SUMMARY
Meiotic chromosome pairing involves not only recognition of homology but also juxtaposition of entire chromosomes in a topologically regular way. Analysis of filamentous fungus Sordaria macrospora reveals that recombination proteins Mer3, Msh4 and Mlh1 play direct roles in all of these aspects, in advance of their known roles in recombination. Absence of Mer3 helicase results in interwoven chromosomes, thereby revealing the existence of features that specifically ensure “entanglement avoidance”. Entanglements that remain at zygotene, i.e. “interlockings”, require Mlh1 for resolution, likely to eliminate constraining recombinational connections. Patterns of Mer3 and Msh4 foci along aligned chromosomes show that the double-strand breaks mediating homologous alignment have spatially separated ends, one localized to each partner axis, and that pairing involves interference among developing interhomolog interactions. We propose that Mer3, Msh4 and Mlh1 execute all of these roles during pairing by modulating the state of nascent double-strand break/partner DNA contacts within axis-associated recombination complexes.
Keywords: Mer3, Msh4, Mlh1, meiosis, pairing/synapsis, bouquet, recombination, interlocking
INTRODUCTION
A central unique feature of meiosis is pairing of homologous chromosomes (“homologs”). Pairing involves recognition of homology plus coming-together of homologs in space. Additionally, juxtaposition must be achieved without high levels of entanglement (Kleckner and Weiner, 1993; von Wettstein et al., 1984; Wang et al., 2009).
In most organisms, pairing involves juxtaposition of homolog structural axes, dependent upon, and mediated via, the biochemical process of DNA recombination. The two aspects are integrated by physical localization of protein/DNA recombination complexes (“recombinosomes”) to their underlying axes (e.g. Henderson and Keeney, 2005; Borner et al., 2004; Moens et al., 2007; Anderson and Stack, 2005; Franklin et al., 2006; Oliver-Bonet et al., 2007; Wang et al., 2009 and references therein). Recombinosome/axis association arises before, or just after, initiation of recombination via programmed double-strand breaks (DSBs) (Blat et al., 2002; Tesse et al., 2003). Thereafter, DNA recombination and recombination-mediated juxtaposition of homolog axes progress in close temporal and functional coordination.
Homologs first become aligned at a distance of ~400nm during leptotene. Except in worm and Drosophila, this process requires DSB formation and is accompanied by appearance of axis-associated foci of RecA homolog(s) (review in Henderson and Keeney, 2005). At the DNA level, alignment is inferred to involve nascent DNA/DNA interactions between a 3′ single-stranded tail at one DSB end and the homologous region on a homolog partner duplex (Hunter and Kleckner, 2001; Hunter, 2006). Then, at zygotene, “synapsis” occurs: axes become progressively linked at a distance of ~100nm by transverse filaments, which, with other proteins, comprise the synaptonemal complex (SC) (review in Page and Hawley, 2004). The leptotene/zygotene transition is also a crucial point for recombination: nascent interactions are differentiated into crossover (CO)-fated and noncrossover (NCO)-fated products, with concomitant onset of stable strand exchange (review in Hunter, 2006). In several organisms, SC formation is nucleated preferentially at sites of CO-designated interactions (Henderson and Keeney, 2005). After full SC has formed, mature CO and NCO products appear at mid-pachytene (e.g. Guillon et al., 2005).
This study analyzes the relationship between recombination and homolog juxtaposition in the filamentous fungus Sordaria macrospora which is a very powerful system for such analysis: continuous axes appear at early leptotene such that spatial relationships among chromosomes, and positions of axis-associated recombination-complexes, can be followed from S-phase onward (e.g. Storlazzi et al., 2003; 2008; Tesse et al., 2003; Zickler, 2006). Additionally, overall progression of meiosis can be followed by progressive increases in the sizes of both the nucleus and its surrounding ascus (meiocyte), thus permitting the detection of temporal delays in the progression of chromosomal events in mutant backgrounds comparatively to wild type (e.g. Storlazzi et al., 2003; 2008).
Interplay between homolog pairing and recombination is probed by analysis of mutants defective in three proteins previously shown to be direct participants in the DNA events of recombination: Mer3, Msh4 and Mlh1. Mer3 is a meiosis-specific 3′-5′ helicase that stimulates Rad51-mediated DNA heteroduplex extension and stabilizes recombinational interactions (Nakagawa and Ogawa, 1999; Nakagawa and Kolodner, 2002; Mazina et al., 2004). Msh4, as part of the Msh4/Msh5 heterodimer, a meiosis-specific homolog of the bacterial mismatch repair protein MutS, encircles and stabilizes branched DNA structures (Snowden et al. 2004). Mlh1/3 are eukaryotic homologs of the bacterial mismatch repair protein MutL and, by analogy, may mediate release of Msh4/5 (Snowden et al., 2004), among other possible roles (Hunter 2006). Mer3 and Msh4 are implicated in ensuring that the fates of CO-designated interactions are faithfully maintained and efficiently implemented during and after the leptotene/zygotene “CO control” transition. In the absence of either gene product, CO formation is specifically abrogated while NCOs form at high levels (e.g. Borner et al., 2004). Mhl1 is implicated at a later stage, in finalization of CO recombinational interactions during pachytene (Hunter, 2006; Franklin et al., 2006). In correspondence with their times of action during recombination, foci of Mer3 and Msh4/5 are observed prior to and during SC formation, whereas foci of Mlh1 are not visible until after completion of synapsis (e.g. de Boer et al., 2006; Oliver-Bonet et al., 2007; Moens et al., 2007; Franklin et al., 2006; Higgins et al., 2004; Jackson et al., 2006, Wang et al., 2009 and references therein).
Our results reveal that each of these three proteins plays a central role in homolog juxtaposition one stage prior its established role in promoting recombination. Mer3 and Msh4 are implicated in leptotene alignment (in advance of the leptotene/zygotene CO control transition) while Mlh1 is implicated in resolution of interlocks at zygotene (in advance of mid-pachytene CO finalization). Mer3 mutant phenotypes suggest the existence of an “entanglement avoidance” process. Further, the patterns and dynamics of Mer3 and Msh4 foci provide new information about the nature of the alignment process and, by implication, the positioning of DSB end(s) during alignment and the CO control transition. Integration of these findings with previous molecular and biochemical information leads to a model for how Mer3 and Msh4 could mediate alignment by stabilizing nascent DSB/partner interactions within axis-associated recombinosomes while Mlh1 mediates interlock resolution by removing Msh4/5 stabilization of these contacts.
RESULTS
Sordaria macrospora Mer3, Msh4 and Mlh1 were cloned and completely null deletion mutations constructed, found to be recessive in heterozygous crosses, and further analyzed. Cytological localization was determined for Msh4-GFP and Mer3-GFP fusion proteins, expressed from their respective promoters at ectopic locations. Both fusion genes complement all meiotic defects of cognate null mutants. See Supplemental Information (SI) for details.
Mer3, Msh4 and Mlh1 Are Not Required for Axis Formation But Are Essential for Normal Pairing, Bouquet Dynamics and Recombination
Chromosomes of the three null alleles were analyzed in leptotene through pachytene nuclei (n = 300 each), by DAPI staining of chromatin and with two previously characterized axis markers: Spo76/Pds5-GFP (van Heemst et al., 1999) and meiosis-specific cohesin Rec8-GFP (Storlazzi et al., 2008). In all three mutants, chromosome axes are indistinguishable from wild-type (WT) axes: Spo76 and Rec8 load at S phase and form complete lines along chromosomes from early leptotene to end pachytene (below). Thus, effects of mutations on spatial relationships can be analyzed without complications from defects in axis development.
All three mutants progress through meiosis and sporulation with the same progressive increase in ascus size as in WT. However, chromosomal progression from leptotene to pachytene is prolonged in all three mutants: by ~10 hrs in mer3Δ or msh4Δ and ~12 hrs in mlh1Δ (24-26 hours instead of 12 in WT). Analogous prolongations occur in Arabidopsis Atmlh3 and Atmsh4 (Higgins et al., 2004; Jackson et al., 2006).
All three mutants exhibit defects in pairing and synapsis (below). They show also defects in the bouquet dynamics. As in WT, chromosome ends transiently cluster in one area of the nuclear periphery at late leptotene and then re-disperse at pachytene onset, but exit from cluster is significantly delayed (Figures S1A-1H).
Recombination is also defective in all three mutants (Figures S2A-2F and details in legends). In mer3Δ and msh4Δ mutants, chiasmata are drastically reduced (4-7) compared to WT (21 ± 3). mlh1Δ confers a milder defect: genetic analysis reveals a ~40% reduction in COs with no reduction in NCOs (see details in legend of Figure S2B). As similar defects were found in other organisms (review in Hunter, 2006), the corresponding proteins likely play the same roles for recombination in Sordaria.
Mlh1 is Required for Interlock Resolution
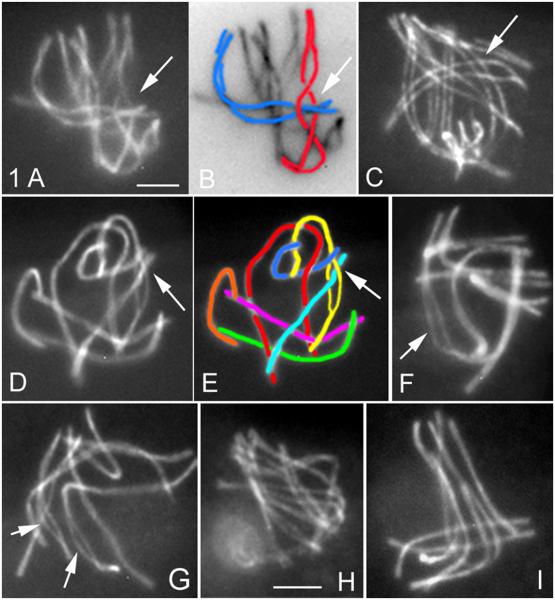
In WT meiosis, unrelated chromosomes are occasionally seen entangled with one another, usually at zygotene. In such “interlocks”, either one chromosome or a pair of chromosomes is located between two aligned homologs, held in place by regular synapsis to either side (Figures 1A and 1B). For Sordaria, interlocked chromosomes are seen in ~20% of WT serially-sectioned zygotene nuclei (n = 121; DZ, unpublished) and in ~15% (n = 300) of nuclei by immunofluorescence using Spo76-GFP as axis marker (Figure 1A). Further, by early pachytene, no interlocks are seen by either method. Thus WT meiosis must include a mechanism for their resolution during zygotene. Resolution of interlocks has been suggested previously (review in von Wettstein et al., 1984; Wang et al., 2009); however, conclusions were often limited by the small number of nuclei examined and by the inability to exclude the possibility that interlocks were absent at pachytene because the corresponding nuclei were absent from the experimental population (e.g. via apoptosis in mammals; reviewed in von Wettstein et al., 1984; Zickler and Kleckner, 1999). In Sordaria, disappearance of interlocks cannot be explained in this way because: (i) there is no apoptosis and (ii) 99% of WT nuclei/asci go on to give eight viable ascospores.
Figure 1. Interlocks in WT and mlh1Δ.
(A-I) Chromosome axes are stained by Spo76-GFP.
(A) WT zygotene bouquet nucleus with a bivalent interlocked in a twisted region of another homolog pair (arrow) plus corresponding cartoon (B).
(C) WT late leptotene nucleus with an aligned homolog pair located between the two homologs of another pair (arrow), thus at risk of being trapped when synapsis (started at ends) elongates.
(D-I) mlh1Δ.
(D) Pachytene nucleus with interlock (arrow) plus (E) corresponding cartoon.
(F and G) Two late pachytene nuclei with either one (arrow in F) or two (arrows in G) bivalents showing non-synapsed regions.
(H) Late leptotene and (I) pachytene nuclei show perfectly synchronous pairing and synapsis. Bars, 2μm.
See S1G and 1H for bouquet formation and S2B for chiasmata.
In Sordaria, potential interlocks are in fact visible prior to onset of synapsis: at late leptotene single chromosomes or aligned pairs are sometimes seen located between another aligned pair of homologs (Figure 1C), thus at high risk of being entrapped at zygotene. Such configurations provide direct evidence that the interlocks defined at zygotene by synaptic patterns reflect the fixation of irregular patterns already present during alignment.
In the absence of Mlh1, in striking contrast to WT, ~45% of all late pachytene nuclei (by ascus size; n = 350) show at least one interlocking (Figures 1D and 1E). Thus, Mlh1 is required for resolution of interlocks. Another ~20% of nuclei present a new configuration, not previously reported, where one or two homolog pairs show non-synapsed regions (variable in length) not involved in interlocks (Figures 1F and 1G). These could represent cases of incomplete resolution and/or left-behind regions that are unable to synapse after resolution. Since Mlh1 is known to be involved in dissociating and/or resolving recombination intermediates (Hunter, 2006), mlh1Δ phenotypes suggest that interlock resolution requires elimination of (cytologically invisible) constraining DNA connections that arise during alignment, with Mlh1 devoted to that role.
Aside from persistence of interlocks, chromosome dynamics in mlh1Δ are morphologically the same as in WT: homologous axes align synchronously, to ~400nm and then ~200nm (Figure 1H), with normal accompanying numbers and patterns of localization for Rad51 foci, which mark the sites of DSBs, and for Mer3 foci (not shown). Importantly, however, while the period of alignment is rather short in WT (10-20% of prophase nuclei show fully aligned homologs), in mlh1Δ, 35% of nuclei (n = 300 for each) show this configuration (Figure 1H). Mlh1 is therefore functionally relevant even prior to its involvement in interlock resolution. The synaptic period is also greatly prolonged in mlh1Δ: while ~10% of WT nuclei are at zygotene, 25% of mutant nuclei exhibit early or intermediate extents of synapsis (n = 300 for each strain). Even so, ultimately, all homologs synchronously synapse (Figure 1I).
Notably, three mlh1Δ phenotypes emerge at zygotene, thus one stage earlier than the defined pachytene role of Mlh1 in recombination (Hunter, 2006): delayed onset of synapsis, delayed bouquet release (above) and persistence of interlocks (resolved during zygotene in WT).
Msh4 is Required to Establish Correct Alignment Distance
Like mlh1Δ, msh4Δ homologs have no problem with initiation or occurrence of presynaptic alignment. An alteration in alignment is nonetheless apparent: msh4Δ homolog axes initially align at a distance of ~600-800nm (Figures 2A and 2B), rather than the ~400nm seen in WT (Figure 2C). This unique phenotype points to direct participation of the recombination complex in determining inter-axis distances. Further, Msh4, like Mlh1, plays a role in homolog pairing at leptotene, thus one stage prior to the time of its defined role in formation of recombinants (at zygotene; Introduction).
Figure 2. Prophase Phenotypes of msh4Δ Compared to WT.
(A-I) All chromosome axes are stained by Spo76-GFP.
(A-C) Mid-leptotene: in msh4Δ homologs are aligned at a greater distance (arrows in A and B) than in WT (arrow in C).
(D) Late leptotene of msh4Δ: one pair of homologs (arrowhead) is aligned at 200nm when others (arrow) remain at 600nm.
(E and F) Contrary to msh4Δ, WT homologs pair (C), start synapsis (E) and synapse (F) synchronously.
(G and H) Synapsis is completely asynchronous in msh4Δ. At zygotene (G) some homologs are half synapsed (arrowhead) while another pair is only synapsed at the telomere region the remainder of the axes remaining widely separated but aligned (arrows). At early pachytene (H) several or, as shown, one pair (arrow) remain(s) unsynapsed.
(I) At late pachytene Rad51-RFP foci (arrow) remain visible on non-synapsed regions of msh4Δ (arrow). Bars, 2μm.
See S1E and S1F for bouquet formation, S2C for chiasmata and Figures S3A-3G for Mer3 foci patterns in msh4Δ.
Once synapsis begins, msh4Δ exhibits also dramatic per-chromosome and per-region defects (Figure 2D). In WT meiosis, all seven chromosome-pairs progress synchronously and efficiently from alignment (Figure 2C) to zygotene (Figure 2E) and synapsis (Figure 2F). In msh4Δ, post-alignment nuclei show a mixture of bivalents at different stages of synapsis (Figures 2G). Finally, 95% of the nuclei that would be in pachytene in a WT ascus of corresponding size still show a mixture of fully and partially synapsed homologs (n = 100; Figure 2H). Since synapsis is tightly coupled to CO-designation and progression during the leptotene/zygotene transition, these defects likely reflect the known role of Msh4 in recombination during this period (Hunter, 2006).
Despite the complexity of synaptic patterns, the dynamics of Rad51 and Mer3 foci remain relatively unperturbed in msh4Δ. Both types of foci appear with WT timing plus location (Figures S3A-3G) and number (respectively 52 ± 8 and 110 ± 9; n = 90). At pachytene (by ascus size) both types of foci remain present in unsynapsed regions while disappearing in synapsed regions in the same nucleus (e.g. Figure 2I for Rad51), as normally seen during synchronous progression through zygotene in WT (below). These results emphasize the tight functional linkage between recombinational progression and pairing/synapsis not only globally, but also locally, on a region-by-region basis.
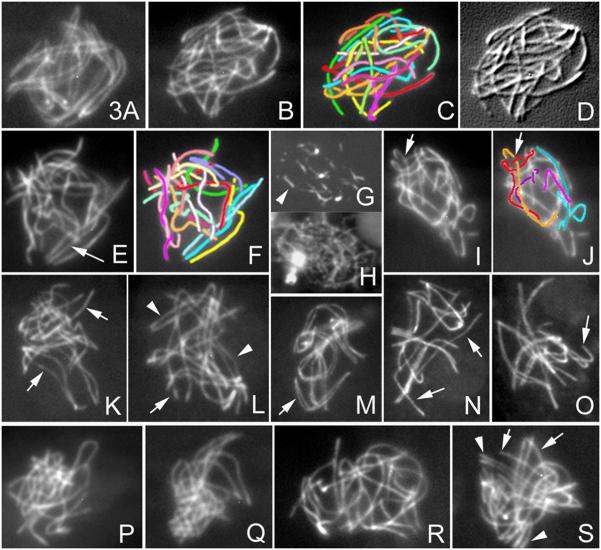
Mer3-dependent Avoidance of Entanglements During Alignment
In WT, at mid-leptotene, all homolog pairs of every nucleus exhibit visible alignment (Figure 3A). At the corresponding ascus size in mer3Δ, chromosomes are still separated and highly “interwoven” (Figure 3B-3D). Further, when alignment does finally occur, at ascus sizes corresponding to zygotene in WT, even more dramatic “interweaving” is seen (Figures 3E and 3F).
Figure 3. Prophase Phenotypes of mer3Δ and mer3 KA Mutants.
(A) In WT mid-leptotene, all homologs are aligned.
(B-D) Example of interwoven chromosomes of mer3Δ at mid-leptotene shown as primary data (B); cartoon (C); and southeast shadowing by Image J (D).
(E) Alignment (arrow) yields even more interweaving; (F) corresponding cartoon.
(G) Rad51 foci form elongated lines (arrowhead) from late leptotene through zygotene (by ascus size); (H) corresponding DAPI.
(I) Telomere regions are often bent and curled around neighbor chromosomes (arrow); (J) Drawing of three pairs sorted by length.
(K and L) Alignment (arrows) and close pairing (arrowheads) are both highly discoordinated in mer3Δ versus WT (M) where all homologs align synchronously (arrow points to one pair).
(N and O) Mixture of synapsing and still unsynapsed (arrows) homologs seen in all late pachytenes of mer3Δ.
(P-S) mer3KA. (P and Q) Late leptotene chromosomes are highly interwoven and entanglements persist at zygotene (R) and pachytene (S) where all nuclei contain a mixture of paired (arrowheads) and widely aligned (arrows) homologs. Bars, 2μm.
See S1C and S1D for bouquet formation and S2D for chiasmata in mer3Δ.
This phenotype reveals that pairing normally includes specific features that preclude or minimize the formation of “interwoven” chromosomes” and, further, that such features require the presence of Mer3. To distinguish this phenotype from interlocks (above), which involve SC formation, we refer to these pre-synaptic configurations as “entanglements”. Occurrence of entanglements is specifically distinct from defects in the process of alignment per se. Sordaria ski8 mutants exhibit reduced levels of DSBs and reduced levels of alignment but do not show entanglements (Tesse et al., 2003). Moreover, in mer3Δ, topological aberrancy is observed in conjunction with a preceding delay in onset of alignment, raising the possibility of a cause-and-effect relationship between these two defects. Avoidance of entanglements may be specifically dependent upon tight temporal coupling between initial identification of a homolog/partner contact and ensuing juxtaposition at that site, with Mer3 being required for such coupling (Discussion).
mer3Δ defects in alignment arise after, and do not reflect defects in, DSB formation. Rad51 foci appear on axes at early leptotene (by ascus size) in a WT-like number (52 ± 7, n = 90). Rad51 morphologies do become aberrant thereafter, with elongated signals occurring and persisting along chromosome axes (Figures 3G and 3H) throughout synapsis, likely related to unrepaired DSBs.
Additional mer3Δ chromosomal defects are notable. (i) Chromosome ends are often bent and curled around neighbor chromosome ends (Figures 3I and 3J). (ii) Contrary to WT where all seven homologs align and synapse synchronously, all mer3Δ nuclei show per-region asynchrony and inefficiency of alignment-related events, including mixtures of partially aligned homologs and completely separated chromosomes (Figures 3K and 3L versus 3M). Unsynapsed or partially synapsed pairs persist through late pachytene by ascus size (Figures 3N and 3O). (iii) mer3Δ and msh4Δ both affect leptotene alignment, but in different ways. Neither the alignment phenotype of mer3Δ nor the pattern of Mer3 foci (described below) is altered by msh4Δ (Figures S3A-H), implying that Mer3 acts independently of Msh4. The severity of the mer3Δ phenotype precludes assessment of whether Msh4 is still relevant when Mer3 is absent.
Since mer3Δ confers delayed onset of homolog pairing at early leptotene, followed by aberrant alignment configurations, Mer3, like Msh4 and Mlh1, plays a role in homolog pairing one stage earlier (leptotene) than its role in promoting progression of recombination (leptotene/zygotene; Introduction).
Entanglement Avoidance Requires Mer3 ATPase/Helicase Activity
Mer3 is a meiosis-specific helicase. In budding yeast, specific abrogation of this helicase activity confers defective progression of CO-fated, but not NCO-fated, DSBs (Nakagawa and Kolodner, 2002). In light of our finding that Sordaria Mer3 is already required during leptotene alignment, we tested whether this earlier role also requires helicase activity. We mutated a conserved amino acid in the motif I of the Mer3 helicase domain (mer3K297A), shown to be required for ATPase activity (and thus helicase activity) in the budding yeast protein (Nakagawa and Kolodner, 2002). This mutant yeast-protein behaves identically to the WT protein in purification (R. Kolodner, personal communication), and the Sordaria Mer3KAGFP protein localizes to chromosomes nearly identically to WT Mer3-GFP (see below). Thus, the mutant protein is apparently unaltered with respect to functions other than ATPase/helicase per se.
A strain carrying the mer3KA gene in a mer3Δ background shows exactly the same pairing defects as the mer3Δ mutant alone: onset of alignment is delayed relative to appearance of Rad51 foci; and when alignment occurs, at ascus sizes corresponding to late leptotene and zygotene in WT, chromosomes are highly “interwoven” (Figures 3P and 3Q). Further, at ascus sizes that should be pachytene in WT, nuclei contain entangled chromosomes (Figure 3R) or/plus a mixture of aligned and partially synapsed homologs, and one or two pairs that are not aligned (Figure 3S). Thus, abrogation of helicase activity is sufficient to generate the mer3Δ alignment and synapsis defects. This result shows that the biochemical activity of Mer3 plays a role in homolog alignment long before it is required to ensure that CO-designated interactions proceed efficiently.
However, Sordaria mer3KA exhibits more normal 8-spored asci (5%, 2 to 8 in each perithecium thus among 150 asci) than mer3Δ alone (<0.1%; one in 4 perithecia). A similar difference is reported for yeast (Nakagawa and Kolodner, 2002). Thus, Mer3 likely has additional roles not strictly dependent on helicase activity.
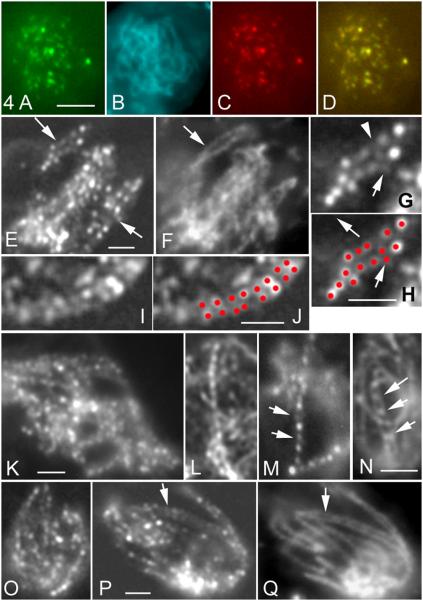
Mer3 Foci Are Evenly Spaced and Occur Transiently in Matched Pairs Along Aligned Homologous Axes
Mer3 appears in foci at early leptotene (Figure 4A), before chromosome alignment (Figure 4B). Co-staining with Rad51-RFP (Figure 4C) shows that the two proteins appear at the same time and mostly colocalize (80-90%, n = 100; Figure 4D). Thus, Mer3, like Rad51, localizes to sites of axis-associated DSBs. Further, Mer3 foci appear at the time corresponding to the alignment defect seen in mer3Δ, and thus well before the Mer3 biochemically-defined role in progression of CO interactions at zygotene (above).
Figure 4. Mer3 and Mer3KA Localization.
(A - D) Colocalization of Mer3-GFP (A) and Rad51-RFP (C) foci at early leptotene with corresponding DAPI (B); (D) Merge of (A) and (C).
(E) Mer3 foci occur at matching sites (arrows) in the two aligned pairs of homologs seen in this optical section. (F) Corresponding pairs (arrows) stained by DAPI.
(G) Double Mer3 foci (arrow) among single foci; arrowhead points to the corresponding “empty” matching site; (H) Cartoon of foci (in red).
(I and J) Another example of Mer3 foci at matching sites with cartoon.
(K-M) Three examples of even-spaced Mer3 foci along leptotene chromosomes. At early leptotene (K and L) all foci are uniformly bright; from mid-leptotene on (M) some foci (arrows) are less bright than others.
(N) At early zygotene, co-staining with Spo76-GFP shows Mer3 foci (arrows) localized between axes in synapsed regions.
(O) Even-spaced Mer3KA foci at mid leptotene.
(P) mer3KA foci occur at matching sites (arrow) in aligned pair of homologs; (Q) corresponding DAPI. Bars, 2μm.
See Figures S3A-3G for patterns of Mer3 foci in msh4Δ.
By the time homolog axes are aligned at ~200nm, foci often appear to occur at matching sites (Figures 4E and 4F). We confirmed this visual impression (Figures 4G - 4J) statistically. We compared the differences in the measured positions of foci along the two axes of 61 pairs of aligned homologs with those predicted by the null hypothesis H0 that foci are positioned independently along the two axes. In all pairs, foci exhibit variable distances on the two homologs and distances between two foci fluctuate from 0.45 to 0.78μm. Our 107 data sets under H0 were obtained by taking the individual focus patterns along the (61 × 2) homolog axes of the experimental data set, and shuffling and repairing them as new randomly chosen partners. Experimental and shuffled data sets were then compared for the extent to which focus positions along partner pairs were (or were not) similar (Experimental Procedures). The probability of H0 (random occurrence) is extremely low (p-value of 0.0026), thus excluding the possibility that Mer3 foci are positioned independently along the two axes of a homolog pair. Since Mer3 is thought to interact specifically with DNA structures at DSB ends (Hunter, 2006), the occurrence of matched pairs of Mer3 foci could imply that the two ends of a DSB are associated with the two-homolog axes (Discussion).
In further support of this interpretation, at leptotene, the total number of Mer3 foci is double that of Rad51 foci (136 ± 10 versus 58 ± 6, n = 200 leptotene nuclei, respectively). Further, the number of Mer3-focus pairs (half of total number, thus 68) and of Rad51 foci (58 ± 6) are three times higher than the number of chiasmata in Sordaria (21 ± 3) and thus correspond to the estimated number of total interactions (Zickler et al., 1992). In some cases (8-10%, n = 50) two Mer3 signals are located one on top of the other along a single axis, with each focus less bright than single foci (arrow in Figures 4G and 4H). Double foci never have a matching partner on the homolog (arrowhead in Figure 4G) and may thus reflect cases in which both DSB ends remain associated with the “donor” homolog and eventually undergo inter-sister recombination.
Mer3 foci exhibit another striking feature: they are evenly spaced along the chromosomes at early leptotene, thus before co-alignment. The clear visual impression to this effect (Figures 4K-4M) is confirmed quantitatively by measurements of interfocus distances (Experimental Procedures). Among 360 interfocus distances (measured along 90 five-foci chromosome segments), minimum and maximum distances vary between 0.54 and 0.7μm with a mean of 0.62 ± 0.03μm. The statistics of interval lengths between adjacent foci were fit to a gamma distribution (Experimental Procedures), leading to ν > 200, where ν is the standard “shape” parameter for this distribution. This high value indicates even spacing and reflects the very small relative fluctuations in the interval lengths (reducing the standard deviation by a factor of two corresponds to an increase of ν by a factor of four). Since Mer3 foci mark the sites of total recombinational interactions (above), these findings further suggest that even spacing arises irrespective of, and likely prior to, CO/NCO differentiation and CO interference.
Mer3 focus patterns further evolve from late leptotene through early zygotene. First, a subset of foci remains bright, while the other foci “fade” at the onset of synapsis (Figures 4M). Then, in early-synapsed regions, Mer3 foci are delocalized from the axes toward the space between the homologs, at which point they are single, not double (Figure 4N). As synapsis progresses, foci disappear from longer synapsed regions and are finally completely absent by mid-pachytene.
The number (122 ± 15, n = 50) and patterns of C-terminally GFP-tagged Mer3KA protein are indistinguishable from those of Mer3-GFP in both WT and mer3Δ backgrounds (compare Figure 4O with Figure 4K and Figures 4P and 4Q with 4E and 4F). The only discernible difference between Mer3KA-GFP and Mer3-GFP foci is a reduction in brightness at all stages (compare Figure 4P with 4E), perhaps reflecting a role for helicase activity in promoting fully normal loading. Thus, Mer3 helicase activity is not required for loading of Mer3 onto chromosome axes. Furthermore, the presence of normal numbers of foci at all stages implies that Mer3 helicase activity is not required for establishment of total recombinational interactions at the DNA level, but only for (ensuing) timely and topologically regular axis juxtaposition during alignment.
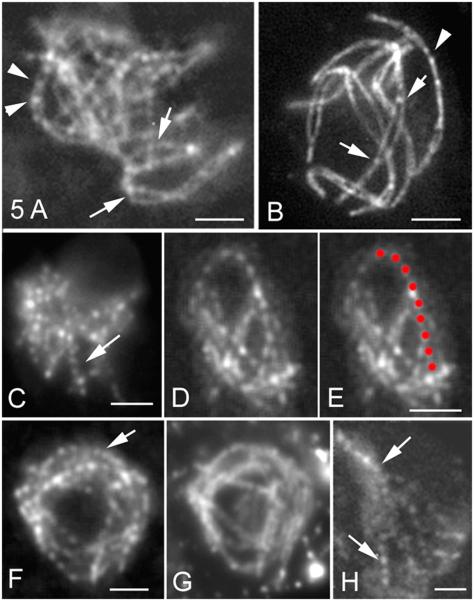
Msh4 Foci Appear After Alignment and Are Evenly Spaced Along Synapsed Homologs
Msh4 foci emerge later than Mer3 foci, first appearing at the transition from alignment to synapsis both on aligned axes (arrows in Figure 5A) and in early-synapsed regions (arrowheads in Figure 5A). They are always single. Further, open regions at sites of interlocks (above) are devoid of foci, although foci do occur on the flanking synapsed regions (arrows in Figure 5B). Thus, Msh4 foci and synapsis are tightly correlated, locally, on a per-region basis. Moreover, appearance of Msh4 foci is closely correlated temporally with the leptotene/zygotene transition and spatially with the presynaptic transition from 200nm to 100nm axis distance. These features correspond temporally to a role of Msh4 in recombination at leptotene/zygotene (Introduction). However, foci appear much later than the time at which we first detect msh4Δ defects (above), implying that important roles are executed by undetectable quantities of protein during alignment.
Figure 5. Msh4 Localization in WT.
(A and B) Nuclei double stained by Spo76-GFP and Msh4-GFP. (A) At zygotene, Msh4 foci are located between synapsed axes (arrowheads) and on the aligned homolog axes (arrows) in non-synapsed regions. (B) Late zygotene nucleus: foci are seen on synapsed homologs (arrowhead) and at the two synapsed ends (arrows) flanking a region that has entrapped another pair of homologs.
(C) At early pachytene, Msh4-GFP foci are evenly spaced along all homologs (arrow). (D) Detail of even spacing along one pair and corresponding cartoon (E). (F) Even spacing of foci persists through early-mid pachytene; (G) corresponding DAPI.
(H) Foci remain evenly spaced (arrows) but decrease in brightness from midpachytene on. Bars, 2μm.
Msh4 foci are dramatically evenly spaced along synapsed chromosomes at early pachytene (Figures 5C-5G). Correspondingly, for 280 inter-focus distances measured along 70 chromosome segments (e.g. Figures 5D and 5E), minimum and maximum distances vary between 0.56 and 0.69μm with a mean of 0.61 ± 0.03μm and Gamma distribution analysis (Experimental Procedures) leads to ν > 200. Even spacing is already obvious on longer synapsed regions at late zygotene (Figure 5A), remains through early pachytene (Figures 5C-5G) and is still seen at mid-pachytene when foci “fade” before disappearing (Figure 5H).
Msh4 foci are maximal in number at late zygotene and early pachytene (81 ± 8; n = 200), diminish in number through mid-pachytene (52 ± 8; n = 100) and are essentially absent from mid-pachytene on. Further, the maximal number corresponds to 3-4 times the number of chiasmata in Sordaria (21 ± 3) and thus corresponds to the estimated number of total interactions (Zickler et al., 1992), again implying that total interhomolog recombinational interactions are evenly spaced, irrespective of CO/NCO differentiation and CO interference.
Discussion
The above results show that three post-DSB recombination proteins are required, each in a different way, for pairing of homologous chromosomes during meiosis. The distinct phenotypes of the three corresponding mutants demonstrate that pairing not only brings homologous chromosomes together but also includes essential features to ensure that homologous pairs are not entangled with unrelated chromosomes. These features are suggested to include an “entanglement avoidance” process that operates during alignment plus a process to resolve residual interlocks that escape this mechanism. Further, recombination is not involved in pairing simply for the purpose of homology recognition but, instead, is an integral mediator of diverse aspects of the pairing process at all stages, with regard to the nature of alignment (i.e. alignment distance via Msh4) as well as topological regularity (via Mer3 helicase and Mlh1). Finally, Mer3, Msh4 and Mlh1 exert their influence on homologous pairing a full stage prior to their known role in mediating the biochemical progression of recombination. Additional insights emerge from analysis of Mer3 and Msh4 foci distribution and integration of these findings with biochemical and physical data regarding the roles of Mer3/Msh4/Mlh1 in recombination. Overall, these results emphasize the power of cytological studies, in combination with other approaches, for dissecting complex processes and the roles of individual molecules in those processes.
Recombination-Mediated Avoidance and Elimination of Topologically Inappropriate Relationships Among Unrelated Chromosomes
Aberrant chromosomal configurations observed in the absence of Mer3 helicase or Mlh1 reveal two new aspects of the way in which the pairing process deals with the threat of entanglements among unrelated chromosomes.
The presence of interwoven chromosomes in mer3 mutants implies that the WT alignment process has specific features designed to ensure that such aberrant configurations do not occur. The mer3 entanglement phenotype is the outcome expected if long-range DNA connections between homologs were made all throughout the genome prior to onset of alignment which, when it does finally occur, locks in the already present meshwork of interactions. In support of this scenario, mer3Δ and mer3 helicase mutants exhibit dramatically delayed onset of alignment. By implication, in WT meiosis, regular pairing of homologs requires tight temporal coupling between the formation of a nascent DSB/partner contact and onset of homolog alignment at the corresponding position. Such coupling would favor neighboring interactions and lead to a per-chromosome pairing process. Homolog pairs would be drawn one-by-one out of the “pairing pool”, promoting topologically regular arrangements as well as facilitating pairing of remaining chromosomes (Figure 6A). Absence of Mer3 helicase activity (e.g. at DSB/partner ends; below) could abrogate this process by triggering a global regulatory surveillance response that delays onset of alignment.
Figure 6. Interlock Avoidance and Recombination-Mediated Axis Alignment.
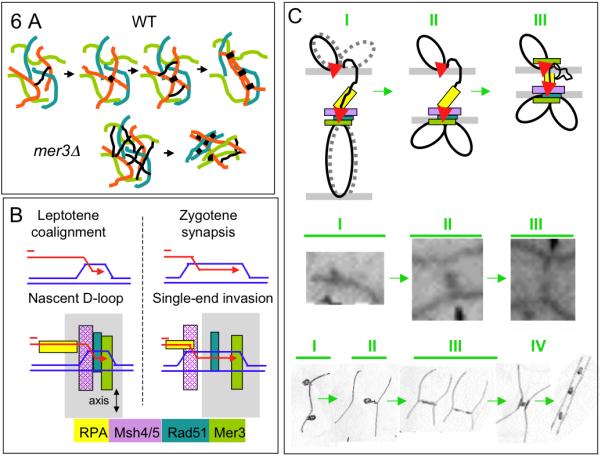
(A) In WT, formation of a connection between homologs at the DNA level (top; black line, left) is rapidly followed by local axis alignment at the corresponding position. This accelerates establishment of connection/alignment nearby (black bars), resulting in propagation of alignment along each chromosome. In mer3Δ (bottom): connections are made throughout the chromosomes (black lines) prior to alignment which, when it does occur, yields highly interwoven chromosomes.
(B) Top: DNA structures are inferred (leptotene) and demonstrated (zygotene) respectively (Hunter and Kleckner, 2001). Bottom: proposed four-layered dispositions of post-DSB proteins on leptotene and zygotene DNA structures.
(C) Possible architecture of recombinosome-mediated homolog pairing. Top: (I) “First” DSB end (red arrowhead) interacts with an homologous chromatin loop, thereby nucleating assembly of a complex containing the four post-DSB recombination proteins as in (B); “second” DSB end (red arrowhead) associates with the axis of the DSB “donor” chromosome. (II) The complex formed with the partner chromosome in (I) becomes axis-associated, thereby bringing donor and recipient chromosomes into closer proximity, with asymmetric evolution of the recipient chromosome complex. (III) Alignment juxtaposes axes to ~400nm. (Middle, Bottom) Corresponding stages (I-III) during Allium alignment as defined by Albini and Jones (1987). At stage IV, recombination “nodule” mediates closer axis juxtaposition for nucleation of SC, analogous to Sordaria progression (above). interwoven chromosomes like mer3Δ. Bars, 2μm.
Any “leftover” problems that escape the entanglement avoidance process are efficiently resolved concomitant with synapsis. Previous considerations of interlock resolution focused on the need to rectify basic topological whole-chromosome relationships. One model invokes movement of the interlocked chromosome out through the ends of the constraining bivalent (Kleckner and Weiner, 1993; Wang et al., 2009) via movements known to occur at the relevant stages (Scherthan et al., 2007; Koszul et al., 2008). Another model invokes DNA topoisomerase II-mediated passage of the trapped chromosomes through the encircling one (von Wettstein et al., 1984).
The finding that Mlh1 is required for interlock resolution brings into focus a completely new aspect. Since Mlh1 is a central player in a set of interrelated DNA transactions (Hunter, 2006), its involvement in interlock resolution suggests that this process requires elimination of constraining DNA connections formed by the recombination process. A classical interlock could arise if recombinational interactions in synapsed regions flanking the entrapped chromosome prevented its movement out the ends of the encircling bivalent. Further, the “leftover” unsynapsed regions in otherwise completely synapsed homologs, also seen at high frequencies in mlh1Δ (above), might reflect “DNA interlocks” in which a stalled recombinational interaction within a previously interlocked region precludes synapsis.
How might the recombination complex sense the presence of a constraining DNA connection to trigger Mlh1-mediated dissolution? The complex might directly sense mechanical tension, resulting from externally-directed movements or ongoing synapsis, with Mlh1 required to sense tension and/or implement a biochemical response. Also, constraining connections might be precluded from normal axis interactions, thus triggering a regulatory checkpoint response that initiates their disassembly.
Communication Among Developing Interhomolog Interactions
Throughout the entire alignment process, DSBs are interacting with homologous partner duplexes, via recombination complexes that are associated with homolog axes (Introduction). Even spacing of Mer3 and Msh4 foci (above) suggests that these recombinational interactions are also evenly spaced. This regularity, in turn, implies communication along chromosomes during the alignment period, independent of and prior to the communication involved in classical CO interference (e.g. de Boer et al., 2006). Moreover, even spacing does not seem compatible with communication occurring independently along each of the two homologs (e.g. during DSB formation), thus suggesting that interference occurs at the level of the interhomolog interaction per se. That is: formation of an interhomolog contact at one position disfavors formation of a contact nearby. The existence of such a process could explain interference of MSH4 and RPA foci in mouse and human (de Boer et al., 2006; Oliver-Bonet et al., 2007) and evidence that COs interfere not only with each other but also with adjacent NCOs in budding yeast (Mancera et al., 2008). Interference at the DSB level has been seen in budding yeast (Wu and Lichten, 1995; Robine et al., 2007). This could be a separate effect; alternatively an interhomolog interaction at one position might preclude subsequent DSB formation nearby. The existence of per-chromosome interference also fits with, and could potentiate, per-chromosome propagation of pairing (above).
Ends-Apart Association of Recombination Complexes with Partner Axes
Colocalization of Mer3 foci with Rad51 foci, plus known biochemical activities of Mer3 (below), suggest that Mer3 foci mark the positions of DSB ends. Our finding that Mer3 foci occur in matching pairs on aligned axes thus suggests that one DSB end is associated with each of the two-homolog axes. This “ends-apart” configuration would correspond to the proposal (Hunter, 2006; Oh et al., 2007) that one DSB end becomes associated with a homolog chromatid while the other DSB end remains associated with its sister. Ends-apart axis association is also supported by observation of matching pairs of axis-associated Rad51 and RPA foci (Franklin et al., 1999; Oliver-Bonet et al., 2007). Early Mer3/Rad51 foci would correspond to formation of nascent DSB/partner interactions on homolog partner axes. Matching Mer3 foci at mid-late leptotene would result from ensuing development of similar complexes between the second DSB end and its sister chromatid. A further implication of the “ends-apart” scenario is that developing recombinosome ensembles would be asymmetric, with one DSB end forming a recombinosome/axis complex de novo with the homolog partner while the other DSB end remains on its original axis in a complex presumably evolved from the initiating DSB recombinosome.
Recombination Proteins Could Mediate Homolog Alignment and Interlock Resolution via Modulation of Nascent DSB/Partner Contacts
Identification of central roles for Mer3 and Msh4 in early homology pairing and for Mlh1 in interlock resolution raises two further questions: how could the known biochemical activities of these molecules be involved in the observed effects; and why do all three molecules have roles for pairing that occur one stage prior to their roles in recombination? Possible answers emerge by considering the fact that throughout the alignment process, DSB/partner interactions involve nascent RecA homolog (Rad51)-promoted DSB/partner contacts (e.g. “nascent D-loops”; Introduction) which are then extended to stable strand invasion intermediates during zygotene as the first molecular landmark of CO/NCO differentiation (Figure 6B top, Hunter et al., 2006).
Mer3 and Msh4/5 are required for efficient progression of CO recombination. In budding yeast, they mediate together the progression of nascent DSB/partner interactions through extension of strand invasion during zygotene (e.g. Borner et al., 2004). The mer3/msh4 recombination defects observed above suggest that the same is likely true in Sordaria. Our analysis reveals that Mer3 and Msh4 are required for homolog pairing during leptotene, prior to their “zygotene role” in recombination. Since alignment initiates with formation of a nascent strand exchange interaction, Mer3/Msh4 could bind to these interactions as they arise, mediate alignment during leptotene and then, already in place, shepherd these intermediates through extension of strand exchange during zygotene. Consistent with this scenario, Mer3 helicase binds at the 3′ single stranded tails of branched structures and blocks “backwards” migration of the branch (Mazina et al., 2004; Hunter, 2006) while Msh4/5 encircles and stabilizes strand exchange junctions (Snowden et al., 2004). Both activities could imply binding to, and conformational modulation of, nascent DSB/partner contacts. Indeed, given these activities, as well as those of other molecules implicated as components of leptotene inter-axis “bridges” (Rad51-mediated strand invasion and RPA binding to single-stranded DNA; Oliver-Bonet et al., 2007; Hunter, 2006), a specific four-layered architecture for nascent DSB/partner complexes emerges (Figure 6B bottom).
By this scenario, Mer3 and Msh4/5 would not directly establish nascent DSB/partner contacts, which are presumably formed by a RecA homolog (e.g. Rad51) but would place the resulting complexes in an appropriate conformation suitable for other events. Such a role is supported by two facts: (i) neither Mer3 nor Msh4 is required for establishment of total DSB/partner interactions for recombination (Borner et al., 2004; Nakagawa and Kolodner, 2002 and references therein); and (ii) Mer3 and Mer3KA foci appear prior to alignment and occur along axes in arrays that correspond in number to total recombinational interactions (above).
Recombinosome Architecture Across the Inter-Axis Space
Absence of Msh4 results in alteration of the alignment distance (above). Thus, Msh4 is clearly present and relevant at this stage, even though corresponding foci do not appear until later. This finding is the first identification of a molecule involved in specification of inter-axis distance and thus raises the general question of how the “architecture” of DSB-mediated inter-axis linkages might be determined. Intriguingly, RPA (and thus single-stranded (ss) DNA) can be seen spanning inter-axis regions during alignment in human (Oliver-Bonet et al., 2007), raising the possibility that the extent of RPA-bound ssDNA determines bridge length. Since Msh4 is thought to bind the strand exchange junction that lies immediately adjacent to single-stranded DNA (above), Msh4 might concomitantly specify alignment distance by influencing the amount of ssDNA available for RPA binding. Figure 6C presents a speculative but synthetic description of DSB-mediated alignment that incorporates existing information (Figure 6C, top I-III). While evolved from the present findings, this description turns out to exactly match the progression seen by EM analysis in Allium (Albini and Jones, 1987; Figure 6C, bottom, I-III).
After alignment, the leptotene/zygotene transition involves closer juxtaposition of homolog axes plus transits of recombinosome foci from on-axis to between-axis localization: Rad51 foci move from axes to inter-axis regions (Tesse et al., 2003); Mer3 foci progress from matched pairs on axes to single foci located between axes; and Msh4 foci appear transiently on aligned axes before moving to inter-axis localization on synapsed regions (above). This progression, also mirrored in the Allium data (Figure 6C bottom, IV), implies a major reorganization of recombination complexes in preparation for (or concomitant with) CO/NCO differentiation.
New Roles for Mlh1
Mlh1 appears to be required for resolution of DNA recombination intermediates that are constraining interlock resolution (above). In WT meiosis, Mlh1 is thought to be required for resolution of double Holiday junction (dHJ) intermediates into CO products at pachytene (Hunter, 2006). Based on analogies with bacterial MutL and MutS complexes, Mlh1 is proposed to mediate removal of Msh4/5 from dHJ intermediates (Snowden et al., 2004). The same activity might resolve interlocks by disassembling nascent DSB/partner interactions in response to entanglement-mediated stalling. Indeed, Mlh1 is already known to mediate the redirection of meiotic recombination intermediates that contain basepair mismatches, which likely occurs at the same stage (Hunter and Kleckner, 2001; Argueso et al., 2003). Moreover, mlh1Δ exhibits a delay in onset of synapsis (above), and thus presumably in progression from nascent DSB/partner complexes to strand invasion extension. Therefore Mlh1 is not recruited de novo to the interlock (or heteroduplex mismatch) sites but is present and “relevant” to nascent DSB/partner contacts even in the absence of any irregularity. In summary: Mlh1, like Mer3 and Msh4/5, is present at the sites of nascent DSB/partner complexes, serving important auxiliary roles for interhomolog interactions, well before it is actually required for efficient/effective progression of recombination per se.
Experimental Procedures
Cloning, Plasmids and Transformation of Sordaria
Sordaria MER3,MSH4 and MLH1 genes were identified by PCR from an indexed genomic library based on Neurospora crassa sequences homology (GenBank accession numbers FJ528581, FJ528583, FJ528582.1). Their sequences predict the conserved domains found in the respective genes of other eukaryotic species. MER3 encodes a predicted protein of 1571 amino acids with 85% identity with Neurospora crassa (hypothetical NCU09793.3), 34% with budding yeast and about 35% with human, mouse and Arabidopsis thaliana. MSH4 encodes a predicted protein of 1284 amino acids with 61% identity with N. crassa (MutS ortholog 4), and about 27% with budding yeast, human, mouse and A. thaliana. MLH1 encodes a predicted protein of 748 amino acids, 93% identity with N. crassa (MutL ortholog-1), 42% with human, mouse, and budding yeast, and 38% with A. thaliana.
Deleted Sordaria strains were obtained by single step gene replacement. A hygromycin resistance cassette replaces the entire ORF in all deletion mutants. Transformants carrying a deleted allele were selected for hygromycin resistance and confirmed by Southern blotting and PCR. The mer3KA mutation (K297A) was created by PCR-based mutagenesis and introduced in Sordaria by cotransformation with a plasmid encoding the nourseothricine resistance cassette (see details in Supplemental Information).
In all analyzed GFP fusions (RAD51, MER3, mer3KA and MSH4) the GFP coding sequence (p-EGFP-1, Clontech) was fused just after the last C-terminal amino acid predicted from the ORF. C-terminally GFP-tagged versions of the genes, under control of their promoters, were introduced in Sordaria at ectopic locations into a WT strain. Msh4-GFP and Mer3-GFP proteins are fully functional: they complement all meiotic and sporulation defects of their cognate null mutants. Further details are provided in Supplemental Information.
Cytology
For cytological analysis, GFP and DAPI (0.5mg/ml) signals were observed, either on living material or after fixation in 4% paraformaldehyde, with a Zeiss Axioplan microscope with a CCD Princeton camera as described (Storlazzi et al., 2003).
Interfocus distances were measured on straight axis segments with 5-12 foci, from the center of the first focus to the center of the next focus (using public domain software ImageJ: http://rsb.info.nih.gov/ij). The ν parameter of the Gamma distribution fitted against interfocus-distances was computed using maximum likelihood fitting with the Wessa Gamma Distribution calculator: http://www.wessa.net/rwasp_fitdistrgamma.wasp/.
Details of the procedures used for measurements of interfocus distances, Gamma distribution and null hypothesis H0 whereby foci positions on two chromosomes are not more correlated on homologs than on non-homologs are given in Supplemental Information.
Supplementary Material
Acknowledgments
We thank Stefanie Pöggeler for the recombinant ku70 plasmid and Olivier Martin for his advice in the statistical analyses. This work, D.Z., A.S. and G.R.-R. were supported by grants from the Centre National de la Recherche Scientifique (UMR 8621) and from the National Institutes of Health (GM025326 and GM044794) to N.K. S.G. was supported by Regione campania Legge 28/03/2002 n.5, and together with A.S. by CNR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albini SM, Jones GH. Synaptonemal complex spreading in Allium cepa and Allium fistulosum I. The initiation and sequence of pairing. Chromosoma. 1987;95:324–338. [Google Scholar]
- Anderson LK, Stack SM. Recombination nodules in plants. Cytogenet. Genome Res. 2005;109:198–204. doi: 10.1159/000082400. [DOI] [PubMed] [Google Scholar]
- Argueso JL, Kijas AW, Sarin S, Heck J, Waase M, Alani E. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol. Cell. Biol. 2003;23:873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA. 2006;103:9607–9612. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, Cande WZ. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin FC, Higgins JD, Sanchez-Moran E, Armstrong SJ, Osman KE, Jackson N, Jones GH. Control of meiotic recombination in Arabidopsis: role of the MutL and MutS homologues. Biochem. Soc. Trans. 2006;34:542–544. doi: 10.1042/BST0340542. [DOI] [PubMed] [Google Scholar]
- Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and Noncrossover pathways in mouse meiosis. Mol. Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. Synaptonemal complex formation: where does it start? Bioessays. 2005;27:995–998. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Springer Berlin; Heidelberg: 2006. pp. 381–442. [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FC. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 2006;25:1315–1323. doi: 10.1038/sj.emboj.7600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harb. Symp. Quant. Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell. 2008;133:1188–1201. doi: 10.1016/j.cell.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51: implications for crossover control in meiotic recombination. Cell. 2004;117:47–56. doi: 10.1016/s0092-8674(04)00294-6. [DOI] [PubMed] [Google Scholar]
- Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes. J. Cell Sci. 2007;120:1017–1027. doi: 10.1242/jcs.03394. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 1999;18:5714–5723. doi: 10.1093/emboj/18.20.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kolodner RD. Saccharomyces cerevisiae Mer3 is a DNA helicase involved in meiotic crossing over. Mol. Cell Biol. 2002;22:3281–3291. doi: 10.1128/MCB.22.10.3281-3291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Bonet M, Campillo M, Turek PJ, Ko E, Martin RH. Analysis of replication protein A (RPA) in human spermatogenesis. Mol. Hum. Reprod. 2007;13:837–844. doi: 10.1093/molehr/gam076. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, Borde V. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:1868–1880. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Gargano S, James F, Kleckner N, Zickler D. Meiotic double strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 2003;17:2675–2687. doi: 10.1101/gad.275203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Pöggeler S, Kleckner N, Zickler D. Coupling meiotic chromosome axis integrity to recombination. Genes Dev. 2008;22:796–809. doi: 10.1101/gad.459308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesse S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. USA. 2003;100:12865–12870. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst D, James F, Pöggeler S, Berteaux-Lecellier V, Zickler D. Spo76p is a conserved chromosome morphogenesis protein that links the mitotic and meiotic programs. Cell. 1999;98:261–271. doi: 10.1016/s0092-8674(00)81020-x. [DOI] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- Wang K, Tang D, Wang M, Lu J, Yu H, Liu J, Qian B, Gong Z, Wang X, Chen J, Gu M, Cheng Z. MER3 is required for normal meiotic crossover formation, but not for presynaptic alignment in rice. J. Cell Sc. 2009;122:2055–2063. doi: 10.1242/jcs.049080. [DOI] [PubMed] [Google Scholar]
- Wang CR, Carlton PM, Golubovskaya IN, Cande WZ. Interlock formation and coiling of meiotic chromosome axes during synapsis. Genetics. 2009;183:905–915. doi: 10.1534/genetics.109.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand-breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Moreau PJ, Huynh AD, Slezec AM. Correlation between pairing initiation sites, recombination nodules and meiotic recombination in Sordaria macrospora. Genetics. 1992;132:135–148. doi: 10.1093/genetics/132.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu. Rev. Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- Zickler D. From early homologue recognition to synaptonemal complex formation. Chromosoma. 2006;115:158–174. doi: 10.1007/s00412-006-0048-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.