Abstract
Background
bHLH transcription factors play many roles in neural development. cousin of atonal (cato) encodes one such factor that is expressed widely in the developing sensory nervous system of Drosophila. However, nothing definitive was known of its function owing to the lack of specific mutations.
Results
We characterised the expression pattern of cato in detail using newly raised antibodies and GFP reporter gene constructs. Expression is predominantly in sensory lineages that depend on the atonal and amos proneural genes. In lineages that depend on the scute proneural gene, cato is expressed later and seems to be particularly associated with the type II neurons. Consistent with this, we find evidence that cato is a direct target gene of Atonal and Amos, but not of Scute. We generated two specific mutations of cato. Mutant embryos show several defects in chordotonal sensory lineages, most notably the duplication of the sensory neuron, which appears to be caused by an extra cell division. In addition, we show that cato is required to form the single chordotonal organ that persists in atonal mutant embryos.
Conclusions
We conclude that although widely expressed in the developing PNS, cato is expressed and regulated very differently in different sensory lineages. Mutant phenotypes correlate with cato's major expression in the chordotonal sensory lineage. In these cells, we propose that it plays roles in sense organ precursor maintenance and/or identity, and in controlling the number of cell divisions in the neuronal branch of the lineage arising from these precursors.
Background
Basic-helix-loop-helix (bHLH) transcription factors are central to neurogenesis in metazoans [1]. The most well known role for such factors in neurogenesis is the so-called 'proneural' function. This function underlies the commitment of neuroectodermal cells to a neural fate, and the term comes originally from the study of proneural genes in Drosophila. In this organism, proneural genes include atonal (ato), amos, scute (sc), and achaete (ac) which are required for the specification of sense organ precursors (SOPs) of the peripheral nervous system [2]. In mutations of these genes, specific subsets of SOPs fail to be formed. For instance, ato is required for the formation of SOPs of chordotonal (Ch) proprioceptive sensory organs [3].
Other members of the bHLH protein family are expressed after neural commitment and play a variety of roles in neural cells leading up to neural differentiation. This is particularly apparent in vertebrates, where for instance the factors NeuroM and NeuroD are required for neuronal migration and differentiation respectively [4,5]. In Drosophila, such 'downstream' neural bHLH factors are represented by asense (ase), cousin of atonal (cato), deadpan (dpn) and target of poxn (tap). These genes are related to sc, ato, hairy/E(spl) and neurogenin respectively. ase, cato and dpn are widely expressed in developing neurons [6-8], whereas tap expression is confined to a small subset of sensory neurons [9]. The functions of these genes are less well known compared with proneural genes. ase is expressed in all neural precursors of both the CNS and PNS [6]. Mutations of ase result in reduced viability but mutant embryos exhibit only subtle PNS defects [10]. In the larval optic lobes, ase participates in the control of mitotic activity in neural precursors [11]. In this process, ase limits proliferation by antagonising dpn. In turn, dpn antagonises dacapo (dap) [12-14]. dap encodes a p21 cyclin-dependent kinase (CDK) inhibitor that is expressed transiently in cells prior to their terminal cell division in order to prevent further divisions [12-14].
Unlike ase and dpn, the expression of cato is confined to the developing PNS, where it was reported to be expressed in all SOPs and their progeny [7]. The function of cato is poorly known. Examination of embryos bearing large deficiencies of the cato region suggested a role in sensory neuron differentiation [7]. We report here the generation and analysis of specific cato mutations. Flies homozygous for cato loss-of-function mutations are viable. Mutant embryos show no gross neuronal differentiation defects, but have a defect in cell proliferation within the Ch sensory lineages. Combination of cato mutation with those of ato and ase reveals a second role for cato in the maintenance of Ch SOP fate or survival.
Results
Expression of Cato differs in Ato/Amos and Sc lineages
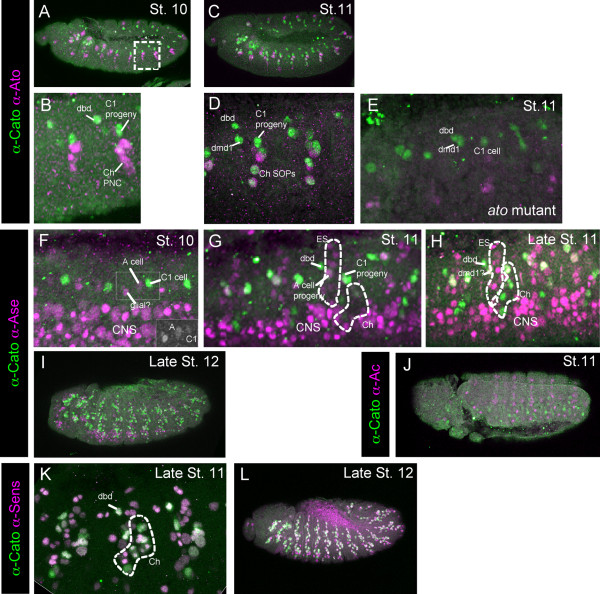
It was previously reported that cato mRNA was initially activated in Ch SOPs, and subsequently it appeared to be expressed generally in all cells of the sensory PNS (both ato-dependent Ch cells and sc-dependent External Sensory (ES) cells) [7]. An anti-Cato antibody confirms and extends these findings. Expression of Cato protein was assessed relative to other markers of early sensory neurogenesis. These include: Amos, which marks the amos-dependent Dorsal Bipolar Dendrite (dbd) and Dorsal Multidendritic 1 (dmd1) neural precursors [15]; Ato, which marks all Ch precursors as well as the dbd and dmd1 cells [3,16]; and Ase and Senseless (Sens), which mark all SOPs and their progeny [6,17]. Cato protein is initially detectable at stage 10 in the first ato-dependent Ch SOP (known as C1 or the 'P' cell [18]), followed shortly by the first amos-dependent precursor, dbd (Fig. 1A, B, F). At this stage it is not expressed in the so-called 'A' cell, which is the first ES SOP to appear (Fig. 1F, G). Shortly after, it is expressed in further Ch SOPs and also in a cell presumed to be the second amos-dependent precursor, dmd1 [16] (Fig. 1C, D, H). At this stage, Cato is still not expressed in ES cells, although numerous ES cells are detectable by Ase and Sens expression (Fig. 1F-H). Eventually, at late stage 11/stage 12, Cato is detectable in the progeny of most SOPs, including ES cells, and overlaps extensively but not completely with Ase and Sens (Fig. 1I-K). In ato mutant embryos, Cato expression is reduced, consistent with loss of most Ch precursors (Fig 1F). However, expression remains in the Amos-dependent cells. It also remains in the C1 cell, consistent with the observation that this Ch precursor usually appears even in the absence of ato function [7,19] (Fig. 1E). In summary, Cato is first expressed in ato- and amos-dependent SOPs. In contrast, it is not expressed in sc-dependent ES cells until after the first division of the SOPs. This difference in expression between the two sensory lineages was also observed in imaginal discs (data not shown).
Figure 1.
Cato protein expression in the embryo. Immunohistochemistry with anti-Cato (green) and a second marker antibody (magenta). (A-E) Expression of Cato relative to Ato. (A, B) Stage 10 embryo. Boxed area is magnified in (B). Cato is expressed in the C1 progeny, the dbd precursor and the probable dmd1 precursor; at this stage Ato is mostly in the proneural cluster (PNC) cells for the remaining Ch precursors. (C) Stage 11 embryo. (D) Magnification of two abdominal segments (different embryo to (C)). Cato is now expressed additionally in further Ch SOPs. (E) Stage 11 ato1 mutant embryo, showing that Cato expression remains in the C1, dbd and dmd1 precursors. Note that non-functional Ato protein is still expressed in these embryos. (F-I) Cato expression relative to the SOP marker, Ase. (F) Stage 10 embryo, with Cato in C1. At this early stage Ase is weakly expressed in C1 and A cells, and a glial cell, and strongly expressed in neuroblasts of the CNS. The green channel for the region boxed is shown in the inset image. (G) Stage 11 embryo, showing more ES precursors (expressing Ase) but these do not express Cato at this stage. (H) Late stage 11 embryos, showing that Cato is still not expressed strongly in most ES cells (Ase-positive cells). (I) Late stage 12 embryo, Cato is now expressed widely in PNS cells. (J, K) Cato expression relative to Sens. At early (J) and late (K) stage 12 there is substantial co-expression of Cato and Sens in PNS cells.
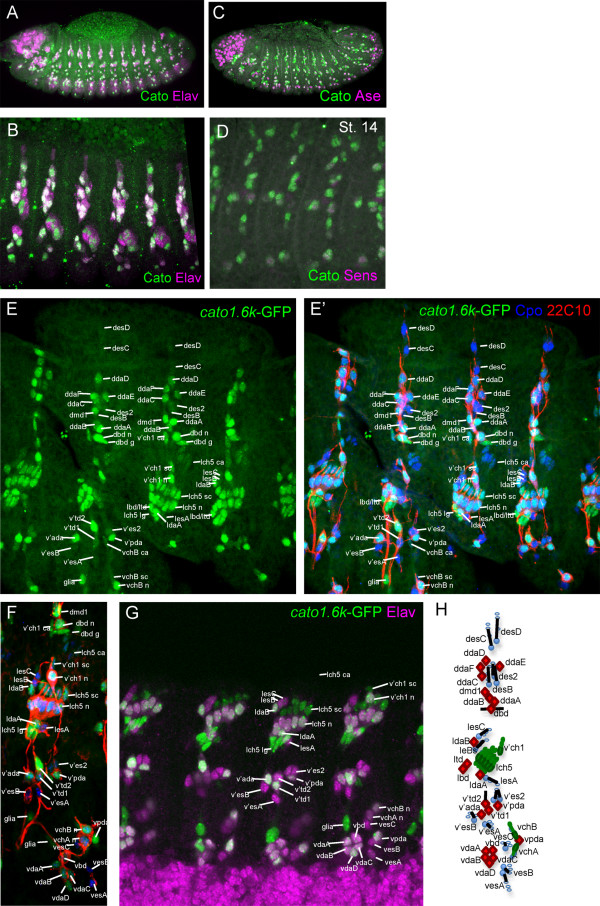
Cato protein expression persists in sensory lineages until terminal differentiation. Expression ceases only after the beginning of terminal differentiation, since there is substantial co-expression of Cato and Elav, a terminal differentiation marker (Fig. 2A, B). The latest expression appears to mark a specific subset of cells that only partially overlaps with the expression of Ase and Sens (Fig. 2B-D). This later expression is recapitulated by a GFP reporter gene construct driven by the genomic region upstream of cato, which contains enhancers for the entire cato expression pattern (see below; S. Cachero, PzL, APJ, submitted). Between stages 10 and 14, GFP expression reflects that of endogenous Cato (except for the delay associated with GFP maturation). In Ch lineages, GFP perdures strongly into neuron, scolopale and ligament cells, but only weakly in cap and attachment cells (Fig. 2E-H). This suggests that whilst cato expression begins in the Ch SOPs, it is subsequently maintained preferentially in daughter pIIb and then pIIIb - the intermediate precursors leading to the neuronal branch of the sense organ lineage [20]. Expression also perdures in the dbd neuron and its glial sib, in one of the v'td neurons, and in a possible peripheral glia cell (Fig. 2E-H). In ES organs, GFP is only weakly expressed in general. However, it perdures strongest in most of the multidendritic (md) neurons that derive from ES lineages (in addition to those derived from Ch lineages) (Fig. 2E-H). This suggests that the late expression of Cato protein noted above is mostly associated with Ch and md neurons.
Figure 2.
Cato protein expression relative to late neural markers. Immunohistochemistry with anti-Cato (green) and a second marker antibody (magenta). (A) Stage 14 embryo showing substantial co-expression of Cato and Elav, a neuronal differentiation marker. (B) Larger magnification view of similar embryo showing dorsolateral regions of several abdominal segments. (C) Stage 13 embryo, showing that Cato expression generally persists longer than Ase in most cells. (D) Ventrolateral view of stage 14 embryo, showing Cato expression generally more persistent that Sens. (E-G) Expression of the cato1.6k-GFP reporter gene. (E, E') Stage 16 embryo, dorsolateral abdominal segments, showing GFP expression (green) relative to Cpo, which marks nuclei of all PNS cells (blue) and 22C10, which marks sensory neurons (red). (F) Ventrolateral view of a similar embryo. (G) Stage 16 embryo showing expression of GFP relative to Elav, ventrolateral view of abdominal segments. (H) Schematic representation of sensory neurons (filled) and support cells (unfilled) of an abdominal segment (based on [44]): Neurons are: Ch (green), ES (blue); md (red). Abbreviations: v = ventral; v' = ventral'; l = lateral; d = dorsal; ch = chordotonal; es = external sensory; da = dendritic arborisation neuron; bd = bipolar dendritic neuron; td = tracheal dendritic neuron; md = multidendritic neuron; n = neuron; sc = scolopale cell; lg = ligament cell; ca = -cap cell; g = glial cell.
Evidence that Cato is directly regulated by Ato and Amos but only indirectly regulated by Sc
Cato is expressed in ato- and amos-dependent SOPs, where its expression overlaps that of Ato and Amos (Fig. 1A-D and data not shown). In contrast, Cato is not expressed until later in the progeny of Sc-dependent ES SOPs. Sc expression in ES SOPs is transient: it is downregulated before SOP division [21]. Cato expression does not begin until after this time. It therefore appears that Cato expression does not overlap with that of Sc in the ES lineage. Consistent with this, at stage 11 ES cells express the direct Sc targets, ase and sens, but not cato (Fig. 1F-H). Moreover, Cato expression does not overlap with the expression of the proneural gene, Ac, which is thought to be coexpressed with Sc [22] (Fig. 1J). Therefore, as a proneural target gene, cato may be directly regulated by Ato and Amos but only indirectly regulated by Sc.
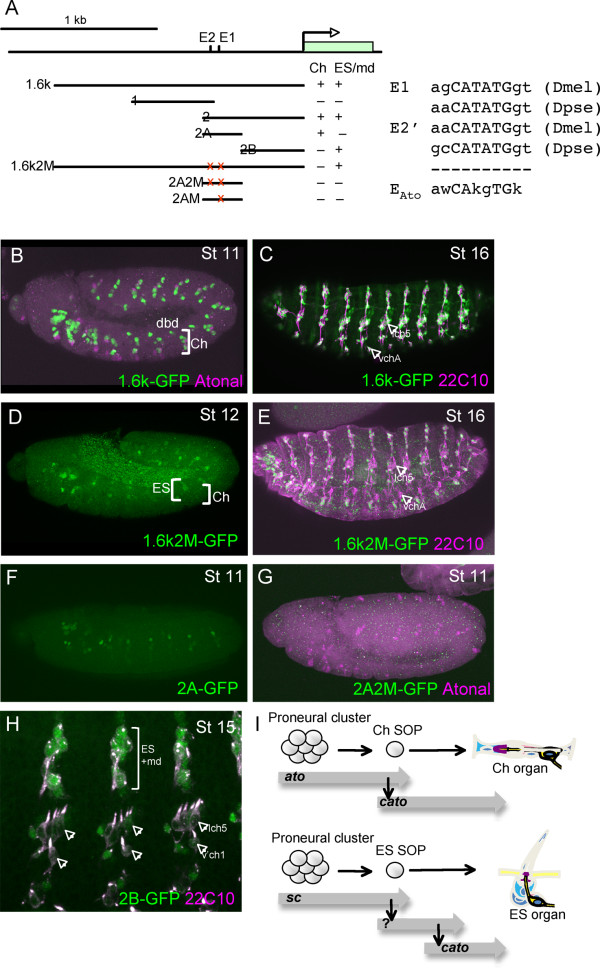
To investigate this, we identified the cis-regulatory elements of cato (Fig. 3A). A 1.6-kb genomic DNA fragment upstream of the cato transcription unit supports GFP reporter expression in a pattern closely resembling that of the endogenous gene. That is, expression is observed initially in Ch cells and later in all PNS cells (Fig. 3B, C). Examination of the sequence of the 1.6-kb region revealed the presence of several E box motifs that resemble the previously identified Ato-specific DNA binding site (EAto)[23](Fig. 3A). When two of these EAto boxes were mutated, the early expression of GFP in Ch cells was abolished but later ES/md cell expression was unaffected (Fig. 3D). Subsequently, we found that the 1.6-kb fragment could be subdivided into at least two enhancers, one that supports early expression in ato- and amos-dependent cells (cato2A; Fig. 3E, F) and one that shows late expression in ES/md cells (cato2B; Fig. 3H). The former contains the two EAto sites identified above. Mutation of both sites or of the E1 site alone in cato2A resulted in loss of GFP expression in Ch cells (Fig. 3G). It is notable that the E1 site is well conserved in Drosophila pseudoobscura (in fact it is an even closer match to the EAto consensus) whereas the E2 site differs markedly in the nucleotides flanking the core E box (Fig. 3A). Thus, it is highly likely that Ato directly regulates cato via the E1 binding site in the 2A enhancer. In contrast, cato is activated only indirectly by Sc in ES lineages. The identity of the factor(s) directly activating cato in ES lineages remains unknown, but it would presumably bind to the 2B enhancer (Fig. 3I).
Figure 3.
Cato has separate Ch and ES/md enhancers. (A) Schematic showing the upstream region of cato, the fragments tested by GFP reporter gene analysis, and a summary of the expression patterns supported (Ch or ES/md). The positions of the two E boxes mentioned in the text are shown, as are their sequences in D. melanogaster and pseudoobscura compared to the known Ato consensus binding site. (B-H) GFP expression in embryos containing different cato reporter genes (green). (B)cato1.6k-GFP in early stage embryo showing early expression in Ch and dbd cells. (C) cato1.6k-GFP in late stage embryo showing expression in all Ch lineages (arrows) and da neurons (costained with 22C10 in magenta). (D) cato1.6k2 M-GFP in stage 12 embryo showing expression just beginning in some ES cells but loss of expression in Ch lineages. (E) cato1.6k2 M-GFP in late stage embryo showing expression in da neurons but loss of expression in Ch lineages (arrows). (F) cato2A-GFP in early embryo showing early expression in Ch precursors (G) cato2A2 M-GFP expression in early embryo, showing lack of Ch expression. (H) cato2B-GFP in late embryo showing expression non-Ch lineages but not in Ch lineages (arrows). (I) Summary of expression pattern and reporter gene evidence that suggests that cato is a direct target of Ato (and Amos), but an indirect target of Sc.
The early Ch enhancer is also expressed in the amos-dependent dbd and dmd1 cells (Fig. 3B). These cells express both Amos and Ato, with the expression of Ato dependent on that of Amos [16]. Thus, cato could be a direct target of Amos, or an indirect target via activation of Ato. We find, however, that cato expression is not affected in these cells by the loss of ato function (Fig. 1E). In contrast, this expression is absent in amos mutant embryos (data not shown). Moreover, expression in amos-dependent cells is also absent when the E1 site is mutated (Fig. 3G). We conclude that cato is likely to be directly activated by both Ato (in Ch cells) and Amos (in dbd and dmd1 cells) via the same E box binding site (E1). This is the first identified direct target gene of Amos.
cato mutation does not cause lethality
The fly stock, KG07568, contains a P-element insertion about 1.5 kb upstream of cato (Fig. 4A). To generate cato mutations, we mobilised the P-element to create imprecise excisions extending in the direction of the cato gene. Two deletions were generated named cato536 and cato513 . cato513 is a protein null: the deletion removes the cato ORF and no Cato expression is apparent in homozygous cato513 embryos (Fig. 4B, C). cato536 deletes the cato upstream region but does not affect the transcription unit. Homozygous embryos show very little protein expression, particularly in the trunk (Fig. 4D, E). This is consistent with the loss of the major regulatory elements upstream of cato. Both mutations are viable, with little sign of gross defects. Adult flies appear normal and active. Thus, cato does not appear to play an essential role in gross neurogenesis.
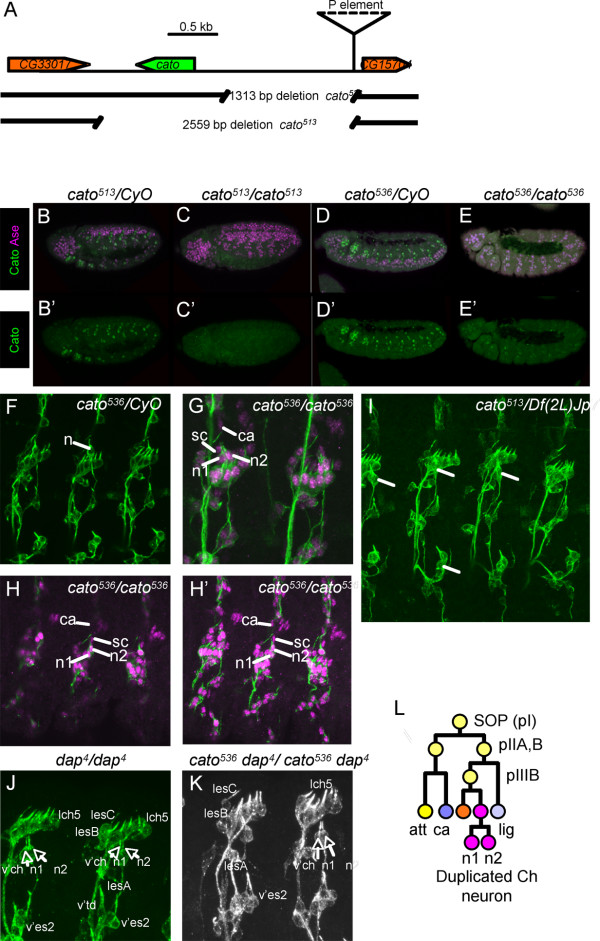
Figure 4.
Mutation of cato results in duplicated v'ch1 neurons. (A) Schematic of cato genomic region showing the location of the P-element and excision deletions. (B-E) Expression of Cato protein (green) is absent or strongly reduced in cato mutant embryos (Ase expression shown in magenta) (B, B') Heterozygous cato513/CyO embryo. (C, C') Homozygous cato513/cato513 embryo. (D, D') Heterozygous cato536/CyO embryo. (E, E') Homozygous cato536/cato536 embryo. (F-K) Late stage embryos stained with 22C10 (green) for sensory neurons and anti-Cpo (magenta) for nuclei of neurons and support cells. Cells of the v'ch1 chordotonal organ are indicated: n = duplicated v'ch1 neurons; sc = unduplicated scolopale cells; ca = unduplicated cap cells. (F) Heterozygous cato536/CyO embryo, showing wild-type 22C10 pattern. (G, H) Homozygous cato536/cato536 embryos, showing duplicated v'ch1 neurons (n1, n2). In (H) several confocal sections are shown. (I) Hemizygous cato513/Df(2L)Jp7 embryo, showing duplicated v'ch1 neurons. (J) Homozygous dap4/dap4 embryo stained with 22C10, showing duplicated v'ch1 neurons. Selected other neurons are labelled, and show no obvious duplication. (K) Double homozygous dap4cato536/dap4cato536 embryos stained with 22C10, showing similar phenotype to each single mutant in which v'ch neurons are duplicated but not adjacent neurons. (L) Schematic summary of the proposed neuronal duplication phenotype.
cato mutant embryos exhibit duplicated chordotonal neurons
Embryos homozygous for either cato mutation show a near normal arrangement of sensory neurons and support cells, as judged by MAb22C10 and anti-Cpo staining (Fig. 4 and data not shown). There is no clear evidence of differentiation or axon defects, in contrast to the phenotypes proposed previously based on the examination of chromosomal deficiencies [7]. A defect was observed, however, in the number of Ch neurons: embryos homozygous for either cato mutation exhibit frequent duplications of Ch neurons (Fig. 4F-I). For cato536 mutant embryos, the v'ch1 neuron is duplicated in 84.5% of abdominal segments (n = 110). Of these, very few (1.8%) seem to be associated with duplicated scolopale cells with the remainder having a single scolopale cell (Fig. 4G, H). Cap cells are more difficult to identify with certainty, but in at least the majority of cases a single cap cell could clearly be associated with the duplicated neuron (Fig. 4G, H). Given this general lack of change in support cell numbers, we conclude that the duplicated neuron most likely results from a defect in cell division number rather than a disruption of asymmetric cell division or an increase in SOP formation (Fig. 4L). Ch neuron duplication is also seen for the vchA and vchB neurons, but only rarely for the lch5 neurons. Similar levels of neuronal duplication were observed in cato513 mutant embryos, and the phenotype is present in embryos expressing a cato RNAi construct (PzL, data not shown), thus ruling out the possibility that it results from an effect of the deletion on the adjacent gene, CG15704.
We explored this neuronal duplication defect by looking for interaction with other genes involved in regulating the number of cell divisions that PNS neural precursors undergo. Cyclin A is known to be required for SOP cell divisions and it has been proposed that proneural factors regulate its expression by binding to an element in its first intron [24]. Loss of one copy of the cyclin A gene resulted in suppression of Ch neuronal duplication in cato homozygote embryos (55% of segments with duplicated v'ch1 neurons, n = 42). This finding supports the conclusion that cato mutants exhibit a specific proliferation defect in Ch neuronal lineages. dap encodes a CDK inhibitor that is required in the epidermis and CNS to terminate cell divisions at the appropriate time [12-14]. In dap mutants, cells typically undergo one extra round of division. We found that this is true for the PNS too: extra sensory neurons are present in dap4 homozygous mutant embryos (Fig. 4J). Interestingly, not all sensory neurons are equally affected: the dap mutant phenotype closely resembles that of cato in that duplication appears most apparent for Ch neurons. In addition, dap/cato double homozygotes do not appear greatly different from either single mutant (Fig. 4K), suggesting that both genes function in the same pathway.
cato expression was shown above to persist particularly in Type II md neurons. These do not appear duplicated. Most of these are dendritic arborisation (da) neurons that have elaborately branched dendrites extending beneath the larval cuticle. We investigated whether differentiated da neurons showed morphological defects in cato mutants by examining dendrite branching patterns in third instar larvae. Da neuronal dendrites were detected using 22C10 (staining all da neuron dendrites) or Gal4447/UAS-mCD8-GFP (a class IV da neuron marker [25]). However, analysis showed no clear defects in the pattern or extent of branching of da neuron dendrites (data not shown).
Redundancy between cato and ato in C1 precursor development
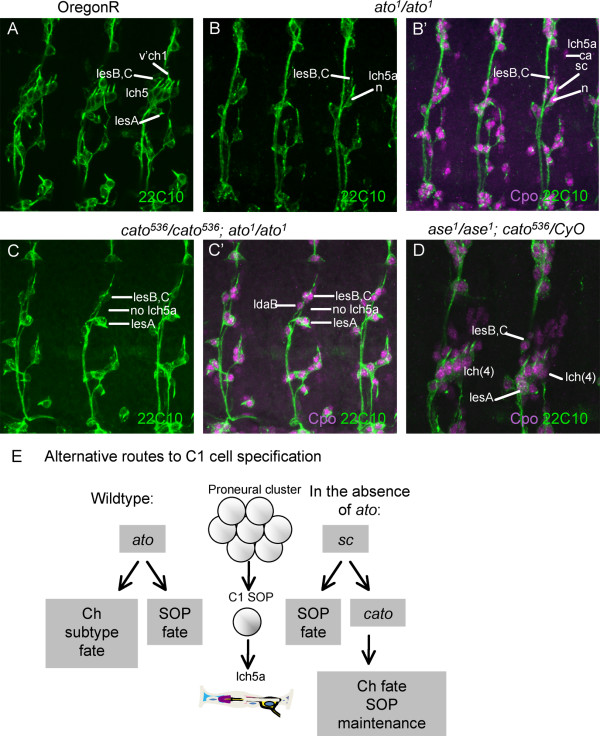
In ato mutant embryos almost all Ch neurons are missing, but one neuron of the abdominal lch5 cluster usually still develops [19]. This is the anterior-most neuron of the cluster (lch5a) and it arises from C1, the first Ch SOP to appear. cato expression in C1 persists in ato mutant embryos (Fig. 1E). Therefore, although cato mutants do not exhibit loss of lch5 neurons (Fig. 4B, C), we speculated that cato might be responsible for C1 formation in the absence of ato function. To test this, we determined whether the lch5a neurons that remain in ato mutant embryos require cato function. In ato1 homozygotes, 90.6% of abdominal segments contained an lch5 neuron (n = 53 segments) (Fig. 5A, B). In cato536; ato1 double homozygotes, however, the presence of this neuron is strongly reduced (Fig. 5C): only 5% of segments contained a possible lch5 neuron (n = 40 segments). Thus, C1 (and the resulting lch5a neuron) can develop when either ato or cato are functional, suggesting at least some redundancy in their function for this cell.
Figure 5.
Embryonic phenotype of double mutants. (A-D) Abdominal segments from late stage embryos stained with 22C10 (green) and anti-Cpo (magenta in B', C', D). (A) OregonR (wild type) embryo with lateral Ch and ES neurons indicated. (B, B') Homozygous ato1 mutant embryo, with neuron (B, B'), scolopale and cap cells (B, B') of lch5a indicated, as well as adjacent ES neurons. (C.C') Double homozygous cato536; ato1 mutant embryo, with ES neurons visible but no lch5a organ. (D) Embryo homozygous for ase1 and heterozygous for cato536, with lch5 Ch clusters reduced from five to four Ch organs. (E) Model for the dependence of C1 cell development on cato, ato and sc. See text for details.
ase and cato cooperate in lch5 development
Like cato, ase is expressed after SOP selection. ase has roles downstream of SOP selection, but its phenotype in sensory lineages is mild considering its wide expression pattern[10]. One phenotype observed in ase1 mutant embryos at a low frequency is the loss of one chordotonal organ from the lateral cluster of five organs (lch5>lch(4); 7% segments showing defect; n = 100 segments). We observed that the frequency of this lch(4) phenotype is greatly enhanced in ase1 mutant embryos with one copy of cato536 (28% segments showing defect (n = 100)) (Fig. 5D). Since support cells are missing in addition to the neuron, it appears that this phenotype results from an early defect in some Ch SOPs and is not linked to the later neuronal duplication defect of cato described above.
Discussion
Although cato is a PNS-specific gene, its expression and function appear to be different in distinct lineages of the PNS. Its expression begins in Ch precursors just after their formation, but appears much later in ES lineages. Correlating with this pattern, we found that cato is directly regulated by ato in Ch SOPs but it is not a direct target of Sc in ES SOPs. This expression pattern, and its underlying regulation, appears to be characteristic of a number of genes, including the transcription factor Rfx and a number of its targets (S. Cachero, PzL, APJ, submitted). We refer to the pattern as 'Ch-enriched' and suggest that such genes mediate part of Ato's subtype specificity in neurogenesis. Interestingly, in different sensory lineages, it seems that cato is regulated by Amos and Ato through the same E box binding site.
The functions we have characterised for cato relate to its major site of expression: the Ch organs. The most obvious defect in cato mutant embryos involves supernumerary cell divisions in the neuronal branch of Ch lineages. This is reminiscent of the known roles of the other non-proneural bHLH proteins, dpn and ase. Thus, in the larval optic lobe, dpn expression maintains proliferation, whilst ase promotes cell cycle exit and neuronal differentiation [14]. The function of cato and ase in limiting cell division resembles the well-known function of vertebrate proneural-like bHLH factors in promoting the cell cycle exit of neuronal progenitors as a prelude to differentiation. This is opposed by HES factors (homologous to dpn), which maintain proliferation [1].
In the case of the larval optic lobe, ase functions in part via the CDK inhibitor, dap [14]. dap itself is generally required for cells to terminate cell division appropriately and cells generally undergo one extra division in dap mutants [12,13]. dap expression is highly dynamic in embryos [26], and it appears that a pulse of dap expression helps to ensure the timely shut down of cyclin function for appropriate cell cycle exit. We show here that dap is similarly required for Ch neurons. Moreover, the PNS phenotype of dap mutant embryos is strikingly similar to that of cato. This suggests that cato regulates dap in Ch neurons. Genetic analysis suggests this might be so, but we see no clear change in dap expression in cato mutant embryos (unpublished data). However, the complex and highly dynamic expression of dap may make small lineage-specific changes in expression difficult to detect. The idea that cato might regulate dap is consistent with previous observations that dap is under the control of multiple developmental regulators rather than of cell cycle regulators themselves [26,27], and also that dap is regulated by Ato in the developing eye [28]. dap is one of several cell cycle regulators (cyclin E [29] and string [30]) that have complex modular cis-regulatory regions. It is notable that cato appears to regulate only the division of the neuron and not support cells. We speculate that this division may require independent regulation from those of the support cells, because the number of neurons within a Ch organ varies in different locations, presumably as a result of extra neuronal cell divisions. For instance, some Ch organs in the adult femur have two neurons, whilst Ch organs in the antenna have three neurons [31].
The other functions detected for cato appear to be unrelated to the neuronal duplication function and show at least some redundancy with other bHLH regulators (ato and ase). In both these cases we suggest that cato plays a partially redundant role in maintaining SOP fate. In the absence of ase and cato, some Ch SOPs fail to form scolopidia. A similar situation applies to C1 in the absence of ato and cato. The apparent redundancy between ato and cato suggests that C1 SOP can form via alternative routes involving ato and cato (Fig. 5E). However, cato is expressed too late to be a proneural gene, and so another factor must supply the proneural function in the absence of ato. It seems likely that this factor is sc, which is expressed in C1 despite being the ES proneural gene [32]. Embryos with a mutation of the achaete-scute complex often show one missing scolopidium in the lch5 cluster [33], while AS-C/ato mutant embryos have no Ch cells at all. Such interchangeability of proneural functions between ato and sc is surprising since sc does not generally have the capacity to direct Ch subtype specification, as shown in misexpression experiments [3,34]. In contrast, ato's subtype specificity function is reflected in its ability to convert ES SOPs to a Ch fate [35]. We suggest that expression of cato in a sc-dependent C1 cell may provide sufficient subtype determination information when ato is absent. It is not clear why such a complicated exception should have arisen. One possibility is that C1 forms a unique neuronal type among Ch organs. Certainly there are a number of genes that are only expressed in, or are only absent from, this one neuron. For instance, MAb49C4 detects an antigen that is expressed in all lch5 neurons except lch5a [36]. Moreover, C1 appears to be functionally unique in that it acts to induce surrounding cells to differentiate as oenocytes via EGFR signalling [37,38]. This function of C1 appears to be 'rescued' by cato function, since the C1 cells present in ato mutants are able to recruit oenocytes [37].
Expression of Cato in ES lineages appears to be mainly as a prelude to late expression in the md/da neurons that derive from both ES and Ch lineages. As yet, no function has been discerned for this late expression, but we speculate that cato mutant larvae may exhibit a physiological defect in da neurons, which are thought to be required for nociception and thermoreception [39].
Conclusions
Characterisation of the first mutations for cato has revealed roles in maintenance and cell division in Ch lineages. These roles are relatively subtle considering that cato is expressed widely in the developing PNS. Moreover, cato orthologues can be readily recognised among Drosophila species and other Diptera (unpublished observations), suggesting strong conservation. It is possible that further functions remain to be uncovered, perhaps in da neuron physiology or in the complex cephalic sense organs.
Methods
Fly stocks
Fly stocks used were y1 w67c23; P{SUPor-P}KG07568 [40], ato1 [41], w*; wgSp1/CyO; ry506 Dr1 P{Δ2-3}99B/TM6, Ubx (Bloomington Stock Centre). They cyclinA stock was: w*; CycAC8LR1/TM3, Sb1P{35UZ}2 and the dacapo stock was dap4/CyO, P{ftz/lacB}E3 (both from Bloomington Stock Centre).
Generation of anti-Cato antibodies
The Cato reading frame was subdivided and the two fragments were each cloned into the pGEX-2T expression vector. The reading frame for the first 100 amino acids, omitting the start codon, was amplified by PCR using the primers 5'-ATCGGATCCTACTACTCGTCTGCC-3' and 5'-GCGCGAATTCCGCTCAATCCAAATCC-3' (added restriction sites are/underlined). The primers for the remaining 89 amino acids were 5'-GGACGGATCCCAGAAAAGGAGACGAC-3' and 5'-GTCAGAATTCCTGGACCGTGGGACTG-3'. Expression of GST fusion proteins was induced in BL21(pLysS) cells using 0.25 mM IPTG. After glutathione affinity purification, proteins were used to raise anti-Cato antibodies in sheep.
Immunohistochemistry
Embryo antibody staining was carried out according to standard procedures. Primary antibodies used were sheep anti-Cato antibody (1:1000), mouse anti-22C10 and mouse anti-Elav (both 1:200; Developmental Biology Hybridoma Bank, Iowa City, Iowa), mouse and rabbit anti-GFP (1:500; Molecular probes, Invitrogen), rabbit anti-Cpo (1:500; [42]), guinea pig anti-Sens (1: 6250; [17] and rabbit anti-Ato (1:4000; [3]. The secondary antibodies (1:500) were from Molecular Probes (Invitrogen). Confocal microscopy was carried out using a Zeiss PASCAL and a Leica TCS SP2 microscope.
P element imprecise excision
The P element insertion P{SUPor-P}KG07568 [40] was mobilised to create imprecise excisions that remove DNA between the insertion site and the cato gene. The KG07568 stock was crossed to a stock carrying transposase (D2-3). Male progeny containing the two elements were then mated individually to females from a CyO balancer stock. After mating, DNA was extracted from each male. DNA from pools of ten males was screened by PCR for deletions. If a shortened PCR product was detected, individual flies from that pool were screened further in individual PCR reactions. The PCR product was subsequently sequenced in order to establish the extent of the deletion. Deletion cato513 was detected using primers P1 (GCTATCTATCGATGTGTAAGC) and 18R (TGTTATGTCCTC) and the smaller deletion, cato536, was detected using primer P1 and eCato2 (TTCACCGCCGTTCTGACC). These indicated deletions of 2559 bp and 1313 bp respectively.
Reporter plasmid constructs
PCR amplified fragments were cloned into the pHStinger vector [43]. Genomic DNA fragments were amplified by PCR, and GFP reporter gene constructs were made in the transformation vector, pHStinger. These were used to make transgenic flies by microinjection into syncytial blastoderm embryos. In general, at least two transgenic lines were examined for each construct. Primers used were: cato1.6k: 5'-GCTGTATCAGGACACGAAGCTCC-3' and 5'-TTCACCGCCGTTCTGACC-3'. cato1: 5'-GTGGAGAAGTATTTGTCAG-3' and 5'-CTGCACCGACCCGACTTTG-3'. cato2: 5'-TCCAGGACCAAAGGC-3' and 5'-TCATTGCAGATCCGAGCG-3'. cato2A: 5'-GACTTTCACGCTCAACG-3' and 5'-TCATTGCAGATCCGAGCG-3'. cato2B: 5'-TCCAGGACCAAAGGC-3' and 5'-GTTGAGCGTGAAAGTC-3'. For site directed mutagenesis of E box sequence motifs, mutagenesis was carried out using the Quik-Change Site-Directed Mutagenesis Kit (Stratagene). Mutations induced were: E1: aacatatgg changed to aaaatattg; E2: agcatatgg changed to agaatattg.
Authors' contributions
PzL co-designed and carried out the experiments and helped analyse the data. APJ co designed the experiments, analysed the data and wrote the paper. Both authors read and approved the final manuscript.
Contributor Information
Petra I zur Lage, Email: petra.zurlage@ed.ac.uk.
Andrew P Jarman, Email: andrew.jarman@ed.ac.uk.
Acknowledgements
We thank Hugo Bellen for antibodies and members of the lab for discussions. This work was supported by a grant from The Wellcome Trust (077266).
References
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Quan X-J, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci. 2005;62:2036–2049. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-W. [DOI] [PubMed] [Google Scholar]
- Lee JE. NeuroD and neurogenesis. Developmental Neuroscience. 1997;19:27–32. doi: 10.1159/000111182. [DOI] [PubMed] [Google Scholar]
- Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter J-M. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- Brand M, Jarman AP, Jan LY, Jan YN. asense is a Drosophila neural precursor gene and is capable of initiating sense organ formation. Development. 1993;119:1–17. doi: 10.1242/dev.119.Supplement.1. [DOI] [PubMed] [Google Scholar]
- Goulding SE, White NM, Jarman AP. cato encodes a basic-helix-loop-helix transcription factor implicated in the correct differentiation of Drosophila sense organs. Developmental Biology. 2000;221:120–131. doi: 10.1006/dbio.2000.9677. [DOI] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Younger-Shepherd S, Jan LY, Jan YN. deadpan, an essential pan-neural gene in Drosophila encodes a helix-loop-helix protein similar to the hairy gene product. Genes and Development. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- Ledent V, Gaillard F, Gautier P, Ghysen A, Dambly-Chaudière C. Expression and function of tap in the gustatory and olfactory organs of Drosophila. International Journal of Developmental Biology. 1998;42:747–750. [PubMed] [Google Scholar]
- Jarman AP, Brand M, Jan LY, Jan YN. The regulation and function of the helix-loop-helix gene, asense, in Drosophila neural precursors. Development. 1993;119:19–29. doi: 10.1242/dev.119.Supplement.19. [DOI] [PubMed] [Google Scholar]
- Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2:1. doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane ME, Sauer K, Wallace K, Jan YN, Lehner CF, Vaessin H. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell. 1996;87:1225–1235. doi: 10.1016/S0092-8674(00)81818-8. [DOI] [PubMed] [Google Scholar]
- de Nooij JC, Letendre MA, Hariharan IK. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell. 1996;87:1237–1247. doi: 10.1016/S0092-8674(00)81819-X. [DOI] [PubMed] [Google Scholar]
- Wallace K, Liu TH, Vaessin H. The pan-neural bHLH proteins DEADPAN and ASENSE regulate mitotic activity and cdk inhibitor dacapo expression in the Drosophila larval optic lobes. Genesis. 2000;26:77–85. doi: 10.1002/(SICI)1526-968X(200001)26:1<77::AID-GENE10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- zur Lage PI, Prentice DRA, Holohan EE, Jarman AP. The Drosophila proneural gene amos promotes olfactory sensillum formation and suppresses bristle formation. Development. 2003;130:4683–4689. doi: 10.1242/dev.00680. [DOI] [PubMed] [Google Scholar]
- Holohan EE, zur Lage PI, Jarman AP. Multiple enhancers contribute to spatial but not temporal complexity in the expression of the proneural gene, amos. BMC Developmental Biology. 2006;6:53. doi: 10.1186/1471-213X-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–362. doi: 10.1016/S0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- zur Lage P, Jan YN, Jarman AP. Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Current Biology. 1997;7:166–175. doi: 10.1016/S0960-9822(97)70087-3. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Sun Y, Jan LY, Jan YN. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030. doi: 10.1242/dev.121.7.2019. [DOI] [PubMed] [Google Scholar]
- Lai EC, Orgogozo V. A hidden program in Drosophila peripheral neurogenesis revealed: fundamental principles underlying sensory organ diversity. Dev Biol. 2004;269:1–17. doi: 10.1016/j.ydbio.2004.01.032. [DOI] [PubMed] [Google Scholar]
- Chang P-J, Hsiao Y-L, Tien A-C, Li Y-C, Pi H. Negative-feedback regulation of proneural proteins controls the timing of neural precursor division. Development. 2008;135:3021–3030. doi: 10.1242/dev.021923. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Rodriguez I, Martinez C, Culi J, FerresMarco D, Beamonte D, Modolell J. Cis-regulation of achaete and scute: Shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes Dev. 1995;9:1869–1882. doi: 10.1101/gad.9.15.1869. [DOI] [PubMed] [Google Scholar]
- Powell LM, zur Lage PI, Prentice DRA, Senthinathan B, Jarman AP. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol Cell Biol. 2004;24:9517–9526. doi: 10.1128/MCB.24.21.9517-9526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda R, Togashi S, Takahisa M, Tsurumura S, Mikuni M, Kondo K, Miyake T. Sensory mother cell division is specifically affected in a Cyclin-A mutant of Drosophila melanogaster. EMBO J. 1992;11:2935–2939. doi: 10.1002/j.1460-2075.1992.tb05363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/S0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Liu TH, Li L, Vaessin H. Transcription of the Drosophila CKI gene dacapo is regulated by a modular array of cis-regulatory sequences. Mech Dev. 2002;112:25–36. doi: 10.1016/S0925-4773(01)00626-8. [DOI] [PubMed] [Google Scholar]
- Meyer CA, Kramer I, Dittrich R, Marzodko S, Emmerich J, Lehner CF. Drosophila p27Dacapo expression during embryogenesis is controlled by a complex regulatory region independent of cell cycle progression. Development. 2002;129:319–328. doi: 10.1242/dev.129.2.319. [DOI] [PubMed] [Google Scholar]
- Sukhanova MJ, Deb DK, Gordon GM, Matakatsu MT, Du W. Proneural basic helix-loop-helix proteins and epidermal growth factor receptor signaling coordinately regulate cell type specification and cdk inhibitor expression during development. Mol Cell Biol. 2007;27:2987–2996. doi: 10.1128/MCB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Richardson H, Saint R. Tissue-specific regulation of cyclin E transcription during Drosophila melanogaster embryogenesis. Development. 2000;127:4619–4630. doi: 10.1242/dev.127.21.4619. [DOI] [PubMed] [Google Scholar]
- Lehman DA, Patterson B, Johnston LA, Balzer T, Britton JS, Saint R, Edgar BA. Cis-regulatory elements of the mitotic regulator, string/Cdc25. Development. 1999;126:1793–1803. doi: 10.1242/dev.126.9.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl DF. Feeling the vibes: chordotonal mechanisms in insect hearing. Current Opinion in Neurobiology. 1999;9:389–393. doi: 10.1016/S0959-4388(99)80058-0. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Brand M, Jan LY, Jan YN. daughterless is essential for neuronal precursor differentiation but not for initiation of neuronal precursor formation in Drosophila embryo. Development. 1994;120:935–945. doi: 10.1242/dev.120.4.935. [DOI] [PubMed] [Google Scholar]
- Chien C-T, Hsiao C-D, Jan LY, Jan YN. Neuronal type information encoded in the basic-helix-loop-helix domain of proneural genes. Proceedings of the National Academy of Sciences, USA. 1996;93:13239–13244. doi: 10.1073/pnas.93.23.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maung SM, Jarman AP. Functional distinctness of closely related transcription factors: a comparison of the Atonal and Amos proneural factors. Mech Dev. 2007;124:647–656. doi: 10.1016/j.mod.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Ahmed I. The specificity of proneural genes in determining Drosophila sense organ identity. Mech Dev. 1998;76:117–125. doi: 10.1016/S0925-4773(98)00116-6. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Barbel S, Shepherd S, Jack JW, Jan LY, Jan YN. Transformation of sensory organs by mutations of the cut locus of D. melanogaster. Cell. 1987;51:293–307. doi: 10.1016/0092-8674(87)90156-5. [DOI] [PubMed] [Google Scholar]
- Elstob PR, Brodu V, Gould AP. spalt -dependent switching between two cell fates that are induced by the Drosophila EGF receptor. Development. 2001;128:723–732. doi: 10.1242/dev.128.5.723. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Cantera R, Urban J, Technau G, Kafatos FC, Barrio R. spalt modifies EGFR-mediated induction of chordotonal precursors in the embryonic PNS of Drosophila promoting the development of oenocytes. Development. 2001;128:711–722. doi: 10.1242/dev.128.5.711. [DOI] [PubMed] [Google Scholar]
- Liu L, Yermolaieva O, Johnson WA, Abboud FM, Welsh MJ. Identification and function of thermosensory neurons in Drosophila larvae. Nature Neuroscience. 2003;6:267–273. doi: 10.1038/nn1009. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, Evans-Holm M, Hiesinger PR, Schulze KL, Rubin GM. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Bellen HJ, Kooyer S, D'Evelyn D, Pearlman J. The Drosophila couch potato protein is expressed in nuclei of peripheral neuronal precursors and shows homology to RNA-binding proteins. Genes and Development. 1992;6:2125–2136. doi: 10.1101/gad.6.11.2125. [DOI] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Orgogozo V, Grueber WB. FlyPNS, a database of the Drosophila embryonic and larval peripheral nervous system. BMC Dev Biol. 2005;5:4. doi: 10.1186/1471-213X-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]