduced fluorescent signal in the gonadal sheath cells (Supplementary Figure S3), providing further evidence that our knockdown was efficient.
It was possible that an alternate FYVE or PX-domain-containing protein (both of which bind to endosomal structures) could be involved in RAB-5 function during corpse removal. Therefore, we focused on the 14 FYVE-domain containing proteins and 12 PX-domain containing proteins in the nematode genome41. Analysis of worms with disruptions in the 14 FYVE domain-containing proteins (either existing mutants in seven genes, or RNAi-mediated knockdown of the other seven candidates) showed no obvious defect in corpse removal in the germ line. Knockdown of two of the twelve PX-domain containing proteins, sorting nexins encoded by Y116A8c.26 and F17H10.3, resulted in increased numbers of germ cell corpses. However, the defects we observed in Y116A8c.26(RNAi) and F17H10.3(RNAi) were relatively weak (potentially due to redundancy of function or poor RNAi efficiency), and we did not see a clear effect on RAB-5 or RAB-7 recruitment to the phagosome (data not shown). We propose that the two sorting nexins may play a redundant role in RAB-5 or RAB-7 recruitment to the phagosome, or affect another aspect of phagosome maturation.
We next tested genes that may regulate RAB-7 function during engulfment. The HOPS complex is a GEF/effector complex that acts on RAB-7 during endocytosis and is composed of the products of seven genes â vps-11, vps-16, vps-18, vps-33, vps-39, vps-41, and vps-4547. RNAi directed against each of these genes [and a vps-16(ok719) mutant, Supplementary Figure S4] resulted in the accumulation of refractile apoptotic cells in the gonad (Figure 2, aâf, Table 1). Since the number of cells undergoing internalization in the gonad, as measured by YFP::actin staining, was similar to wild type (Supplementary Figure S2), these refractile corpses appeared to represent internalized but undegraded apoptotic cells. To place these genes within the pathway for phagosome maturation, we assayed RAB-5 and RAB-7 recruitment to the phagosome (Figure 2, g, hâs). Interestingly we found that RNAi-mediated disruption of five of the members of the VPS/HOPS complex (vps-11, vps-16, vps-18, vps-33 and vps-41) results in apoptotic cells arrested in RAB-7-positive phagosomes. These proteins are predicted to interact with SNAREs and syntaxins and may function as a âtethering complexâ for the docking of vesicles onto the phagosome48, 49, suggesting that the delivery of some component may be required for resolution of the RAB-7(+) stage.
Two additional HOPS complex members, vps-41 and vps-45, also appeared to function in maturation of phagosomes containing corpses. VPS-45 is a member of a family of proteins (Sec1 homologues) that mediate targeting of vesicles in the lysosomal degradation pathway. Knockdown of vps-45 showed only a mild increase in the number of RAB-7 or RAB-5 phagosomes, while robustly generating increased number of germ cell corpses. This suggests that VPS-45 might be dispensable for RAB-5/RAB-7 recruitment and resolution, potentially functioning at later stages downstream of RAB-7 (see Discussion). Additionally, vps-41(RNAi) resulted in a mild increase in the number of RAB-5-positive phagosomes, suggesting VPS-41 may influence RAB-5 release from the phagosome. These data further extended our linear pathway for phagosome maturation, with HOPS complex members functioning at multiple steps of RAB-5/RAB-7 function (Figure 2, t).
An unbiased genetic screen identifies additional genes required for efficient corpse removal
In our targeted gene analyses above, we were unable to identify upstream regulators of RAB-5 function that would be required for maturation of apoptotic cell-containing phagosomes. We thus undertook an unbiased, reverse genetic screen38 to systematically identify genes involved in both corpse internalization and processing during germ cell apoptosis. We previously showed that the vital dye acridine orange selectively stains internalized apoptotic cell corpses at late stages of degradation29, 38 (Figure 3, aâd). Corpses in rab-7(RNAi) worms, which are arrested in RAB-5(+) phagosomes, weakly stained with acridine orange (Figure 3, j, k), suggesting that RAB-5(+) phagosomes are acidified. Using a feeding RNAi library50, we screened for genes on chromosomes I, III, IV and X that suppress acridine orange staining of apoptotic cells in the adult hermaphrodite gonad (see Methods).
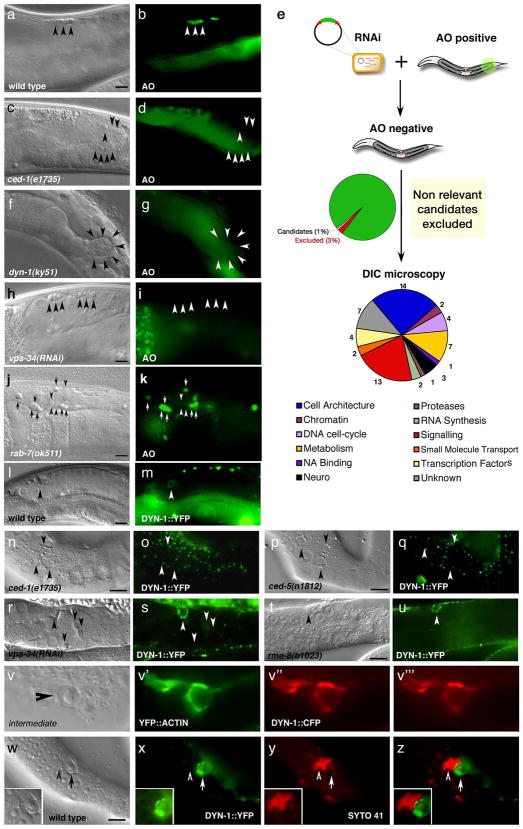
Figure 3. A reverse genetic screen identifies DYN-1, which is required for efficient corpse removal and localizes around apoptotic cells in the adult hermaphrodite gonad.
Bright field images represent DIC micrographs. Arrows/arrowheads indicate refractile cell corpses in the DIC images or the indicated protein for localization around apoptotic cells in the fluorescence images. dyn-1(ky51) mutant worms are shown at the nonpermissive temperature. Scale bar, 10 Îm.
(aâd) In wild-type worms (a, b), acridine orange (AO) preferentially stains engulfed apoptotic cells present in acidic compartments. In worms mutant for genes required for efficient removal of apoptotic cells, refractile cell corpses persist but do not stain with AO [ced-1(e1735), c, d].
(e) Schematic of reverse genetic screen. Worms were fed bacteria expressing dsRNA as at the L1 larval stage, then stained with AO as adults. AO negative candidates were then compared to previous RNAi screens to exclude false negatives due to sterility or other gonadal defects. Genes identified were assigned to categories based on proposed function (e.g., cell architecture). Finally, refractile cell corpses were scored in the 12-hour adult hermaphrodite gonad by DIC microscopy (Supplementary Table S1).
(fâk) Inactivation of candidate genes identified in the screen, e.g. dyn-1(ky51) (f, g) and vps-34(RNAi) (h, i) showed persistent corpses without AO staining (g, i, arrowheads). rab-7(ok511) showed corpses arrested in AO-staining phagosomes (j, k).
(lâu) DYN-1 is recruited around the apoptotic cell during phagocytosis (l, m, arrowhead), but not when the engulfment process is disrupted, as in ced-1 (n, o) and ced-5 (p, q) deficient worms. In vps-34 (r, s) and rme-8 (t, u) deficient worms, DYN-1 is still recruited, suggesting DYN-1 may function upstream of VPS-34 and RME-8 during corpse removal. Quantitation of the data presented here is shown in Supplementary Table S2.
(v) DYN-1 is recruited to the phagocytic cup and associated with early phagosomes. DYN-1::CFP colocalizes with YFP::actin around early/intermediate stage apoptotic cell corpses (v and the appropriate subpanels).
(wâz) DYN-1 is recruited around SYTO 41-negative (early) apoptotic cell corpses (w, x, z, overlay, inset, arrow) and is not present on late, SYTO41-positive corpses (y, overlay, inset, arrowhead). SYTO 41, like acridine orange, preferentially stains late-stage, internalized apoptotic cells.
By design, we expected to identify candidate genes whose disruption caused defects in phagosome maturation or acidification of phagosomes and phagolysosomes (candidates most relevant to this manuscript), as well as genes affecting programmed cell death or corpse recognition/internalization (manifesting as a decrease in the number of AO positive corpses). Following elimination of all genes previously shown to cause developmental delay or sterility41, we identified 61 genes whose knockdown reproducibly resulted in decreased AO staining (Figure 3e, Supplementary Table S1). In addition to the known genes required for corpse recognition (ced-1) and internalization (ced-5 and ced-6) present in the RNAi library50, we also identified components of the Arp2/3 complex in C. elegans, arx-3 and arx-551. Since the Arp2/3 complex plays a key role during actin reorganization and corpse removal in mammalian systems52, the inactivation of the Arp2/3 complex in C. elegans may similarly interfere with actin-dependent corpse internalization. Additionally, we identified wip-1 and C24A1.3 (a tyrosine kinase), which may regulate activity of the small GTPase Cdc42 during corpse removal. Among the remaining genes, homologues of several candidates, phi-25, dyci-1, rab-10, and rme-8, suggest these genes may play a role during corpse processing and phagosome maturation41. Further, we isolated vha-15, a vacuolar ATPase, which may be required for acidification of the lysosome and hence onset of AO staining.
Among our candidates causing accumulation of refractile corpses were VPS-34 and DYN-153 (Figure 3, fâi). VPS-34 is the nematode homologue of the PI-3-kinase Vps34, and is a known Rab5 effector54 involved in maturation of endosomes into acidic lysosomes. DYN-1 is the nematode homologue of mammalian dynamin, and belongs to a family of large GTPases that have been linked to both organization of the actin cytoskeleton55â58 and internalization of antibody-opsonized cells59. Dynamin has also been shown to play a role in receptor-mediated endocytosis and protein trafficking60 and to form an endogenous complex with Arp2/3 and other actin binding proteins55. However, the exact function of dynamin during remodeling of the actin cytoskeleton in the context of engulfment is unclear60. In the nematode, DYN-1 is involved in cytokinesis and is required for synaptic vesicle transport53, 61. We decided to focus on DYN-1 for further study, as corpses in dyn-1(lf) worms appeared arrested in abnormal phagosome structures and a role for dynamin in phagosome maturation had yet to be described. DYN-1 has also been linked to corpse internalization62; however, as described below, our careful analyses in C. elegans and mammalian cells suggest no obvious role for dynamin in internalization of apoptotic cells per se, rather an important role for dynamin in phagosome maturation subsequent to internalization of the corpse.
DYN-1 is recruited to the phagosome downstream of corpse internalization
dyn-1(RNAi) resulted in an accumulation of refractile corpses in the adult hermaphrodite gonad (Figure 3f, g and Supplementary Table S1, S2). A dyn-1 mutant, ky51, with a proline to serine substitution (residue 70) in the GTPase domain has been described; this mutant dynamin protein is inactivated within 2 minutes at the non-permissive temperature (25ÂC)53. dyn-1(ky51) mutant worms also showed increased AO negative apoptotic corpses at 25ÂC, suggesting that DYN-1 function is required early during corpse removal. To further investigate the role of DYN-1 in engulfment, we generated YFP- or CFP-tagged DYN-1 constructs and assayed localization of DYN-1 during corpse removal. We detected enrichment of DYN-1 in punctate structures surrounding early apoptotic cells at the stage during which active phagocytosis occurs29 (Figure 3, l, m, and Supplementary Figure S5). These DYN-1::CFP structures colocalized with the YFP::actin meshwork that forms around early apoptotic cells (Figure 3, v) during corpse internalization. We could not detect enrichment of DYN-1 adjacent to the apoptotic cell in ced-1 mutant worms (Figure 3, n, o), suggesting that DYN-1 is recruited at a stage following corpse recognition. Finally, in ced-5 or ced-6 mutant worms (Figure 3, p, q), and other genetic backgrounds deficient for corpse internalization (Supplementary Figure S5), DYN-1 enrichment adjacent to apoptotic cells was also greatly decreased compared to wild type. These results suggested that DYN-1 is recruited to the apoptotic cell following recognition and at a late stage during or following internalization of the corpse. Earlier studies have proposed that the ced-2, -5, -12 pathway was dispensable for DYN-1 recruitment during embryogenesis62; however, this work utilized a group of cells that require a single pathway (comprised of ced-1, -6, -7) for efficient corpse internalization, whereas the adult hermaphrodite gonads uses both pathways for corpse removal30, 35. Our more rigorous analysis suggests that the coordinated activity of both engulfment pathways results in recruitment of DYN-1 to the phagosome. We also noted that DYN-1 recruitment to apoptotic corpses was transient: DYN-1 is not found on SYTO41-staining phagosomes, which contain âlateâ apoptotic cell corpses (Figure 3, wâz and Supplementary Figure S2), suggesting DYN-1 is rapidly removed from the phagosome membrane following corpse internalization and actin disassembly.
DYN-1 is required for phagosome maturation and maintenance of the actin cytoskeleton
One of the best known roles for dynamin during endocytosis is in pinching off vesicles from the plasma membrane (membrane scission)60. A similar but phenotypically and molecularly distinct process occurs when the phagocytic cup fully extends around the apoptotic cell and then fuses to completely encompass the apoptotic cell. To test whether apoptotic cells are engulfed in dyn-1(ky51) mutant worms and to monitor phagocytic cup closure, we monitored CED-1::GFP and YFP::CED-6 localization, which are enriched at the cell membrane around apoptotic cells during corpse internalization20, 29. Both CED-1::GFP and YFP::CED-6 were efficiently recruited around apoptotic cells in dyn-1(ky51) mutant worms (Figure 4, aâd, quantitated in Supplementary Table S3), and the phagocytic cup appeared to be completely closed around the apoptotic cell (Figure 4, d, inset), suggesting DYN-1 is not required for pseudopod extension or phagosome closure. In wild-type worms, CED-1 is rapidly removed from the phagosome following corpse internalization (Figure 4b)20, 29. However, in dyn-1(ky51) mutants the total number of CED-1- and CED-6-staining phagosomes was increased compared to wild type, consistent with early defects in phagosome maturation (Figure 4, d and Supplementary Table S3). We also monitored actin recruitment to the phagocytic cup: worms deficient in DYN-1 showed similar numbers of corpses undergoing internalization (as measured by actin recruitment to the phagocytic cup) as wild type nematodes (Figure 4, eâh, quantitated in q). In contrast, internalization was not seen in ced-1(lf) (Figure 4, i, j), which has previously been linked to actin rearrangement during corpse recognition29. Taken together, these results suggest that the refractile corpses in dyn-1(ky51) mutant worms were arrested in phagosomes at a stage following corpse internalization.
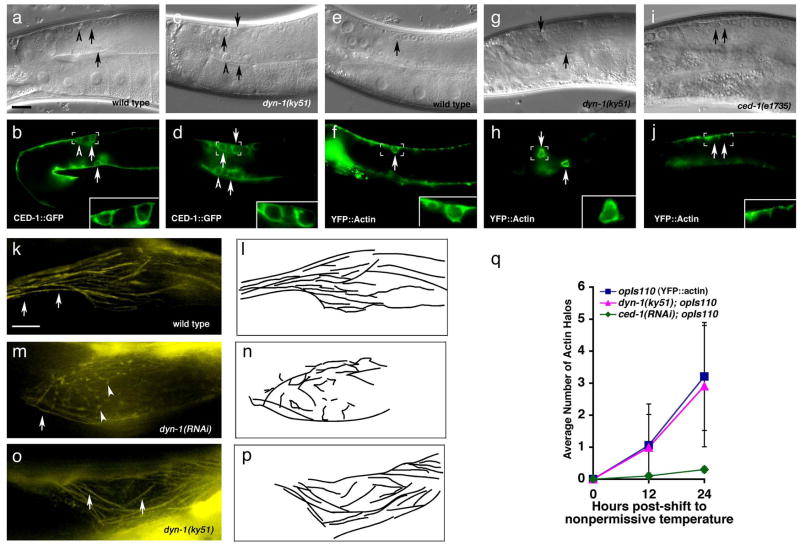
Figure 4. Apoptotic cells are efficiently internalized in dyn-1 mutant worms.
Bright field images represent DIC micrographs. dyn-1(ky51) mutant worms are scored at the nonpermissive temperature (25ÂC) unless otherwise stated. Arrows, arrowheads indicate apoptotic cells. Scale bar, 10 Îm. Error bars represent s.d.
(aâj) Normal recruitment of CED-1 and actin polymerization around corpses in dyn-1(ky51) mutant worms at 25ÂC. Both CED-1::GFP recruitment, which marks the phagocytic cup (b, inset), and actin reorganization (e, arrows) during engulfment appear normal in dyn-1(ky51) worms at the non-permissive temperature (d, h, arrows), while ced-1(e1735) mutant worms display a severe defect in actin polymerization and internalization of the apoptotic cell (j, arrows). Residual staining in the sheath cells represents cortical actin filaments or G-actin. Quantitation of this data is presented in (q).
(kâp) In wild type worms, actin stress fibers tend to run longitudinally in the proximal gonad (k and camera lucida, l). However, after long-term knockdown with dyn-1(RNAi) (48 hours), animals show distorted, disoriented actin fibers (m, n), which may have resulted in the phagocytic detected at these time points (Supplementary Figure S5). In dyn-1(ky51) mutant worms, incubated for 24 hours at 25ÂC, the actin fiber pattern appears as wild type (o,p).
In a recent report, Yu et al. showed that approximately 40% of the corpses present in the gonad of dyn-1(RNAi) worms persist un-engulfed62. However, our studies suggested no obvious defect in corpse internalization per se but instead that DYN-1 may act during phagosome maturation. Our studies use the ky51 temperature sensitive allele, whereas those of Yu et al use dyn-1(RNAi), which would inactivate DYN-1 function over a much longer timeframe. We addressed whether the time point when the animals were scored may be the source of the discrepancy between our two studies. In fact, we did observe a potential phagocytic defect in 24- and 48-hour adult gonads in dyn-1(ky51) and dyn-1(RNAi) worms, as measured by decreased numbers of observed actin halos (Figure 4, q and data not shown). However, the caveat with scoring at this time point is that loss of dynamin function has been associated with defects in the actin cytoskeleton in mammalian cells55â58, 60, thus the phenotype seen could have been a secondary effect. In our analysis, loss of DYN-1 in C. elegans also seems to affect integrity of the actin cytoskeleton at late timepoints (24- and 36-hours post-shift to 25ÂC), with gross alterations in F-actin staining [Figure 4, m and camera lucida, n compared to wild type (k, l) or dyn-1(ky51) (o, p)]. At these later times, we cannot say whether DYN-1 plays a âdirectâ role in corpse internalization (which would be less sensitive to gene inactivation by RNAi and temperature shift) or whether this might be a secondary effect of accumulated defects in the actin cytoskeleton. Thus, to avoid potential unintended secondary defects, we have performed all our nematode assays here at the 12-hour adult stage following dyn-1(ky51) inactivation.
Dynamin function during phagosome maturation is evolutionarily conserved
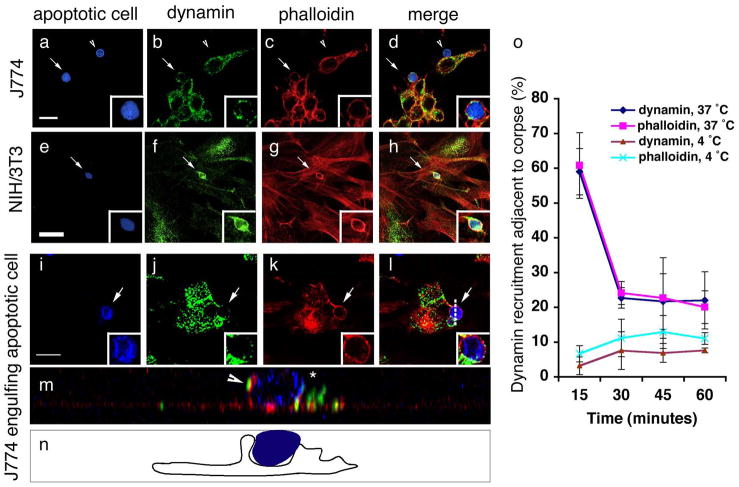
We next addressed the role of dynamin in phagosome maturation in the mammalian context. There are three mammalian dynamins: dynamin-2 is ubiquitously expressed, while dynamin-1 and dynamin-3 show a more restricted expression (brain and testes, respectively63). Since most cell types possess the ability to engulf, we chose to focus on the role of dynamin-2. In both J774 macrophages and NIH3T3 fibroblasts (models for professional and nonprofessional phagocytes, respectively) we observed recruitment of endogenous dynamin around apoptotic cells (Figure 5, aâh) with kinetics similar to those observed for actin polymerization (Figure 5o, Supplementary Figure S6). Observed dynamin staining originated from phagocytes and not from targets being engulfed, as apoptotic cells not undergoing internalization showed no obvious dynamin staining (Figure 5, a, b, arrowhead). Recruitment of dynamin into the phagocytic cup was also specific, and not due to a general recruitment of proteins involved in actin organization or proteins involved in endocytosis; for example, cortactin, a dynamin-interacting protein60, was excluded from the phagocytic cup (Supplementary Figure S7).
Figure 5. Dynamin in phagocytes is recruited around apoptotic cells coincident with corpse internalization.
CFSE/TAMRA stains the cytoplasm of apoptotic cells, frequently resulting in a lunette of staining around the nucleus. Arrows and asterisks indicate apoptotic cells. Scale bar, 10 Îm. Error bars represent s.d.
(aâh) J774 macrophages (aâd) or NIH/3T3 fibroblasts (eâh) were incubated with apoptotic thymocytes or apoptotic Jurkat cells, respectively, and localization of endogenous dynamin (green) and polymerized actin (red) were monitored. Dynamin was recruited to the phagocytic cup with actin in J774 (15 min) (d, merge) and NIH/3T3 cells (30 min) (h, merge) (indicated by arrow), but was not recruited around bound apoptotic cells (d, arrowhead).
(iân) Confocal z-sections were reconstructed to generate yz planes (m, n). J774 macrophages incubated with apoptotic cells at 4 ÂC did not show phagocytic cup formation or enrichment of endogenous dynamin around the apoptotic cell (Supplementary Figure S6). Cells incubated at 37 ÂC showed dynamin localized in the phagocytic cup (m, arrowhead and n, camera lucida) adjacent to the apoptotic cell in a punctate pattern (j, k, inset). Dotted lines (l) indicate plane of yz reconstruction.
(o) Time course of endogenous dynamin recruitment around the apoptotic cell corpse in J774 macrophages. Cells that had bound apoptotic cells were scored positive for dynamin recruitment if a âhaloâ of dynamin was seen to surround the apoptotic cell. 4 ÂC, n=100 (4 experiments); 37 ÂC, n=200 (8 experiments).
To better address dynamin localization within the phagocytic cup, we acquired confocal z-stacks and reconstructed xz- or yz-planes. In the absence of phagocytic cup formation, bound apoptotic cells (at either 4 ÂC or 37 ÂC) showed no observable dynamin staining (Supplementary Figure S6). Apoptotic cells being internalized by phagocytes showed punctate staining within the phagocytic cup (Figure 5, j, l, m and camera lucida, n). Dynamin staining appeared to be excluded from the leading edge, suggesting that dynamin is likely not involved in membrane extension around the apoptotic cell. Consistent with these observations, we could not detect any defect in internalization using either a dominant negative Dyn2K44A construct, which has impaired GTPase activity64, or siRNA-mediated knockdown of dynamin (Supplementary Figure S8). As in the nematode, dynamin recruitment during engulfment was transient, as staining was rapidly lost from the phagosome following corpse internalization (Figure 5o and Figure 4).
We next addressed whether the role of mammalian dynamin, similar to DYN-1 in C. elegans, would also regulate phagosome maturation. While apoptotic thymocytes appear to be internalized normally by NIH3T3 cells, corpses within NIH3T3 cells expressing dominant negative Dyn2K44A or treated with dyn2 siRNA did not mature into acidic endosomes, as determined by Lysotracker Red staining (Figure 6aâh, quantitated in u, v). This phenotype was unlikely due to general defects in endocytosis, as Dyn2siRNA did not affect clathrin pit resolution (Supplementary Figure S8). Taken together, these data suggested a critical role for mammalian dynamin during phagosome maturation after internalization of apoptotic cells. Since this function of dynamin appeared to be roughly at a step when Rab5 localization to the phagosome would occur, this suggested an exciting possibility that dynamin and/or Vps34 could provide the missing upstream link to Rab5 recruitment/retention to nascent phagosomes during apoptotic cell processing.
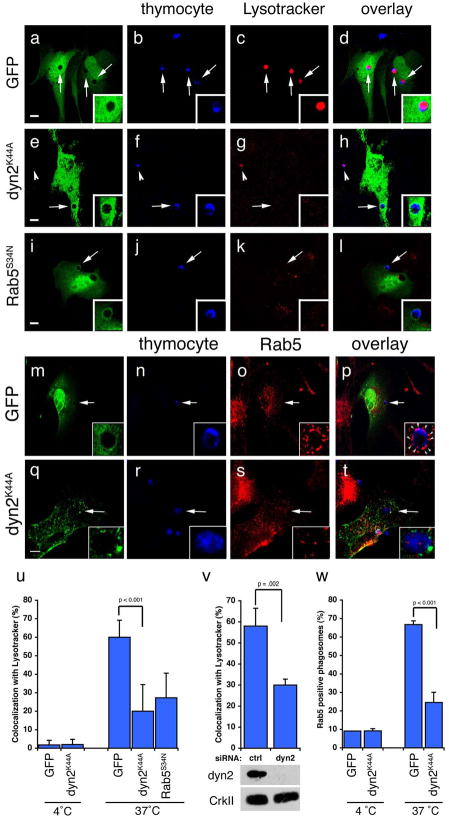
Figure 6. Dynamin is required for maturation of engulfed apoptotic cells into Rab5-coated endosomes.
Arrows and arrowheads indicate apoptotic cells or protein recruited around engulfed apoptotic cells in phagosomes. Neither Dyn2K44A nor Dyn2RNAi had any obvious effect on staining of endogenous endosomal/lysosomal structures with Lysotracker Red or Rab5. Error bars represent s.d. Scale bar, 10 Îm.
(aâl) NIH/3T3 fibroblasts transfected with GFP (aâd), HA-Dyn2K44A (eâh), or GFP-Rab5S34N (iâl) were incubated with apoptotic thymocytes in the presence of Lysotracker Red to determine the efficiency of phagosome maturation. In the majority of GFP-transfected cells, internalized apoptotic thymocytes (arrows) co-stained with Lysotracker red (d, inset); cells transfected with Dyn2K44A (h, inset) or Rab5S34N (l, inset) showed decreased numbers of engulfed thymocytes co-staining with Lysotracker. Apoptotic cells incubated at 4 ÂC with phagocytes did not stain strongly with Lysotracker Red (u, Supplementary Figure S9). HA-Dyn2K44A expressing cells were stained with anti-HA and an Alexa 488-conjugated secondary antibody for visualization of transfected cells.
(mât) Apoptotic cells were incubated with NIH/3T3 fibroblasts, transfected with either GFP alone (as control) or HA-Dyn2K44A and the localization of endogenous Rab5 was monitored. The majority of engulfed apoptotic thymocytes inside GFP-transfected cells were in endosomes coated with Rab5 (o, arrowheads, p, inset). In Dyn2K44A transfected cells, phagosome maturation was disrupted and decreased numbers of engulfed thymocytes were in Rab5 coated endosomes (s, t, arrowhead, inset). Apoptotic cells incubated at 4 ÂC with phagocytes did not stain strongly for Rab5 (w, Supplementary Figure S9)
(u) Quantitation of experiment shown in (aâl). 4 ÂC n=20 (2 experiments), 37 ÂC, n=60 (3 experiments).
(v) NIH/3T3 fibroblasts were electroporated with a control siRNA or dynamin-2 (dyn2) siRNA and assayed as in (aâl). Dyn2siRNA cells showed decreased co-localization with Lysotracker as compared to controlsiRNA cells. n=50 (2 experiments).
(w) Quantitation of experiment shown in (mât). 4 ÂC n=20 (2 experiments), 37 ÂC, n=60 (4 experiments).
Dynamin regulates recruitment/retention of Rab5 to the nascent phagosome
Phagosomes in dynamin-deficient cells appear arrested in abnormal, unacidified lysosomal structures. Unlike control transfected NIH/3T3 cells, in which Rab5 was efficiently recruited to apoptotic cell-containing phagosomes (Figure 6, mâp, quantitated in w), NIH/3T3 cells transfected with dominant negative Dyn2K44A showed a significant decrease in the percent of apoptotic cells in Rab5-positive phagosomes (p < 0.001) (Figure 6, qât, w). This suggests that dynamin is required for efficient maturation of a nascent phagosome containing internalized apoptotic cells into Rab5-positive phagosomes. Consistent with this hypothesis, cells expressing dominant negative Rab5S34N had decreased numbers of internalized apoptotic cells staining with Lysotracker Red (Figure 6, iâl, quantitated in u), confirming that phagosomes must become Rab5-positive before becoming acidified.
We next asked whether C. elegans DYN-1 would also regulate RAB-5 or RAB-7 recruitment. In dyn-1(ky51) worms at the nonpermissive temperature, phagosomal RAB-5 staining was decreased, suggesting a defect in recruitment of RAB-5 to the maturing phagosome (Figure 7, e, f vs. wild type, a, b; quantitated in q). In addition, phagosomes in dyn-1(ky51) mutant worms were abnormally large compared to those seen in wild-type worms (Figure 7, e vs. a, arrowheads, quantitated in s); these enlarged phagosomes resemble structures observed in mammalian cells that are the result of abnormal fusion events65. In contrast, uninternalized cell corpses in ced-1 or ced-5 mutant worms did not show a similar increase in corpse volume (Figure 7, s, Supplementary Figure S1). RAB-7 halos were also greatly decreased in dyn-1(ky51) mutant worms, consistent with a block in phagosome maturation (Figure 7, g, h vs. wild type, c, d, quantitated in q). It is noteworthy that RAB-5 and RAB-7-positive endosomes, likely resulting from receptor-mediated endocytosis, appeared normal (Supplementary Figure S1), suggesting a more stringent requirement for DYN-1 in the recruitment of RAB-5 and RAB-7 to phagosomes containing apoptotic cells. Taken together, these data suggest a specific and evolutionarily conserved role for dynamin homologues in regulation of maturation of phagosomes containing apoptotic cells upstream of RAB-5.
Figure 7. DYN-1 is required for efficient recruitment of RAB-5 and RAB-7 to phagosomes containing engulfed apoptotic cells in the C. elegans gonad.
Bright field images represent DIC micrographs. Arrows and arrowheads indicate apoptotic germ cells or protein localized around apoptotic germ cells. dyn-1(ky51) worms are shown at the nonpermissive temperature (25 ÂC) unless otherwise stated. Scale bar, 10 Îm. n >10 for each transgenic. Error bars represent s.e.m.
Compared to wild type (aâd), CFP::RAB-5 (e,f) and YFP::RAB-7 (g,h) halos are decreased in the gonad of dyn-1(ky51) mutant worms (f,h, arrowheads); quantitated data is presented in (q). Further, phagosomes appear larger in dyn-1(ky51) mutant worms (e, g, arrowhead and volume quantitated in (s). Nematodes deficient in vps-34 (iâl) or rme-8 (mâp) showed similar decreases in the number of RAB-5 or RAB-7 corpses (quantitated in r).
(t) Schematic of the genetic pathway for phagosome maturation containing apoptotic cells, based on the data presented above and in earlier figures.
VPS-34 functions downstream of dynamin and is required for Rab5 recruitment to the phagosome
We next asked whether DYN-1 functions upstream or downstream of VPS-34. In vps-34(RNAi) worms, DYN-1::YFP was still efficiently recruited to apoptotic corpses (Figure 3, râs), suggesting that DYN-1 likely functions upstream of (or in parallel to) VPS-34 during engulfment. Since our localization studies suggested that DYN-1 might function upstream of VPS-34, we next looked at recruitment of RAB-5 in vps-34-deficient nematodes (Figure 7, iâl). Knockdown of vps-34 also resulted in defects in RAB-5 recruitment or stabilization on the nascent phagosomes (Figure 7, r), essentially similar to dyn-1(ky51) mutant worms. This suggested that DYN-1 and VPS-34 function at a step upstream of RAB-5 recruitment to the nascent phagosome. However, how these proteins might function together to mediate RAB-5 recruitment was unclear (Figure 7t).
During endocytosis, Vps34 is thought to function downstream of Rab5, mediating recruitment of Rab5 effectors. However, our genetic studies above place Vps34 upstream of Rab5 recruitment. To further investigate the timing of Vps34 function, we transiently transfected NIH3T3 cells with a FLAG-tagged Vps34 construct and monitored Vps34 recruitment to the forming phagosome. Consistent with our genetic studies, we found that Vps34 was recruited to the phagocytic cup, where it co-localized with endogenous Dyn2 (Figure 8, aâd). Since both DYN-1 and VPS-34 seemed to function genetically at the same stage, we next asked whether Vps34 localization might be affected by decreased dynamin activity. Cells expressing either GFP (as transfection control, Figure 8, o) or Dyn2WT (Figure 8, eâi, quantitated in o) showed robust recruitment of Vps34 to forming phagosomes. However, in cells expressing dominant-negative Dyn2K44A (Figure 8, jân, o), Vps34 recruitment was significantly reduced (Figure 8, o, p < 0.001). Taken together with our previous studies showing that DYN-1 is properly localized in vps-34(RNAi) worms, this data strongly suggests that Vps34 functions downstream of dynamin, but upstream of RAB-5 recruitment.
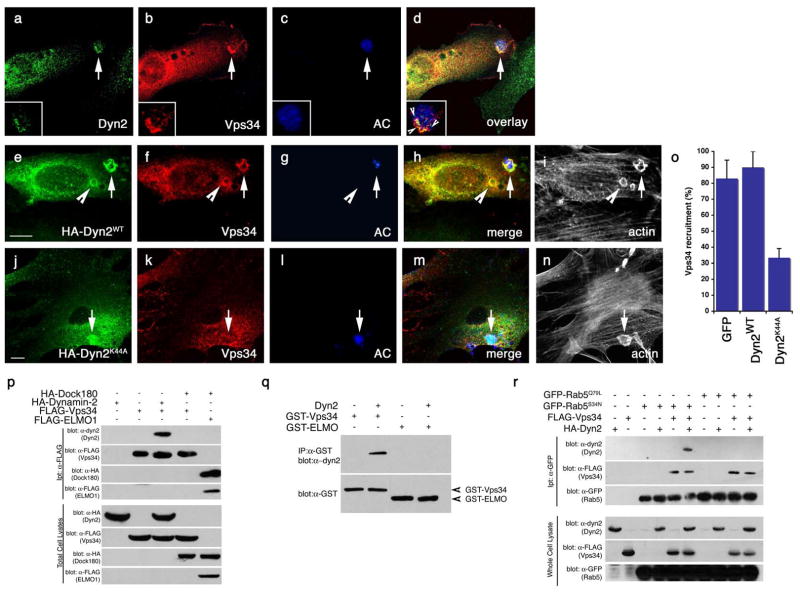
Figure 8. Vps34 functions downstream of Dyn2 and mediates the interaction between Dyn2 and Rab5.
Arrows indicate localization of phagocytic cup or co-localizaiton of indicated proteins. Size bar, 10 Îm. Apoptotic cells were stained with CMHC, whose fluorescence becomes less detectable when the apoptotic cells are engulfed (g, arrowhead).
(aâd) Apoptotic Jurkat cells were incubated with NIH/3T3 fibroblasts transiently transfected with FLAG-Vps34 and the localization of endogenous Dyn2 (a) and FLAG-Vps34 (b) in phagocytic cups containing apoptotic cells (c) were monitored. Both proteins co-localize in the phagocytic cup (d, overlay, arrow)
(eân) Apoptotic Jurkat cells were incubated with NIH/3T3 fibroblasts transfected with either GFP alone (as control, not shown) or HA-tagged Dyn2WT (e) or Dyn2K44A (j) and the localization of FLAG-tagged Vps34 (f, k) on phagocytic cups containing apoptotic cells (g, l) was monitored. In Dyn2K44A transfected cells, recruitment of FLAG-Vps34 to the forming phagosome was disrupted (k, arrow), while Vps34 was efficiently recruited to the phagocytic cup in Dyn2WT (f) or GFP-transfected cells (not shown). Phalloidin-stained cells are included to better discriminate phagocytic cups (i, n), which are enriched in actin. Quantitation of these data are presented in (o).
(p) 293T cells were transiently transfected with the indicated proteins, lysed and immunoprecipitated using anti-Flag conjugated agarose beads and assessed by immunoblotting for the indicated proteins.
(q) GST-tagged Vps34 or ELMO1 proteins were expressed in bacteria and purified using GST-sepharose. Approximately 10 Îg purified His-Dyn2 was added to approximately 2 Îg of each GST-tagged protein. Protein interaction was assessed by immunoblotting.
(r) 293T cells were transiently transfected with the indicated proteins, lysed and immunoprecipitated using anti-GFP conjugated agarose beads and assessed by immunoblotting for the coprecipitating proteins. Rab5Q79L and Rab5S34N are considered to mimic for GTP and GDP-bound Rab5, respectively.
A Dyn2-Vps34 complex mediates Rab5 recruitment to the nascent phagosome
Mammalian Vps34 has previously been detected in a complex with Rab566; thus, it was possible that Vps34 served as the link between dynamin and Rab5. We first observed that Dyn2 could bind Vps34 when transiently overexpressed in 293T cells (Figure 8, p). Under these conditions Vps34 did not associate with other proteins involved in the engulfment of apoptotic cells, such as Dock180 (Figure 8, p), ELMO1, GULP, or CrkII (data not shown). To determine whether Dyn2 physically binds Vps34 or if the two are part of a larger complex, we produced GST-Vps34 in bacteria and asked if this could bind purified Dyn2. In this case, Dyn2 could be precipitated with GST-Vps34 (but not GST-ELMO1), suggesting that Dyn2 and Vps34 directly interact (Figure 8, q and Supplementary Figure S10).
We next tested whether Dyn2, Vps34 and Rab5 could form a complex and made several interesting observations. First, we could detect a complex containing Dyn2, Vps34, and Rab5S34N, which mimics the GDP-bound form of Rab5 (Figure 8, r). Dyn2 did not interact with Rab5 in the absence of Vps34, suggesting a bridging role for Vps34 in linking dynamin and Rab5. Consistent with our biochemical studies, we found that DYN-1::YFP and CFP::RAB-5 can co-localize on phagosomes in the nematode (Figure 9, aâd, arrow). Importantly, DYN-1::YFP did not co-localize to a significant extent with CFP::RAB-7 (Figure 9, mâp, arrows vs. arrowheads). Knockdown of vps-34 disrupted co-localization between DYN-1 and RAB-5, supporting a role for VPS-34 in mediating DYN-1-RAB-5 interaction and recruitment of RAB-5 to the phagosome (Figure 9, iâl, arrows).
Figure 9. DYN-1 co-localizes with RAB-5 and is required for VPS-34 activity.
Bright field images represent DIC micrographs. Arrows and arrowheads indicate apoptotic germ cells or protein localized around apoptotic germ cells. dyn-1(ky51) worms are shown at the nonpermissive temperature (25 ÂC) unless otherwise stated. Scale bar, 10 Îm. n >10 for each transgenic. Error bars represent s.e.m.
(aâp) DYN-1::YFP (b, arrow) and CFP::RAB-5 (c, arrow) co-localize on phagosomes containing apoptotic cells (a, arrow and d, overlay) at similar times. In rab-7(RNAi) worms, phagosome-containing corpses (e) are arrested at the RAB-5(+) stage (g, arrows and arrowheads); a population of these stain with DYN-1::YFP (f, h, arrows), suggesting proteins transiently stain the same phagosome as it matures. vps-34(RNAi) inhibits co-localization of DYN-1::YFP (j, l, arrowheads) and CFP::RAB-5 (k, l, arrowheads). DYN-1::YFP and CFP::RAB-7 did not co-localize on apoptotic cell containing phagosomes (mâp, arrowheads).
(qâv) The YFP::2xFYVE construct specifically binds domains enriched in PtdIns(3)P. YFP::2xFYVE is recruited to phagosomes containing early apoptotic corpses (q, pre-refractile corpse, arrow), while it is excluded from phagosomes containing late corpses (refractile, arrow head). 2xFYVE::YFP is recruited to phagosomes (q, qâ, arrows) in wild type worms; this recruitment is greatly decreased in ced-6(n1813) (r, râ), dyn-1(ky51) (s, sâ), and rab-5(RNAi) (u, uâ) worms. Recruitment to phagosomes is greatly decreased in vps-34(RNAi) worms (t, tâ), consistent with a role for VPS-34 in generation of PtdIns(3)P. 2xFYVE::YFP is still recruited in rab-7(RNAi) (v, vâ). These data were quantitated in (w).
(x, y) Potential model for maturation of phagosomes containing apoptotic cells based on cell biological (x) and genetic studies (y) in C. elegans and mammals presented here.
Our studies suggest that DYN-1 is transiently recruited to the phagosome and may define an intermediate structure prior to RAB-5 recruitment. Notably, in rab-7(RNAi) worms, where phagosomes were arrested at the RAB-5(+) stage, we also found that although most phagosomes stain with CFP::RAB-5, only a subset stain with DYN-1::YFP (Figure 9, eâh). Thus, DYN-1 and RAB-5 appear to mark distinct, but overlapping stages during phagosome maturation.
VPS-34 kinase activity is regulated by RAB-5
The function of many proteins is modified following recruitment to their sites of action, providing an additional level of regulation beyond protein recruitment. Our data suggests VPS-34 is required for RAB-5 recruitment; in vitro studies have suggested an additional level of protein regulation, where Rab5GTP promotes optimal VPS34 activity67. To further narrow down the mechanism of Vps34 function, we created transgenic nematodes expressing a YFP::2xFYVE fusion construct. The FYVE domains have been shown to specifically bind PtdIns(3)P generated by VPS-34 on endosomes68. We found that this marker was recruited to phagosomes containing apoptotic cells in wild type (Figure 9, q, qâ, arrows, quantitated in w), but not in worms deficient in ced-1 or ced-6 (Figure 9, r, râ and w), suggesting that staining occurs only following corpse internalization. Staining was also greatly reduced in vps-34(RNAi) animals (Figure 9, t, tâ), consistent with a known role for Vps34 in generation of PtdIns(3)P. The YFP::2xFYVE construct was not recruited around apoptotic cells in dyn-1(ky51) mutant worms (Figure 9, n, nâ), confirming that VPS-34 functions downstream of DYN-1. Interestingly, in rab-5(RNAi) treated worms, YFP::2xFYVE recruitment to phagosomes was greatly reduced (Figure 9, u, uâ), suggesting that RAB-5 is required for VPS-34 activity. Knockdown of rab-7 resulted in the accumulation of YFP::2xFYVE-positive phagosomes (Figure 9, v, vâ), confirming that VPS-34 functions upstream of RAB-7 during phagosome maturation. Taken together, these data define an evolutionarily conserved pathway in mammals and worms, wherein dynamin and Vps34 localize to phagosomes containing internalized apoptotic corpses and mediate recruitment of Rab5. This is followed by a Rab5-dependent activation of Vps34 kinase activity. subsequent recruitment of FYVE-domain containing proteins, and the maturation of this phagosome to a RAB-7 positive state coupled with acidification of the phagosome.
Discussion
In this report, through a combination of genetic, biochemical and cell biological studies in C. elegans and mammalian cells, we have identified several players important for maturation of phagosomes containing apoptotic cells and corpse degradation. Through in-depth analysis of the candidate genes identified in the unbiased genetic screens, we now define a novel mechanism wherein the large GTPase DYN-1 (dynamin) and the PtdIns(3)-kinase VPS-34 (Vps34) are recruited to the nascent phagosome and then regulate recruitment of Rab5 (Figure 9, x). We identify for the first time a novel and direct physical interaction between dynamin and Vps34 and the binding of Rab5GDP to this complex. Our work also suggests that an as-yet unidentified GEF would convert Rab5 to Rab5GTP, resulting in activation of Vps34 kinase activity, and subsequent recruitment of FYVE-domain-containing proteins that bind PtdIns(3)P, the product of Vps34 enzymatic activity67, 69. We also identify further maturation steps, leading to RAB-7 recruitment and the importance of HOPS complex proteins in regulating phagosome maturation during apoptotic cell clearance. Taken together, our studies identify a number of new evolutionarily conserved players involved in corpse processing subsequent to internalization. This work also identifies the temporal order of recruitment/function for many of these components, and highlights the importance of these heretofore unappreciated downstream components for proper corpse processing (Figure 9, x, y).
Surprisingly, we also find that many proteins commonly used as markers for phagosome structures (such as EEA-1 and other Rab5 effectors) are not required for corpse degradation; however, that does not mean that these genes do not play a minor (or potentially redundant) role in phagosome maturation. It was also possible that RNAi (when a mutant was not available) was not efficient enough to produce a phenotype. We were also not able to identify a GEF that would regulate Rab5 activation. All known Rab5 GEFs contain a canonical Rab5-GEF domain, termed the Vps9 domain, after the yeast Rab5 homologue40. It is then somewhat surprising that none of the three predicted Vps9-domain-containing proteins in the nematode, rme-6, rabx-5, or tag-333, were required individually or redundantly for corpse removal. We are not comfortable suggesting that there is no GEF for Rab5 during this process, we are compelled to suggest that there may exist a class of Rab5 GEFs with a novel architecture, lacking the Vps9 domain, that may specifically regulate phagosomes maturation.
It has been loosely assumed that once the corpse is internalized, phagosomes containing the âapoptotic cargoâ would feed into common endocytic machinery. This is not necessarily the case for bacterial pathogens; our work clearly highlights several features of phagosome maturation during apoptotic cell clearance that are distinct from classical endocytosis. For example, worms deficient in clathrin light and heavy chains (clc-1 and clhc-1, respectively), show increased germ cell apoptosis, but no defects in corpse engulfment (Supplementary Table S4). Additionally, we found that members of the exocyst complex were not required for corpse removal (data not shown). Moreover, knockdown of nsf-1 and mutations in eea-1 and rabs-5 did not inhibit corpse removal, even though they have been shown to regulate fusion pore formation in yeast and mammalian cells, and proper receptor-mediated endocytosis, These results, combined with our other observations that the Rab5 GEFs rme-6 and rabx-5 are not required for corpse clearance, suggest that classical endocytosis and apoptotic cell clearance not only use distinct internalization processes but likely use different post-internalization maturation pathways.
We also assessed DYN-1 localization after disruption of RME-8, another candidate that was isolated in the screen as having increased numbers of refractile corpses in the gonad. rme-8 was initially identified in a screen for genes defective in receptor-mediated endocytosis; RME-8 localizes to the early endosome, though its exact mechanism of function is unknown70â72. In rme-8(b1023ts) worms, DYN-1::YFP was efficiently recruited around corpses, further suggesting that DYN-1 may function between corpse internalization and an early phagosome structure. Additionally, rme-8 was also required for recruitment of RAB-5 and RAB-7 to the phagosome (Figure 3, t, u and 7, mâp). Little is known about how rme-8 functions in endocytosis, but our studies suggest that rme-8 (and our other identified candidate proteins) play a role during an early stage of phagosome maturation.
We also identified members of the HOPS complex, a multifunctional protein complex that regulates multiple stages of endocytosis, in our targeted screen. vps-16, vps-18, vps-33, and vps-41 are components of a SNARE complex, which function with syntaxins to direct vesicles to their appropriate organelle48, 49; vps-39 is a likely GEF for Rab7. The yeast orthologue of vps-11 has been shown to interact with vps-39, though the significance of this is uncertain73. The HOPS complex has been implicated in S. cerevisiae as being part of a tethering complex, which would direct the fusion of vesicles to the endosome. Here, we show that knockdown of vps-11, vps-16, vps-18, vps-33, vps-39 and vps-41 result in phagosome arrest at the RAB-7(+) stage. This is consistent with studies suggesting these proteins likely function as a tether for site of vesicular fusion, though it is uncertain whether fusing vesicles would be derived from existing lysosomes or would be targeted from the Golgi to create de novo lysosomes. Loss of the HOPS complex would then render RAB-7(+) phagosomes unable to mature into later lysosomal structures. Other studies have shown that the recruitment of Rab7 to endosomes is linked to Rab5 activity; hence, HOPS complex members can also be found in a complex39 with Rab5GTPÎS. We find that knockdown of one complex member, vps-41 resulted in an increase in the number of corpses in RAB-5(+) phagosomes, suggesting that in the nematode, VPS-41 may be involved in the recruitment of the HOPS complex to maturing RAB-5(+) phagosomes. Our current studies could not identify a role for the final member of the HOPS complex, VPS-45 (a Sec1-family member39), as this gene appears to function downstream of RAB-7 loss from the phagosome. Interestingly, studies on endocytosis have suggested that the HOPS complex primarily functions to mediate âRab conversionâ of Rab5(+) phagosomes39, again highlighting that while phagosome maturation utilizes a subset of the endocytic machinery, it appears to use them in a novel fashion.
Recent studies have also suggested a controversial role for the fusion of phagosomes with the endoplasmic reticulum (ER) during phagosome maturation74. Other groups have suggested that this may be due to contamination of phagosome preparations with ER-derived membrane75. rab-5(RNAi) has also been shown to affect integrity of the ER, making it formally possible that our knockdown may have identified a role for the ER is corpse removal. RET-1 and YOP-1 (nematode homologues of RTN4a/NogoA and DP1/REEP5, respectively) are required for new tubulation of the ER76, which would be required for entry of apoptotic cells into an ER structure. ret-1(gk242); yop-1(RNAi) worms did not show defects corpse removal, suggesting that, at least in the worm, the ER may not play a direct role in phagosome biogenesis or corpse degradation.
In both mammalian cells and in C. elegans, a feedback regulation between internalization and degradation of the apoptotic cell corpse have been implied5, 77â80. However, the protein machinery that connects these events is unknown. Here, we have identified dynamin, which is recruited to the phagocytic cup at the early stages of engulfment, but whose function is required later for maturation of the internalized corpse into an acidic, Lysotracker/SYTO staining compartment. We also identified several homologues of sorting nexins in our screen, including lst-4, which appears to function in the internalization phase of corpse removal (data not shown). Intriguingly, dyn-1 mutants may also show a potential feedback into corpse internalization, and lst-4 may represent a point of intersection between internalization of apoptotic cells and their degradation. Further study of these proteins should provide interesting insights into the control of complex processes.
In addition to a simple âgarbage disposalâ function, phagosome maturation is essential for the proper function of the cellular immune system. Apoptotic cells are not trafficked into MHC class II-containing phagosomes32, and the inability to properly degrade apoptotic cells has also been linked to autoimmune disease. This opens the possibility that blocking corpse degradation via the âstandardâ pathway results in inappropriate presentation of antigens derived from apoptotic cells4. Thus, the components of the phagosome maturation pathway described here, along with several previously unrecognized players identified through the reverse genetic screens in C. elegans (Supplementary Table 1) may allow further characterization of the process of apoptotic cell clearance with significant implications for tolerance to apoptotic cell-derived self antigens and autoimmunity.
Methods
Nematode Strains and Reagents
Nematode strains were cultivated as previously described12. Mutations used were as follows: LGI: gla-3(op216), ced-12(k149), ced-1(e1735); LGIII: ced-6(n1813), ced-7(n1996); LGIV: ced-2(n1994), ced-10(n3246), opIs110[Plim-7::yfp::act-5]; LGV: bcIs39[Plim-7::ced-1::gfp], LGX: dyn-1(ky51). opIs160[Pced-6::yfp::ced-6, unc-119(+)], Integration sites of opIs220[Peft-3::dyn-1::yfp, unc-119(+)], opIs223[Peft-3::yfp::rab-7, unc-119(+)], opIs282[Pced-1::yfp::rab-5, unc-119(+)], and opIs334 [Pced-1::yfp::2xFYVE, unc-119(+)] were not mapped. Unless noted otherwise mutations were previously described41. opEx1278[Peft-3::yfp::rab-7, unc-119(+)], opEx1279[Peft-3::cfp::rab-5, unc-119(+)], opEx1304[Pced-1::yfp::rab-5, unc-119(+), opEx1303[Pced-1::yfp::rab-7, unc-119(+)], vfEx130 [Pced-1::cfp::rab-7, unc-69(+)], vfEx123 [Pced-1::dyn-1::yfp, unc-69(+)], vfEx129 [Pced-1::dyn-1::yfp, Pced-1::cfp::rab-5, unc-69(+)] and vfEx128 [Pced-1::yfp::nsf-1; rol-6(su1006)] are extrachromosomal arrays.
Reagents used in this study were Alexa-647 phalloidin, Hoechst 33342, SYTO41, SYTO59, acridine orange (Invitrogen), anti-FLAG clones M2 (Western) and M5 (immunofluorescence) (Sigma), anti-clathrin light chain CON-1, anti-cortactin, anti-dynamin-2 C-18 (used for Western analyses), anti-HA F-7 (Santa Cruz), anti-dynamin Hudy1 (Upstate), anti-dynII (BD Bioscience) (used for immunofluorescence), Alexa 488, 555 anti-mouse, and Alexa 555 anti-rabbit (Invitrogen); all secondary antibodies were highly cross-adsorbed to minimize cross reactivity between species. Plasmids pcDNA-HA-Dyn2WT and pcDNA-HA-Dyn2K44A were a gift from Dorothy Schaffer, pcDNA5-FLAG-Vps3481 were a gift of Jae Jung, and pGreen Lantern-Rab5S34N and pGreen Lantern-Rab5Q79L were a gift of Jim Casanova.
Reverse genetic screening
Feeding RNAi was performed as described50 with the following modifications. Plates containing NGM-agarose and 1â5mM IPTG (âRNAi platesâ) were inoculated with 300 ÎL of appropriate bacterial cultures (transformed with constructs for generation of double stranded RNA under the control of the T7 promoter) were incubated for 8â12 hours before addition of worms. Between 30 and 60 synchronized gla-3(op216) L1 stage worms (which was used to increase the number of apoptotic germ cells) were placed on each RNAi plate and left for 72h at 20ÂC. Worms on plates were then stained with Acridine Orange38 and scored for percent of worms showing AO staining of apoptotic cell corpses under an M2Bio epifluorescence dissecting microscope (Zeiss). Candidate genes were grouped into functional classes as previously described50. To eliminate genes that reduce the number of apoptotic cell corpses in the germ line (either directly or indirectly), we compared our candidates to the results of other RNAi screens and removed candidates whose knockdown resulted in delayed morphogenesis or sterility. Number of germ cell corpses was then scored by DIC microscopy for each candidate where possible (Supplementary Table S1).
The following dsRNA-synthesizing bacteria were used as positive controls for each RNAi experiment: bir-1(RNAi), which gives rise to high embryonic lethality, unc-22(RNAi), which results in an Uncoordinated phenotype, and ced-3(RNAi), which potently suppresses germ cell apoptosis. Worms fed with HT115(DH3) bacteria transformed with the original L4440 RNAi vector containing no insert were used as a reference strain.
DIC and Fluorescence microscopy (Nematode)
Worms were placed on 2% agarose pads in M9, anaesthetized with 3â5mM levamisole (Sigma) and mounted under a cover slip for observation using a Leica DM-RA or Zeiss Axiovert 200 microscope equipped with DIC (Nomarski) optics and standard epifluorescence with filtersets appropriate for visualization of YFP, CFP/SYTO 41, SYTO 59 or GFP. Images were false-colored in OpenLab or Adobe Photoshop 7.0, which was also used to optimize brightness/contrast.
Staining of worms with Acridine Orange (Molecular Probes) was performed as previously described29. To visualize engulfed cells, gonads were dissected in PBS supplemented with 12.5ÎM of SYTO 41 or SYTO 59, then incubated in the dark for 10 minutes prior to observation.
Synchronization and C. elegans phagocytosis assays
To score apoptotic corpses in the hermaphrodite germ line, clean worms were synchronized by picking hermaphrodites at the L4 larval stage (Christmas tree vulva). These worms were incubated for 24 hours at 20 ÂC then scored for persistent cell corpses and fluorescent halos where appropriate unless otherwise noted. Inactivation of DYN-1 in dyn-1(ky51) mutant worms occurs in less than a minute following shift to the nonpermissive temperature53. For all experiments using the temperature sensitive dyn-1(ky51) and rme-8(b1023) allele worms were shifted to 25 ÂC as L4s and scored after 12, 24 and 36 hours for persistent cell corpses and fluorescent haloes. At the nonpermissive temperature, dyn-1(ky51) mutants exhibits pleiotropic phenotypes and only worms exhibiting a gonad morphology as close to wild-type as possible were chosen for scoring. For all experiments using the temperature sensitive rme-8(b1023) worms were grown at 15ÂC and were shifted to 25ÂC as L4s. For most animals only one gonad arm was scored, as the other arm was concealed by the intestine.
Immunofluorescence in mammalian cells
Cells were incubated overnight in Lipofectamine 2000 as previously described82, then washed and incubated in DMEM + 10% serum ~8 hours before engulfment assay was conducted. For siRNA experiments, cells were transfected using Amaxa program U-30 and Kit R for NIH/3T3 cells or T-21 and Kit V for J774 macrophages (Amaxa, Germany) with an siRNA SMARTpool containing 4 siRNAs targeting mouse dynamin-2 (Dharmacon cat # M-044919-01), elmo1 (M-041254-00) or a noncoding SMARTpool (Dharmacon cat # D-001206-13) using 1.2 Îg of total siRNA (0.3 Îg of each individual siRNA) as previously described83, then incubated 48 hours to recover. Images were acquired using a Zeiss 510-UV laser scanning confocal microscope with 405, 488, 543, and 633 lasers (Zeiss AG, Germany). For z reconstruction experiments, confocal z sections were acquired every 0.3 Îm; z-axis was reconstructed in LSM and subsequently deconvolved.
Apoptotic thymocytes and Jurkat cells were generated as previously described84; apoptotic thymocytes (5 x 105 cells per condition) were stained with either CMFDA, TAMRA, or BODIPY 630 (Invitrogen) and added to NIH/3T3 cells in 4-well Labtek II culture chambers followed by a brief centrifugation to pellet cells onto the slide. Of the cells that bind to the phagocyte monolayer, ~98% are apoptotic by anti-active caspase-3 staining (data not shown). Thymocytes were allowed to be engulfed for 30 minutes; unbound apoptotic thymocytes were gently washed off DMEM + 10% FBS, and then subsequently incubated for 2 hours. Jurkat cells were induced to die with etoposide (30 ÎM). For Lysotracker staining, cells were incubated with apoptotic cells in DMEM + 10% FBS containing 1/10,000 dilution of Lysotracker Red. Cells were then fixed with 3% paraformaldehyde (Sigma) in PBS for 30 minutes, permeabilized with 0.1% Triton X-100 (Sigma) and blocked with 5% milk that had been clarified by high speed centrifugation. Antibody staining was then done as previously described82.
Generation of transgenic nematodes
Transgenic worms were obtained by microparticle bombardment in a Biolistic PDS-1000 (Bio-Rad) transformation (opEx and opIs alleles) or microinjection as previously described85, 86. unc-119 or unc-69 was used as a transformation marker. opIs220 [Peft-3::dyn-1::yfp] was found to rescue the temperature sensitive persistent cell corpse phenotype of the dyn-1(ky51) mutants. For opEx1278 [Peft-3::yfp::rab-7] and opEx1279 [Peft-3::cfp::rab-5] we observed an accumulation of the fluorescent proteins into vesicular structures suggesting that the fusion proteins are functional (Supplementary Figure S1). Similar vesicular structures were observed in worms carrying opEx1304[Pced-1::yfp::rab-5; unc-119(+)], opEx1303[Pced-1::yfp::rab-7; unc-119(+)], opIs282[Pced-1::yfp::rab-5; unc-119(+)], opIs334 [Pced-1::yfp::2xFYVE, unc-119(+)], vfEx130 [Pced-1::cfp::rab-7, unc-69(+)], vfEx123 [Pced-1::dyn-1::yfp, unc-69(+)], vfEx129 [Pced-1::dyn-1::yfp, Pced-1::cfp::rab-5, unc-69(+)] and vfEx128 [Pced-1::yfp::nsf-1; rol-6(su1006)].
Immunoprecipitations
293T cells (in 10 cm dishes) were transiently transfected with 1â4 Îg of each appropriate constructs. After 36 h, the cells were lysed in a 4ÂC environment room (in 1% Triton X-100, 50mM Tris and 150mM NaCl, 10mM MgCl2) and immunoprecipitated using anti-FLAG (clone M2, Sigma) or anti-GFP (clone B-2, Santa Cruz) antibody directly coupled to sepharose. Vps34 was cloned as a KpnI-NotI fragment from pcDNA5-FLAG-Vps34 into pGEX4T-2, then transformed into Arctic Express competent cells, which were induced with 0.1 mM IPTG at 10 ÂC overnight. Bacteria were lysed in cell lysis buffer, sonicated, and GST-Vps34 protein was purified from the lysate using glutathione sepharose (GE Healthcare). Approximately 10 Îg of purified Dyn2 (gift of D Shafer) was incubated with GST-Vps34 or GST-ELMO187 in IP buffer (0.5% Triton X-100, 50 mM Tris, 150 mM NaCl, 10mM MgCl2, 0.1% milk), then washed (0.1% Triton X-100, 50 mM Tris, 150 mM NaCl, 10mM MgCl2) and processed for Western blotting.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Ravichandran and Hengartner laboratories for helpful input and suggestions on this manuscript. We would also like to thank Dorothy Schafer for dynamin plasmids and purified Dyn2 protein, David Castle, Jim Casanova and Heidi McBride for Rab5 expression constructs, Jae Jung for Vps34 expression constructs and Lukas Neukomm for providing pLN022 and pLN019 plasmids, and Jan Redick and Christie Davis from the Advanced Microscopy Facility for help with confocal microscopy. Some strains used in this work were obtained from the Caenorhabditis Genetics Center (CGC). This work was supported by grants from the National Institutes of Health (USA) to KSR, and from the EU Project Apoclear, the Ernst Hadorn Foundation, the University of Zurich and the Swiss National Science Foundation to MOH. JMK is an Arthritis Foundation Postdoctoral Fellow.
Footnotes
The authors declare no competing financial interests.
References
- 1.Scott RS, et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411:207–11. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- 2.Savill J, Fadok V. Corpse clearance defines the meaning of cell death. Nature. 2000;407:784–8. doi: 10.1038/35037722. [DOI] [PubMed] [Google Scholar]
- 3.Hanayama R, Miyasaka K, Nakaya M, Nagata S. MFG-E8-dependent clearance of apoptotic cells, and autoimmunity caused by its failure. Curr Dir Autoimmun. 2006;9:162–72. doi: 10.1159/000090780. [DOI] [PubMed] [Google Scholar]
- 4.Kawane K, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 5.Schrijvers DM, De Meyer GR, Kockx MM, Herman AG, Martinet W. Phagocytosis of apoptotic cells by macrophages is impaired in atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25:1256–61. doi: 10.1161/01.ATV.0000166517.18801.a7. [DOI] [PubMed] [Google Scholar]
- 6.Gardai SJ, Bratton DL, Ogden CA, Henson PM. Recognition ligands on apoptotic cells: a perspective. J Leukoc Biol. 2006;79:896–903. doi: 10.1189/jlb.1005550. [DOI] [PubMed] [Google Scholar]
- 7.Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 2006;27:244–50. doi: 10.1016/j.it.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Tibrewal N, Birge RB. Phosphatidylserine recognition by phagocytes: a view to a kill. Trends Cell Biol. 2006;16:189–97. doi: 10.1016/j.tcb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Franz S, et al. Apoptosis and autoimmunity: when apoptotic cells break their silence. Curr Rheumatol Rep. 2006;8:245–7. doi: 10.1007/s11926-006-0001-y. [DOI] [PubMed] [Google Scholar]
- 10.Gaipl US, et al. Inefficient clearance of dying cells and autoreactivity. Curr Top Microbiol Immunol. 2006;305:161–76. doi: 10.1007/3-540-29714-6_8. [DOI] [PubMed] [Google Scholar]
- 11.Horvitz HR. Worms, life, and death (Nobel lecture) Chembiochem. 2003;4:697–711. doi: 10.1002/cbic.200300614. [DOI] [PubMed] [Google Scholar]
- 12.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- 14.Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–6. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- 15.Wu YC, Horvitz HRC. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–4. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 16.Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NYC. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell. 2001;1:477–89. doi: 10.1016/s1534-5807(01)00058-2. [DOI] [PubMed] [Google Scholar]
- 18.Gumienny TL, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 19.Brugnera E, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–82. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 21.Su HP, et al. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP) J Biol Chem. 2002;277:11772–9. doi: 10.1074/jbc.M109336200. [DOI] [PubMed] [Google Scholar]
- 22.Hamon Y, et al. Cooperation between Engulfment Receptors: The Case of ABCA1 and MEGF10. PLoS ONE. 2006;1:e120. doi: 10.1371/journal.pone.0000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luciani MF, Chimini G. The ATP binding cassette transporter ABC1, is required for the engulfment of corpses generated by apoptotic cell death. Embo J. 1996;15:226–35. [PMC free article] [PubMed] [Google Scholar]
- 24.Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998;93:951–60. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- 25.Jehle AW, et al. ATP-binding cassette transporter A7 enhances phagocytosis of apoptotic cells and associated ERK signaling in macrophages. J Cell Biol. 2006;174:547–56. doi: 10.1083/jcb.200601030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu QA, Hengartner MO. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell. 1998;93:961–72. doi: 10.1016/s0092-8674(00)81202-7. [DOI] [PubMed] [Google Scholar]
- 27.Henson PM. Engulfment: ingestion and migration with Rac, Rho and TRIO. Curr Biol. 2005;15:R29–30. doi: 10.1016/j.cub.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Kinchen JM, Hengartner MO. Tales of cannibalism, suicide, and murder: Programmed cell death in C. elegans. Current Topics in Developmental Biology. 2005;65:1–45. doi: 10.1016/S0070-2153(04)65001-0. [DOI] [PubMed] [Google Scholar]
- 29.Kinchen JM, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–9. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- 30.Kinchen JM, Ravichandran KS. Journey to the grave: signaling events regulating removal of apoptotic cells. J Cell Sci. 2007;120:2143–9. doi: 10.1242/jcs.03463. [DOI] [PubMed] [Google Scholar]
- 31.Stuart LM, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 32.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–8. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 33.Blander JM. Signalling and phagocytosis in the orchestration of host defence. Cell Microbiol. 2007;9:290–9. doi: 10.1111/j.1462-5822.2006.00864.x. [DOI] [PubMed] [Google Scholar]
- 34.Hall DH, et al. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–23. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- 35.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–22. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 36.Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–6. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 37.Gruenberg J, van der Goot FG. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 38.Lettre G, et al. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 2004;11:1198–203. doi: 10.1038/sj.cdd.4401488. [DOI] [PubMed] [Google Scholar]
- 39.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 40.Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Bieri T, et al. WormBase: new content and better access. Nucleic Acids Res. 2007;35:D506–10. doi: 10.1093/nar/gkl818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato M, et al. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nat Cell Biol. 2005;7:559–69. doi: 10.1038/ncb1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews R, Ahringer J. Asymmetry of Early Endosome Distribution in C. elegans Embryos. PLoS ONE. 2007;2:e493. doi: 10.1371/journal.pone.0000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gengyo-Ando K, et al. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–7. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayakawa A, et al. The WD40 and FYVE domain containing protein 2 defines a class of early endosomes necessary for endocytosis. Proc Natl Acad Sci U S A. 2006;103:11928–33. doi: 10.1073/pnas.0508832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas A. NSF--fusion and beyond. Trends Cell Biol. 1998;8:471–3. doi: 10.1016/s0962-8924(98)01388-9. [DOI] [PubMed] [Google Scholar]
- 47.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–7. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horazdovsky BF, Emr SD. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J Biol Chem. 1993;268:4953–62. [PubMed] [Google Scholar]
- 49.Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell. 2000;6:661–71. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 50.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 51.Sawa M, et al. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J Cell Sci. 2003;116:1505–18. doi: 10.1242/jcs.00362. [DOI] [PubMed] [Google Scholar]
- 52.Tosello-Trampont AC, Nakada-Tsukui K, Ravichandran KS. Engulfment of apoptotic cells is negatively regulated by Rho-mediated signaling. J Biol Chem. 2003;278:49911–9. doi: 10.1074/jbc.M306079200. [DOI] [PubMed] [Google Scholar]
- 53.Clark SG, Shurland DL, Meyerowitz EM, Bargmann CI, van der Bliek AM. A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans. Proc Natl Acad Sci U S A. 1997;94:10438–43. doi: 10.1073/pnas.94.19.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christoforidis S, et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1:249–52. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 55.Krueger EW, Orth JD, Cao H, McNiven MA. A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol Biol Cell. 2003;14:1085–96. doi: 10.1091/mbc.E02-08-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schafer DA. Coupling actin dynamics and membrane dynamics during endocytosis. Curr Opin Cell Biol. 2002;14:76–81. doi: 10.1016/s0955-0674(01)00297-6. [DOI] [PubMed] [Google Scholar]
- 57.Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5:463–9. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 58.Schafer DA, et al. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr Biol. 2002;12:1852–7. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gold ES, et al. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med. 1999;190:1849–56. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orth JD, McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15:31–9. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 61.Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12:2111–7. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu X, Odera S, Chuang CH, Lu N, Zhou ZC. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–57. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci U S A. 1997;94:377–84. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reubold TF, et al. Crystal structure of the GTPase domain of rat dynamin 1. Proc Natl Acad Sci U S A. 2005;102:13093–8. doi: 10.1073/pnas.0506491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duclos S, et al. Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J Cell Sci. 2000;113(Pt 19):3531–41. doi: 10.1242/jcs.113.19.3531. [DOI] [PubMed] [Google Scholar]
- 66.Shin HW, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol. 2005;170:607–18. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roggo L, et al. Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. Embo J. 2002;21:1673–83. doi: 10.1093/emboj/21.7.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gillooly DJ, et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. Embo J. 2000;19:4577–88. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chang HC, Hull M, Mellman I. The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J Cell Biol. 2004;164:1055–64. doi: 10.1083/jcb.200311084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Girard M, Poupon V, Blondeau F, McPherson PS. The DnaJ-domain protein RME-8 functions in endosomal trafficking. J Biol Chem. 2005;280:40135–43. doi: 10.1074/jbc.M505036200. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Grant B, Hirsh D. RME-8, a conserved J-domain protein, is required for endocytosis in Caenorhabditis elegans. Mol Biol Cell. 2001;12:2011–21. doi: 10.1091/mbc.12.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–62. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gagnon E, et al. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell. 2002;110:119–31. doi: 10.1016/s0092-8674(02)00797-3. [DOI] [PubMed] [Google Scholar]
- 75.Touret N, et al. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell. 2005;123:157–70. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 76.Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol. 2007;178:43–56. doi: 10.1083/jcb.200701139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–48. [PMC free article] [PubMed] [Google Scholar]
- 78.Parrish JZ, Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol Cell. 2003;11:987–96. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 79.Krieser RJ, et al. Deoxyribonuclease IIalpha is required during the phagocytic phase of apoptosis and its loss causes perinatal lethality. Cell Death Differ. 2002;9:956–62. doi: 10.1038/sj.cdd.4401056. [DOI] [PubMed] [Google Scholar]
- 80.Erwig LP, et al. Differential regulation of phagosome maturation in macrophages and dendritic cells mediated by Rho GTPases and ezrin-radixin-moesin (ERM) proteins. Proc Natl Acad Sci U S A. 2006;103:12825–30. doi: 10.1073/pnas.0605331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 82.Grimsley CM, Lu M, Haney LB, Kinchen JM, Ravichandran KS. Characterization of a novel interaction between ELMO1 and ERM proteins. J Biol Chem. 2006;281:5928–37. doi: 10.1074/jbc.M510647200. [DOI] [PubMed] [Google Scholar]
- 83.Tosello-Trampont AC, et al. Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ. 2007 doi: 10.1038/sj.cdd.4402094. [DOI] [PubMed] [Google Scholar]
- 84.Tosello-Trampont AC, Brugnera E, Ravichandran KS. Evidence for a conserved role for CRKII and Rac in engulfment of apoptotic cells. J Biol Chem. 2001;276:13797–802. doi: 10.1074/jbc.M011238200. [DOI] [PubMed] [Google Scholar]
- 85.Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–26. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu M, et al. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nat Struct Mol Biol. 2004;11:756–62. doi: 10.1038/nsmb800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.