SUMMARY
Accumulating evidence implicates heterogeneity within cancer cell populations in the response to stressful exposures, including drug treatments. While modeling the acute response to various anti-cancer agents in drug-sensitive human tumor cell lines, we consistently detected a small subpopulation of reversibly “drug-tolerant” cells. These cells demonstrate >100-fold reduced drug sensitivity, and maintain viability via engagement of IGF-1 receptor signaling and an altered chromatin state that requires the histone demethylase RBP2/KDM5A/Jarid1A. This drug-tolerant phenotype is transiently acquired and relinquished at low frequency by individual cells within the population, implicating the dynamic regulation of phenotypic heterogeneity in drug tolerance. The drug-tolerant subpopulation can be selectively ablated by treatment with IGF-1 receptor inhibitors or chromatin-modifying agents, potentially yielding a therapeutic opportunity. Together, these findings suggest that cancer cell populations employ a dynamic survival strategy in which individual cells transiently assume a reversibly drug-tolerant state to protect the population from eradication by potentially lethal exposures.
Keywords: cancer, KDM5A, IGF-1R, chromatin, drug resistance, bacterial persisters
INTRODUCTION
The relatively rapid acquisition of resistance to cancer drugs remains a key obstacle to successful cancer therapy. Substantial efforts to elucidate the molecular basis for such drug resistance have revealed a variety of mechanisms, including drug efflux, acquisition of drug binding-deficient mutants of the target, and engagement of alternative survival pathways (Redmond et al., 2008). Such mechanisms are generally believed to reflect the existence of rare, stochastic, resistance-conferring genetic alterations within a tumor cell population that are selected during drug treatment.
However, more recent findings have also revealed non-mutational mechanisms of drug resistance. For example, accumulating evidence suggests that a small population of “cancer stem cells” are intrinsically more refractory to the effects of a variety of anti-cancer drugs, possibly via enhanced drug efflux (Trumpp and Wiestler, 2008). Other studies have implicated epigenetic mechanisms, suggesting that acquired drug resistance does not necessarily require a stable heritable genetic alteration (Glasspool et al., 2006). Indeed, an increasingly observed phenomenon in cancer therapy is the so-called “re-treatment response”. For example, some non-small cell lung cancer (NSCLC) patients who respond well to treatment with EGFR (epidermal growth factor receptor) tyrosine kinase inhibitors (TKIs), and who later experience therapy failure, demonstrate a second response to EGFR TKI re-treatment after a “drug holiday” (Kurata et al., 2004; Yano et al., 2005). Similar re-treatment responses are well established for several other anti-cancer agents (Cara and Tannock, 2001). Such findings suggest that acquired resistance to cancer drugs may involve a reversible “drug-tolerant” state, whose mechanistic basis remains to be established. Furthermore, while specific resistance-conferring mutations have indeed been identified in many cancer patients demonstrating acquired drug resistance—especially following treatment with selective TKIs—the relative contribution of mutational and non-mutational mechanisms to drug resistance, and the role of tumor cell subpopulations remains somewhat unclear.
While modeling the acute response to various anti-cancer agents in several different drug-sensitive human cancer cell lines, we consistently detected a small subpopulation of reversibly “drug-tolerant” cells that maintain viability under conditions where the vast majority of the cell population is rapidly killed. Significantly, the emergence of these “drug-tolerant persisters” (DTPs) is observed even following expansion of single drug-sensitive cancer cells, and they are detected at a frequency much higher than would be expected due to mutational mechanisms, implicating epigenetic regulation. Interestingly, we found that this drug-tolerant phenotype is transiently acquired and relinquished by individual cells within the population at a low frequency, implicating the dynamic regulation of phenotypic heterogeneity within a cancer cell population in drug resistance. Taken altogether, our findings suggest that cancer cell populations exhibit reversible tolerance to drugs (and possibly other stressful stimuli) via the maintenance of a phenotypically distinct subpopulation of cells that can protect the population from eradication by potentially lethal exposures.
RESULTS
Detection of a drug-tolerant cancer cell subpopulation
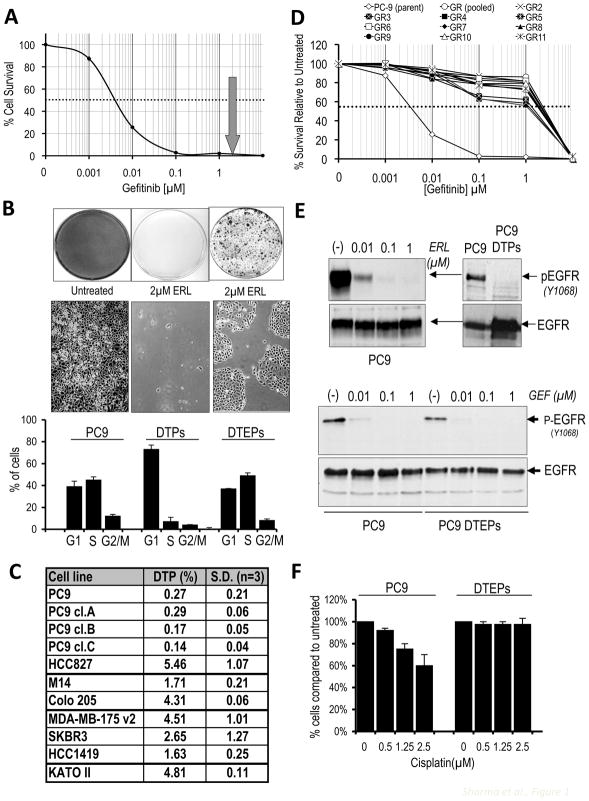
While modeling the response to EGFR TKIs in an EGFR-mutant NSCLC-derived cell line (PC9) that demonstrates exquisite EGFR TKI sensitivity, we observed that while the vast majority of cells are killed within a few days of exposure to a drug concentration 100-fold greater than the IC50 value, a small fraction of viable, largely quiescent cells could still be detected 9 days later (Fig. 1A,B). We refer to this subpopulation of cells as “drug-tolerant persisters” (DTPs), and they constitute ~0.3% of the starting PC9 population (Fig. 1C). An analogous subpopulation of DTPs was detected in several other cancer cell lines with established drug sensitivity (Fig. 1C). Similarly, PC9 cells treated with the chemotherapy drug cisplatin also yielded a small percentage of DTPs (not shown). Thus, a drug-tolerant cancer cell subpopulation is broadly present within tumor-derived cell lines.
Figure 1. Detection of a drug-tolerant subpopulation of cancer cells.
(A) Survival curve describing the viability of PC9 NSCLC cells treated with the indicated concentrations of the EGFR TKI gefitinib for 72 hours (similar results were observed with erlotinib; Fig S1). Each data point represents the average value from 4 samples, and is expressed as a percentage of surviving cells relative to untreated controls. The dashed line corresponds to 50% cell killing. The grey arrow indicates the concentration of EGFR TKI (2μM) used to generate DTPs.
(B) PC9 cells were plated and left either untreated (left), or treated with 2μM erlotinib (ERL) for 9 days (middle) or re-treated with drug every 3 days for 33 days (right). At the appropriate times, cells were fixed and stained with Giemsa (upper panel), or representative microscopic images were photographed (middle panel). In the lower panel, cells from the three different conditions were replated at equal densities, and 20 hours post-plating, cells were analyzed by FACS to determine the percentage of cells in the indicated cell cycle phases. Data are based on two independent experiments performed in duplicate.
(C) NSCLC-derived cell lines PC9 and HCC827 (EGFR mutation ΔE746-A750) were treated with erlotinib (2μM). The melanoma-derived cell line M14 and the colorectal cancer-derived line Colo-205 (BRAF mutation V600E) were treated with the RAF kinase inhibitor AZ628 (2μM). The breast cancer cell lines MDA-MB175v2, SKBR3 and HCC1419 (activated Her2) were treated with Lapatinib (2μM). The gastric cancer-derived cell line KATO II (MET amplification) was treated with the MET inhibitor PF-2341066 (1μM). Single cell-derived PC9 clones are designated PC9 cl.A,B,C. Following 9 days of treatment (fresh drug was added every 3 days), surviving DTPs were quantified. Each experiment was performed in triplicate.
(D) Survival curves describing PC9 cells and several PC9-derived DTEP clones generated by selection in 2μM gefitinib, and then treated with the indicated gefitinib concentrations for 72 hours. Data represent average values determined from four identically treated samples. Data are expressed as a percentage of surviving cells relative to untreated controls. The dashed line corresponds to 50% cell killing.
(E) (Upper panels) Lysates from PC9 cells treated with increasing concentrations of erlotinib (0.01, 0.1 and 1μM) (left panel) or parental PC9 cells and PC9-derived DTPs in 2μM erlotinib (right panel) were analyzed by immunoblotting to detect phosphorylated EGFR (pEGFR) and total EGFR. (Lower panel) Lysates from PC9 cells and gefitinib-tolerant DTEPs after 3 passages in drug-free medium, and subsequently treated for 2h with the indicated gefitinib concentrations, were analyzed by immunoblotting to detect phospho-EGFR and total EGFR. (F) PC9 cells and PC9-derived DTEPs were either untreated (0) or treated with the indicated concentrations of cisplatin for 72h, after which cell numbers were determined. Each experiment was performed in triplicate, and the percentage of DTPs (relative to untreated controls) is presented. Error bars represent standard deviations from the mean.
While DTPs are largely quiescent, approximately 20% of them eventually resume normal proliferation in the presence of drug, yielding colonies of cells referred to as “drug-tolerant expanded persisters” (DTEPs), which can be propagated in drug indefinitely (Fig. 1B). To elucidate mechanisms underlying drug tolerance, the PC9 model was further explored. PC9-derived DTEPs are ~500-fold less drug-sensitive than parental PC9 cells (Figs. 1D and S1A), and can be maintained indefinitely in erlotinib. In DTPs and DTEPs, EGFR TKIs suppress EGFR kinase activity, indicating that drug efflux does not account for their ability to survive treatment (Fig. 1E). PC9-derived DTEPs retain the activating EGFR mutation (Fig. S1B), confirming that they did not arise from contaminating cells. Moreover, they have not acquired the EGFR T790M mutation or MET gene amplification often associated with acquired EGFR TKI resistance in NSCLC patients (Fig. S1C and data not shown), suggesting a distinct state of drug insensitivity. The cell subpopulation demonstrating EGFR TKI tolerance also exhibits reduced sensitivity to cisplatin, suggesting that the observed drug tolerance is not pathway-specific (Fig. 1F).
Drug tolerance is associated with heterogeneity within a cancer cell population
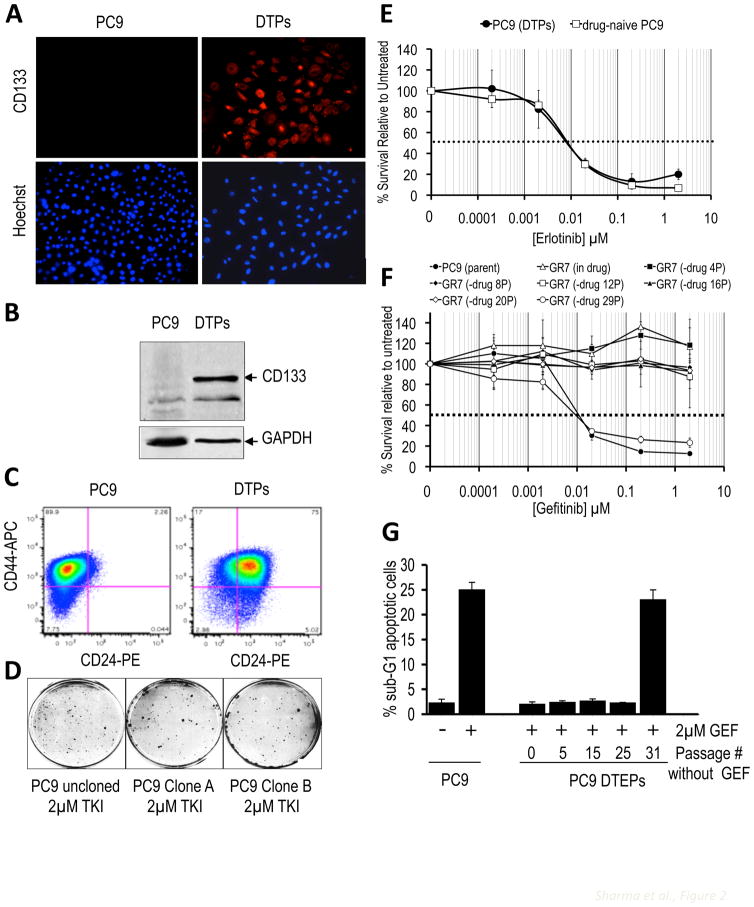
Considering reported links between drug resistance and a “cancer stem cell” (CSC) phenotype, we examined CSC markers. The putative CSC marker CD133 (Hemmati et al., 2003) is expressed in all DTPs, but only in approximately 2% of the parental PC9 population (Figs. 2A,B). DTPs were also highly enriched (relative to parental PC9 cells) for expression of CD24, another CSC marker in some settings (Vermeulen et al., 2008), whereas another CSC marker, CD44 was equally represented in both populations (Figs. 2C and S2A). Thus, DTPs correspond to a small subpopulation of cancer cells that can survive a high concentration drug exposure that kills the vast majority of cells, reflecting phenotypic heterogeneity within the population. Significantly, DTEPs exhibit a CD133 and CD24 expression profile resembling parental PC9 cells (Fig. S2B, C), indicating that the conversion of DTPs (uniformly CD133 positive) to DTEPs involves the re-establishment of heterogeneity with respect to surface markers.
Figure 2. Heterogeneity and reversibility in the drug-tolerant cell populations.
(A) PC9 cells and PC9-derived DTPs were processed for immunofluorescence using anti-CD133 and counterstained with Hoechst to visualize nuclei. Magnification, 20X.
(B) Cell lysates from PC9 cells and PC9-derived DTPs were analyzed by immunoblotting with anti-CD133, and anti-GAPDH as loading control.
(C) PC9 cells and PC9-derived DTPs were labeled with CD24 antibody conjugated to PE and a CD44 antibody conjugated to APC and analyzed by FACS. Note the enrichment of cells with surface expression of CD24 in DTPs (quantitation is shown in Fig. S4).
(D) Uncloned PC9 cells or cells from two different single cell-derived PC9 clones (A and B) were treated with 2μM erlotinib for 33 days and then Giemsa stained.
(E) Survival curves of PC9 cells and PC9-derived DTPs following recovery and re-expansion in drug-free medium and subsequent exposure to the indicated erlotinib concentrations for 72 hours, demonstrating the reversibility of drug tolerance. Each data point represents the average value determined from four samples. Data are expressed as percent surviving cells relative to untreated controls. Error bars represent standard deviations from the mean. The dashed line corresponds to 50% cell killing.
(F) Survival curves of PC9 cells and a PC9-derived DTEP line, GR7, after gefitinib withdrawal for the indicated number of passages (P) and subsequent exposure to the indicated gefitinib concentrations for 72 hours. Each data point represents the average value determined from four samples. Data are expressed as percent surviving cells relative to untreated controls. Error bars represent standard deviations from the mean. The dashed line corresponds to 50% cell killing.
(G) PC9 cells or PC9-derived DTEPs after gefitinib withdrawal for the indicated number of passages were either untreated or treated with 2 μM gefitinib (GEF) for 48 h, after which cells were subjected to FACS analysis. Presented in the graph is the percentage of dying cells in the population (sub-G1 DNA content). Error bars represent standard deviations from the mean from duplicate plates.
The drug-tolerant phenotype can emerge de novo and is reversible
PC9 cells plated at low density yield clones with high efficiency (~50–75%; not shown), and all tested single cell-derived PC9 clones also yield DTPs and DTEPs at a frequency similar to that of “uncloned” PC9 cells (Fig. 2D and 1C), indicating that the drug-tolerant subpopulation can emerge de novo at low frequency from a largely drug-sensitive population. DTEPs derived from clonal PC9 cells similarly demonstrate a low percentage of CD133-positive cells, consistent with the spontaneous emergence of heterogeneity within the population (Fig. S2D). Similar findings were made in several of the other tested cancer cell lines following clonal expansion from single cells (data not shown).
The relatively high percentage of DTPs detected within these various cancer cell populations (0.3–5%; Fig. 1C) is consistent with a non-mutational, and therefore, possibly reversible mechanism. Indeed, DTPs propagated in drug-free media resume growth and rapidly reacquire EGFR TKI sensitivity (within 9 doublings) (Fig. 2E). The same reversibility was seen with DTPs isolated from several other tested cell line models (data not shown). Notably, restoration of drug sensitivity in DTEPs occurs abruptly around passage number 30 (Fig. 2F,G), suggesting a temporal requirement to “unlock” the drug-tolerant state. Proliferating DTEPs (from NSCLC, colorectal, or melanoma cells) can be similarly drug-resensitized by drug-free passaging, although it requires ~90 doublings (or 20–30 passages) to restore sensitivity (Figs. 2F,G and S2E,F and G), suggesting that the drug-tolerant state becomes stabilized over time.
Establishment of drug tolerance requires the histone demethylase KDM5A/RBP2/Jarid1A
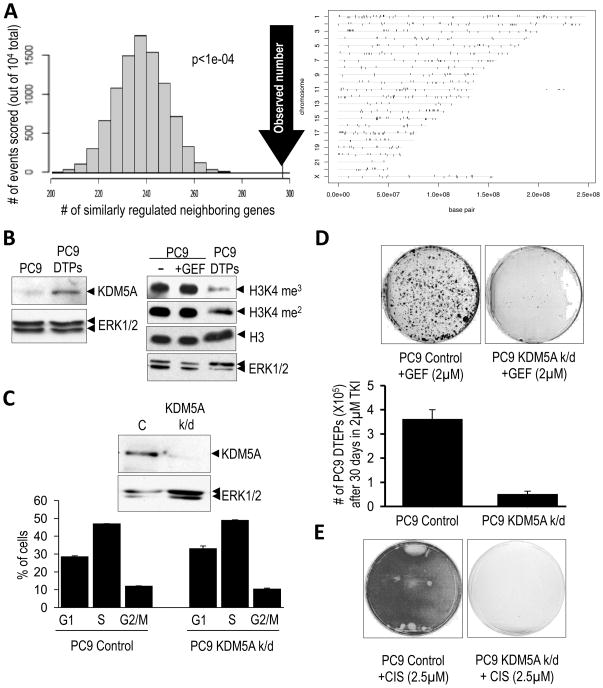
To identify mechanisms underlying reversible drug tolerance, we first undertook a comparative genome-wide gene expression analysis of PC9 cells and PC9-derived DTPs and DTEPs. This comparison revealed striking differences in the overall expression profiles of all three populations (not shown), and in DTEPs, the distribution of differentially expressed genes along chromosomes was highly non-random, implicating global chromatin changes in these cells (Fig. 3A). Indeed, we observed differential nuclease sensitivity when comparing chromatin purified from parental PC9 cells and PC9-derived DTEPs (Fig. S3A), as well as in DTEPs from colorectal cancer (Fig. S3A), and melanoma cells (not shown).
Figure 3. Chromatin alterations and a requirement for the KDM5A histone demethylase in the drug-tolerant state.
(A) Non random expression differences between PC9 cells and PC9-derived DTEPs, indicative of global chromatin alterations. (Left panel) Non-random chromosomal arrangement of differentially up-regulated and down-regulated genes from an array analysis of PC9 cells versus PC9-derived DTEPs was revealed by counting the number of neighboring differentially expressed genes that are either both up-regulated or both down-regulated (104 total pairs examined). The distribution on the graph indicates the expected frequency of similarly regulated neighboring genes, assuming no relationship between expression status and chromosomal configuration, and the arrow indicates the observed number of neighboring genes that are similarly regulated. The p-value reflects the statistical likelihood that the observed number is different than the expected number. (Rght panel) Vertical ticks show the locations along different chromosomes of genes differentially expressed between PC9 cells and PC9-derived DTEPs (tolerant to either gefitinib or erlotinib). Genes whose expression is elevated in both the gefitinib- and erlotinib-tolerant PC9 cells (relative to untreated PC9) are shown above the horizontal lines corresponding to the chromosomes and genes whose expression is attenuated in these cells are shown below the horizontal lines. Individual genes are represented by vertical ticks of equal width. Consequently, ticks that appear wide represent a closely spaced cluster of genes.
(B) The left panel shows increased expression of KDM5A in PC9-derived DTPs, relative to ERK1/2 as a loading control. The right panel shows the decrease in histone H3K4 methylation (H3K4 me3/2) in PC9-derived DTPs compared to parental PC9 cells either untreated (−) or treated for 20 hours with 2μM gefitinib (GEF), with total histone H3 and ERK1/2 serving as loading controls.
(C) (Upper panel) Immunoblot demonstrating reduced expression of KDM5A in PC9 cells lentivirally-infected with shRNA targeting KDM5A (KDM5A k/d) compared to PC9 cells infected with a control shRNA (C). (Lower panel) Cell cycle profiles of these cells as assessed by FACS. Bar graphs represent 2 experiments performed in duplicate.
(D) PC9 cells infected with control or KDM5A (KDM5A k/d) shRNAs were plated at equal density and treated with 2μM gefitinib (GEF) for 30 days with media/drug changes every 3 days. Surviving cells were counted (lower panel) or Giemsa stained and photographed (upper panel). Graphs represent two independent experiments performed in duplicate.
(E) PC9 cells transduced with control or KDM5A (KDM5A k/d) shRNAs were plated at equal density and then treated with 2.5μM cisplatin with media changes ever 2 days. Plates were Giemsa stained 12 days post-treatment.
To identify proteins that potentially contribute to the chromatin alterations required for drug tolerance, we focused on genes whose expression was elevated in both DTPs and DTEPs. Among a small group of genes demonstrating elevated expression in both cell populations was a single gene that encodes a chromatin-modifying enzyme, KDM5A/RBP2/Jarid1A (hereafter referred to as KDM5A). KDM5A was initially discovered as a retinoblastoma-binding protein (Defeo-Jones et al., 1991; Fattaey et al., 1993) and was found to exhibit histone H3K4 demethylating activity (Christensen et al., 2007; Iwase et al., 2007; Klose et al., 2007; Secombe et al., 2007). Consistent with elevated KDM5A in DTPs, they display reduced H3K4me3/2 (Figs. 3B and S3B). To exclude the possibility that the reduced histone methylation in DTPs reflects quiescence, we fractionated PC9 cells after a brief exposure to EGFR TKI on the basis of CD133 surface expression and determined that they are all quiescent, as expected; however, only the drug-tolerant CD133-positive fraction demonstrated reduced H3K4 methylation (Fig. S3C).
To test a specific requirement for KDM5A in drug tolerance, we depleted KDM5A expression in PC9 cells using RNA interference (Fig. 3C and S3D). In parental PC9 cells, KDM5A knockdown does not detectably affect proliferation (Fig. 3C and S3E), but significantly reduces the number of DTPs as well as DTEPs upon EGFR TKI treatment (Figs. 3D and S3F). Ablation of KDM5A also reduced the yield of DTEPs following cisplatin treatment (Fig. 3E). Furthermore, transient expression of KDM5A in PC9 cells decreased the sensitivity of the population to EGFR TKI treatment (Fig. S3G). Together, these observations suggest that the histone demethylase KDM5A is required to establish a metastable chromatin state that underlies reversible drug tolerance.
Drug-tolerant cancer cells are selectively ablated by HDAC inhibitors
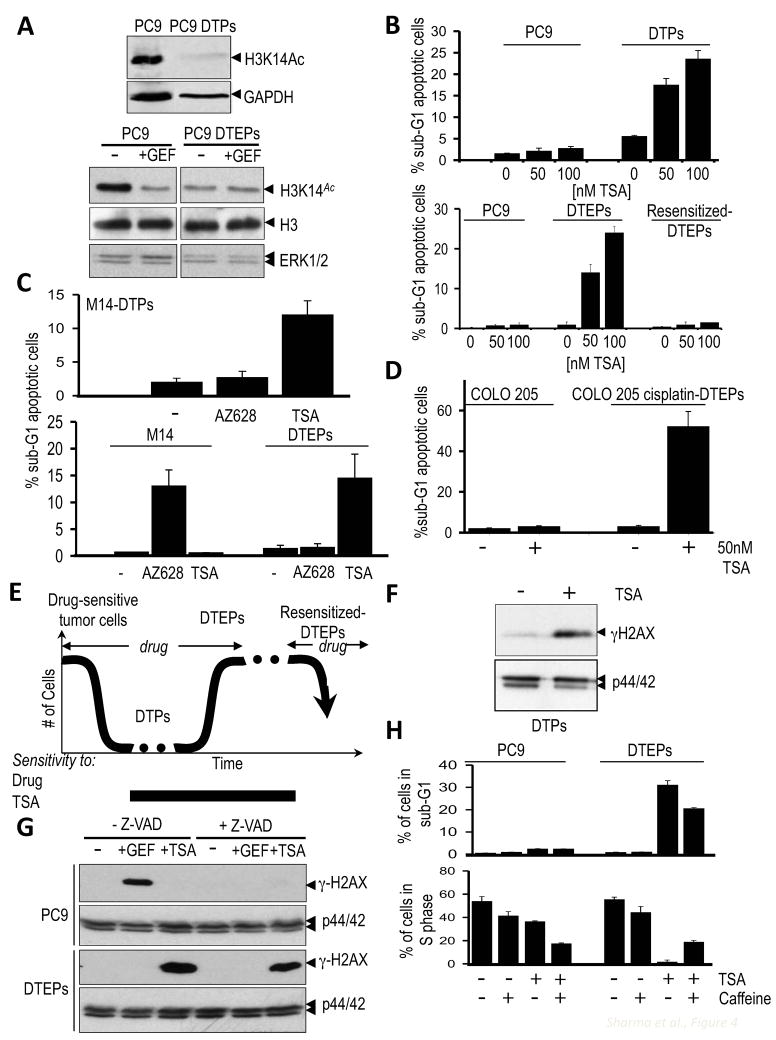
The requirement for KDM5A in drug tolerance could yield a therapeutic opportunity to prevent the development of resistance to cancer drugs; however, pharmacologic inhibitors of KDM5A have yet to be developed. KDM5A associates with histone deacetylases (HDACs) (Klose et al., 2007); reviewed in (Cloos et al., 2008), and a yeast genetic screen to identify suppressors of DNA damage revealed the KDM5A ortholog Msc1, which was found to promote reduced histone H3K9/K14 acetylation (Ahmed et al., 2004). Acetylation of histone H3 K14 is substantially reduced in PC9-derived DTPs and DTEPs, consistent with up-regulation of KDM5A and altered HDAC activity in the drug-tolerant subpopulation (Fig. 4A). We therefore reasoned that HDAC inhibition might phenocopy KDM5A knockdown. Indeed, trichostatin A (TSA), an inhibitor of class I/II HDACs (reviewed in (Bolden et al., 2006), caused the rapid death of DTPs and DTEPs, but did not detectably affect parental PC9 cells or TKI re-sensitized DTEPs (Figs. 4B and S4A, B). Similarly, drug-tolerant populations of M14, Colo205, HCC827, and HT-29 cancer cell lines also demonstrated TSA hypersensitivity (Figs. 4C, D, S4C, D and data not shown), suggesting a vulnerable HDAC-dependent state within the drug-tolerant population in all cases (Fig. 4E).
Figure 4. Drug-tolerant cancer cells are sensitive to HDAC inhibition.
(A) (Upper panel) Immunoblot demonstrating reduced H3K14 acetylation in PC9-derived DTPs treated with 2μM gefitinib. GAPDH serves as a loading control. (Lower panel) Immunoblots to detect histone H3K14 acetylation in PC9 cells and PC9-derived DTEPs either untreated (−) or treated for 24h with 1μM of gefitinib (+GEF). Total histone H3 (H3) and ERK1/2 serve as loading controls.
(B) (Upper panel) Sensitivity of PC9 cells and PC9-derived DTPs to Trichostatin A (TSA). Cells were plated at the same density and either untreated or treated with the indicated TSA concentrations for 20 hours, after which cells were subjected to FACS analysis. Graphs indicate the percentage of dying cells in the population (sub-G1 DNA content), based on 2 independent experiments. (Lower panel) TSA sensitivity of PC9 cells, PC9-derived DTEPs and re-sensitized DTEPs (cells that re-acquired sensitivity to EGFR TKIs following 30+ passages in drug-free medium). Cells were either untreated or treated with the indicated TSA concentrations for 24 h, after which they were subjected to FACS analysis. Presented in the graph is the percentage of dying cells with sub-G1 DNA content, based on 3 independent experiments. Error bars represent standard deviations from the mean.
(C) (Upper panel) TSA sensitivity of M14-derived DTPs. Parental M14 melanoma cells and M14-derived DTPs were plated at equal densities and were either untreated (−), treated with 2μM AZ628 or 100nM TSA for 36 hours, after which they were subjected to FACS analysis. Indicated in the graph is the percentage of cells that display a sub-G1 DNA content indicative of apoptotic cells. The values presented are the average of 2 separate experiments. Error bars represent standard deviations from the mean. (Lower panel) TSA sensitivity of M14 and M14-derived DTEPs to AZ628 and TSA. M14 cells or M14-derived DTEPs were plated at equal density and were either untreated (−), treated with 2μM AZ628 or 100nM TSA, and then analyzed as described above. The values presented are the average of 2 separate experiments. Error bars represent standard deviations from the mean.
(D) COLO 205 colorectal cancer cells or cisplatin-tolerant COLO 205 cells were plated at equal density and were either untreated (−) or treated with 50 nM TSA. 36 hours post-plating cells were subjected to FACS analysis. Indicated in the graph is the percentage of cell with a sub-G1 DNA content indicative of apoptosis. The values presented are the average of 2 separate experiments. Error bars represent standard deviations from the mean.
(E) Model describing the emergence and reversibility of drug-tolerant cancer cells from a largely drug-sensitive population.
(F) Lysates from PC9-derived DTPs either untreated (−) or treated with 50nM TSA for 20 hours were analyzed by immunoblotting using antibodies directed against γH2AX and ERK1/2 as a loading control.
(G) PC9 cells and PC9-derived DTEPs were either untreated (−), treated for 24h with 1μM gefitinib (+GEF) or 50 nM TSA (+TSA), in the presence or absence of the caspase inhibitor, Z-VAD, and lysates were analyzed by immunoblotting using antibodies directed against γ-H2AX. p44/42 ERK was immunoblotted as a loading control.
(H) PC9 cells and PC9-derived DTEPs were either untreated, treated with 2mM caffeine, 75nM TSA alone or, 75nM TSA + caffeine for 36h, after which cells were subjected to FACS analysis. Indicated on the plot is the percentage of cells displaying sub-G1 DNA content (apoptotic cells; top histogram) and the percentage of cells in S phase (bottom histogram).
An altered DNA damage response underlies TSA sensitivity of drug-tolerant cancer cells
Although the effects of HDAC inhibition on gene transcription are well established, accumulating evidence suggests that genomic instability of cancer cells plays a role in their response to HDAC inhibitors (Eot-Houllier et al., 2009). Significantly, the histone variant H2AX (γH2AX) (Lavin, 2008), a marker of DNA damage, is dramatically induced by TSA in PC9-derived DTPs and DTEPs, but not in parental PC9 cells (Fig. 4F, G). TSA-induced γH2AX was similarly observed in M14-derived DTEPs, but not in parental M14 melanoma cells (Fig S4E). γH2AX accumulation can arise as an indirect consequence of DNA fragmentation. However, whereas γH2AX in EGFR TKI-treated PC9 cells is indeed a consequence of DNA fragmentation, as indicated by its attenuation in cells co-treated with a caspase inhibitor, the increased γH2AX in TSA-treated DTEP cells is unaffected by caspase inhibition, consistent with a distinct mechanism of cell death in the TSA-treated drug-tolerant subpopulation (Fig. 4G). Furthermore, treatment of DTEPs with the checkpoint override drug caffeine partially rescues these cells from TSA-induced death and promotes S-phase entry (Fig. 4H), suggesting a checkpoint-dependent mechanism of cell death of DTEPs by TSA. Thus, the distinct chromatin state within the drug-tolerant subpopulation renders these cells hypersensitive to a TSA-induced DNA damage response, resulting in cell death.
Inhibition of HDAC activity prevents the development of drug resistance
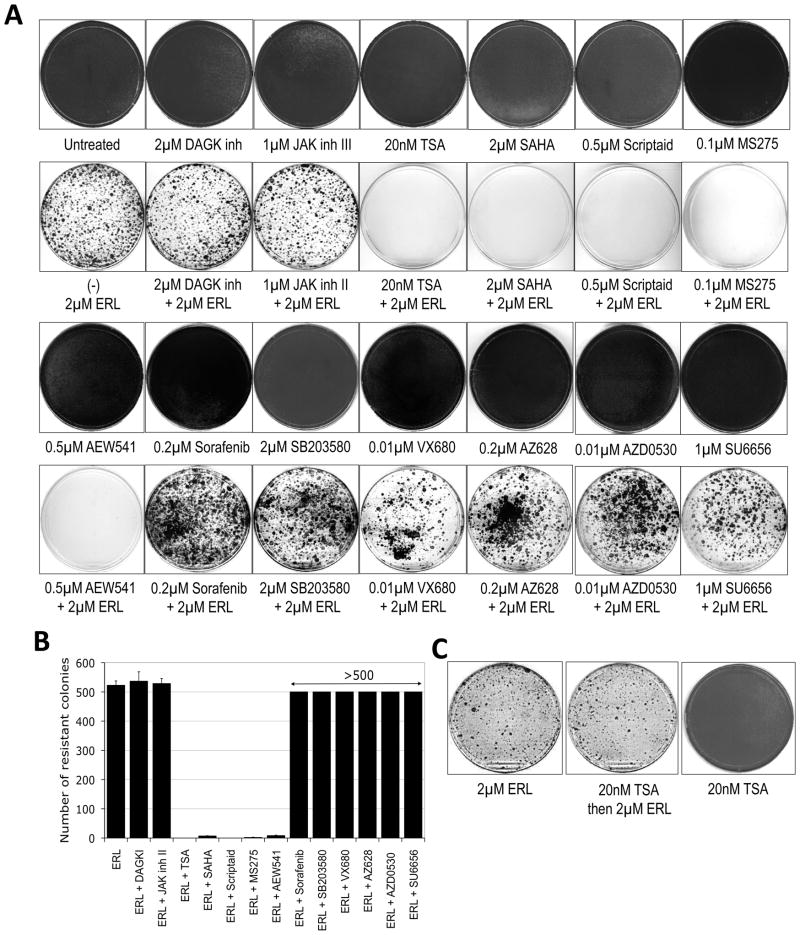
The detection of a reversibly drug-tolerant state prompted us to determine whether pharmacologic disruption of this potentially “intermediate” state could prevent acquired drug resistance. We examined the ability of 13 putative anti-cancer compounds to prevent the emergence of EGFR TKI-tolerant PC9 colonies by co-treating cultures continuously with TKI and these additional compounds. Among the tested compounds, 4 different HDAC inhibitors (TSA, SAHA, MS-275 and Scriptaid) (Bolden et al., 2006), as well as AEW541, a selective inhibitor of the insulin-like growth factor 1 receptor (IGF-1R) kinase (Garcia-Echeverria et al., 2004), virtually eliminated the emergence of DTEP clones, whereas 8 other tested agents had no detectable effect on colony formation in the presence of erlotinib (Figs. 5A, B). Importantly, none of these agents, when tested individually, demonstrated any significant effects on the growth and survival of parental PC9 cells (Fig. 5A).
Figure 5. Preventing the establishment of drug-tolerant clones.
(A) PC9 cells were either untreated or treated singly with the indicated pharmacological agents for 6 days (top rows) or with erlotinib (ERL) alone or the combination of the indicated pharmacological agent with erlotinib for 33 days (bottom rows). Fresh media with drugs was provided every three days. Following treatment, plates were fixed and Giemsa stained. All experiments were performed in triplicate and representative plates are shown.
(B) Individual colonies were counted and the quantified results were graphed. In some cases the colonies were too numerous to count (indicated as >500 colonies). Results reflect the mean and standard deviation from triplicate samples.
(C) PC9 cells were either treated with 2μM of erlotinib (ERL) for 25 days or pre-treated for 9 days with 20nM of TSA and then treated with 2μM of erlotinib for 25 days. In parallel, PC9 cells were also treated continuously with 20nM TSA for 25 days (~ 4 passages). After 25 days, plates were fixed and Giemsa stained. The experiment was performed in triplicate and representative plates are shown.
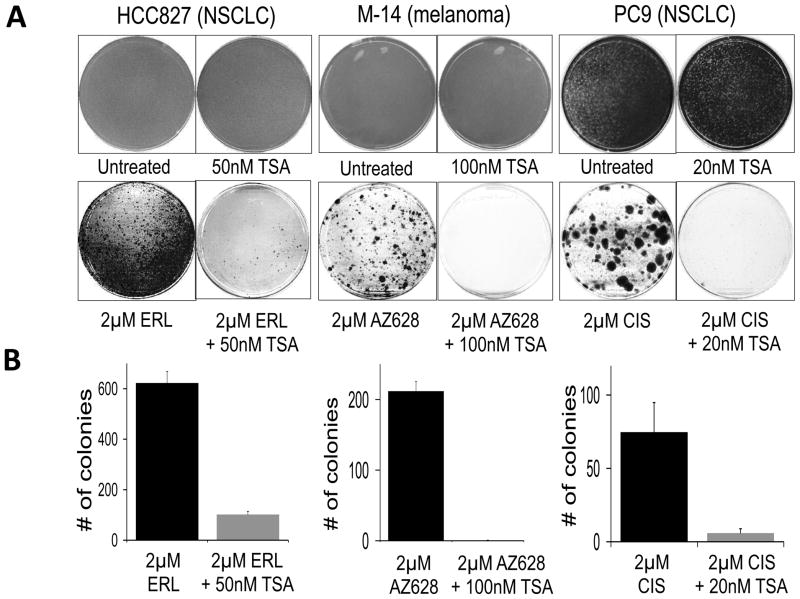
Significantly, HDAC inhibitors must be continuously present to prevent EGFR TKI resistance. Thus, PC9 cells treated for 9 days with TSA prior to erlotinib treatment still yield DTEPs when TSA is withdrawn, suggesting that drug-tolerant cells are continuously replenished in the absence of agents that eliminate them (Fig. 5C). This is consistent with our finding that DTPs arise de novo within single cell-derived drug-sensitive populations. The ability of TSA co-treatment to disrupt DTEP formation was extended to several additional cancer cell lines (Figs. 6A,B and S5), suggesting that an HDAC-dependent drug-tolerant state is broadly relevant in the acute response of cancer cell populations to a lethal drug exposure.
Figure 6. Inhibition of HDAC activity disrupts tolerance to a variety of cancer drugs.
(A) The EGFR-mutant NSCLC cell line NCI-HCC827 was either untreated, treated with TSA (50nM TSA), erlotinib (2μM ERL), or erlotinib plus TSA (2μM ERL + 50nM TSA). After 6 days, the untreated and TSA-treated plates had reached confluence and were Giemsa stained. The erlotinib and erlotinib + TSA-treated plates were similarly fixed and stained after 19 days of treatment. BRAF-mutant M14 melanoma cells were either untreated, treated with TSA (100nM TSA), treated with AZ628 (2μM AZ628), or treated with AZ628 + TSA (2μM AZ628 + 100nM TSA). After 6 days, the untreated and TSA-treated plates had reached confluence and were Giemsa stained. The AZ628 and AZ628 + TSA treated plates were similarly fixed and stained after 39 days of treatment. PC9 cells were either left untreated, treated with TSA (20nM TSA), cisplatin (2μM CIS), or cisplatin plus TSA (2μM CIS + 20nM TSA). After 6 days, the untreated and TSA treated plates had reached confluence and were Giemsa stained. The CIS + TSA treated plates were similarly fixed and stained after 45 days of treatment. Drug treatments were repeated every three days. The experiment was performed in triplicate and representative stained plates are shown.
(B) Quantification of the lower panels in (A) expressed as the number of drug-tolerant colonies observed, with erlotinib-, AZ628- and cisplatin-only treated cells shown in black and the combination of these drugs with TSA shown in grey. Error bars represent the standard deviation from the mean (n=3).
IGF-1R signaling is required for the emergence of reversible drug tolerance
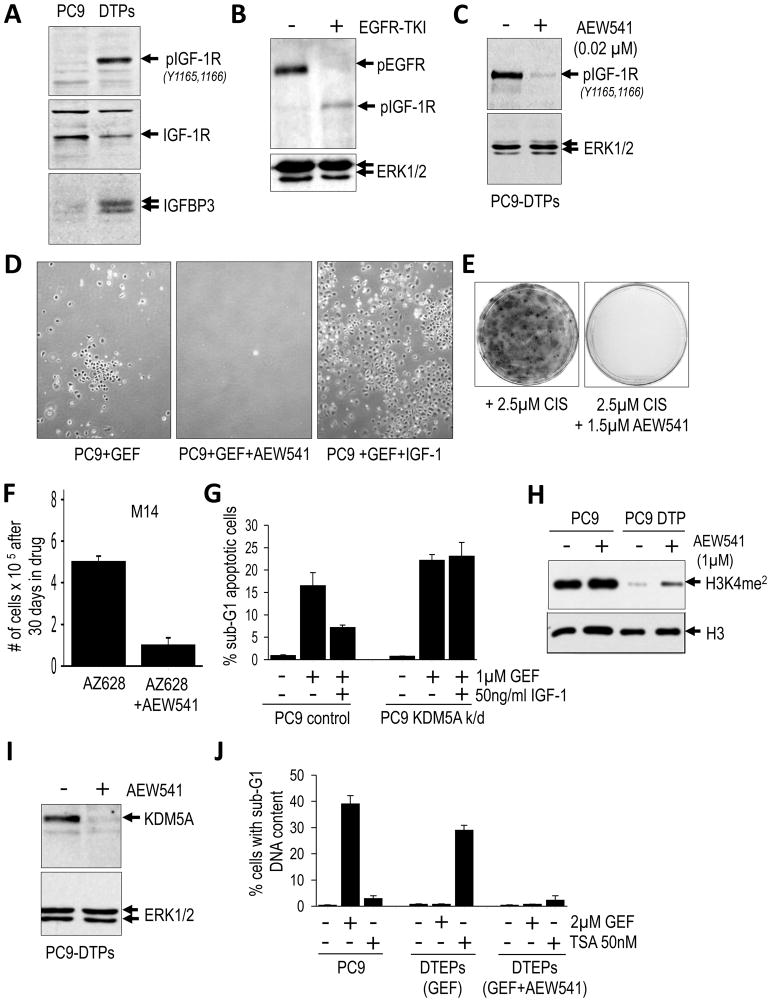
As described above, among many tested kinase inhibitors, only an IGF-1R inhibitor, AEW541, could prevent the emergence of EGFR TKI-tolerant DTEPs. IGF-1R phosphorylation is indeed increased specifically in PC9-derived DTPs (Fig 7A). The IGF-1R signaling pathway is regulated at multiple levels (expression of ligand, receptor, and IGF-binding proteins), and we determined that IGFBP3 (IGF-binding protein 3) levels were specifically up-regulated in the drug-tolerant PC9 cells (Fig. 7A). Previous reports have demonstrated that IGFBP3 promotes IGF-1R signaling (Baege et al., 2004; Ernst and Rodan, 1990), suggesting a potential mechanism by which IGF-1R is activated in the drug-tolerant cells. A similar analysis was performed in a genetically engineered mouse tumor model (Ji et al., 2006). Shortly following erlotinib treatment of transgenic mice bearing EGFR mutant lung tumors, which results in rapid tumor regression, the residual tumor material demonstrates increased phospho-IGF-1R levels, relative to untreated tumors, consistent with the emergence of an IGF-1R-dependent drug-tolerant state in vivo (Fig 7B). Furthermore, AEW541 treatment of DTPs completely suppressed IGF-1R phosphorylation (Fig 7C), and co-treatment of parental PC9 cells with EGFR TKI plus AEW541 prevented the emergence of DTPs, whereas addition of the IGF-1R ligand IGF-1 to the culture media of drug-naïve PC9 cells increased the yield of DTPs (Fig. 7D). IGF-1R signaling is similarly required for the drug-tolerant phenotype observed in other tested cancer cell lines. Thus, drug-tolerant cells derived from cisplatin-treated PC9 cells, as well as melanoma cells treated with the BRAF inhibitor, were efficiently killed by AEW541 (Figs. 7E,F). Together, these results suggest that the viability of the DTP subpopulation requires IGF-1R activation.
Figure 7. IGF-1R activity is required for the chromatin state in drug tolerant cells.
(A) Immunoblot demonstrating phosphorylation of IGF-1R and expression of IGFBP3 in DTPs versus parental PC9 cells. Total IGF-1R is shown as a loading control.
(B) Immunoblot of extracts from an erlotinib-treated (or untreated) mouse lung tumor from transgenic mice expressing an activating EGFR mutant. After 2 days of treatment, EGFR phosphorylation is reduced, and phospho-IGFR is detected in residual tumor material. ERK1/2 is shown as a loading control.
(C) Immunoblot demonstrating reduced phosphorylation of IGF-1R following treatment with 1μm of the IGF-1R inhibitor AEW541 for 2 hours. ERK1/2 is used as a loading control.
(D) Photomicrographs of PC9-derived DTPs following 4 rounds of treatment (12 days) with 1μM gefitinib (GEF), 1μM gefitinib plus 1μM AEW541, or 1μM gefitinib plus 50ng/ml human IGF-1. Representative fields are shown.
(E) PC9 cells were treated with 2.5μM cisplatin or 2.5μM cisplatin in combination with 1μM AEW541. After 30 days the plates were Giemsa stained. A representative example is shown.
(F) M14 melanoma cells were treated with 2μM AZ628 or 2 μM AZ628 in combination with 1μM AEW541 in triplicate. After 30 days cells were counted and the bar graphs reflect the number of remaining cells per dish. Error bars represent standard deviations from the mean.
(G) PC9 cells and PC9 cells in which KDM5A was stably depleted by shRNA (k/d) were either untreated or treated with gefitinib (GEF) or gefitinib plus IGF-1 for 48 hours and then analyzed by FACS to determine the percentage of cells with a sub-G1 content (apoptotic). Graphs reflect the average of two independent experiments performed in duplicate. Error bars represent standard deviations from the mean.
(H) Immunoblot of extracts from parental PC9 cells or PC9-derived DTPs plated at equal density and treated with 1μM of AEW541 for 16 hours. Note that H3K4me2 (dimethylated) levels increase upon addition of AEW541 to PC9-derived DTPs.
(I) Immunoblot of lysates from PC9-derived DTPs either untreated or treated with 1μM of AEW541 for 24 hours. ERK1/2 is shown as a loading control.
(J) PC9 cells, PC9-derived DTEPs (gefitinib-treated), or PC9-derived DTEPs (gefitinib- and AEW541-treated) were either untreated or treated with gefitinib (GEF) or TSA for 48 hours and then BrdU labeled and analyzed by FACS to determine the percentage of cell with a sub-G1 content (apoptotic). Graphs reflect the average of two independent experiments performed in duplicate. Error bars represent standard deviations from the mean.
To determine whether IGF-1R-mediated drug tolerance requires the chromatin-modifying activity of KDM5A, we examined the requirement for KDM5A in IGF-1R-mediated DTP formation. Whereas exposing PC9 cells to IGF-1 increased the yield of DTPs upon treatment with EGFR-TKI, this effect is suppressed by KDM5A ablation (Figs. 7G and S6A). Furthermore, IGF-1R inhibition partially restores the reduced histone H3K4 methylation seen in DTPs (Fig. 7H), suggesting that IGF-1R signaling required for drug tolerance is mediated by the histone demethylating activity of KDM5A. Significantly, IGF-1R inhibition leads to a substantial reduction in KDM5A expression, suggesting a direct link between IGF-1R signaling and KDM5A function (Fig. 7I). When PC9 cells were co-treated for several weeks with EGFR TKI plus AEW541, a very small number of DTEPs eventually emerged, and these were found to harbor EGFR T790M alleles, indicating that this model can yield a clinically established genetic mechanism of drug resistance. Significantly, the rare DTEPs that emerge following co-treatment with EGFR TKI plus AEW541 are TSA-insensitive (Fig. 7J) and do not demonstrate evidence of chromatin alterations, as revealed in nuclease sensitivity assays (Fig. S6B). Taken together, these findings suggest that a transient chromatin state, dependent on IGF-1R signaling and engagement of KDM5A activity, mediates the emergence of drug tolerance.
DISCUSSION
Our analysis implicates a reversible drug-tolerant state in the acute response of cancer cell populations to a lethal drug exposure. The collective findings reveal a subpopulation of cancer cells that transiently exhibit a distinct phenotype characterized by the engagement of IGF-1R activity, hypersensitivity to HDAC inhibition, altered chromatin, and an intrinsic ability to tolerate drug exposure, which does not involve drug efflux (Fig. S6C). Reversible drug tolerance appears to reflect dynamic heterogeneity within a cancer cell population that can be established even following the clonal expansion of single drug-sensitive cells. Such phenotypic heterogeneity has been observed in some clonally-derived normal mammalian cells, such as stem cells (Chang et al., 2008; Stewart et al., 2006), and has been implicated in cancer cell fates following drug exposure in culture (Cohen et al., 2008; Gascoigne and Taylor, 2008).
The ability of the drug-tolerant subpopulation to maintain viability following an otherwise lethal drug exposure appears to involve IGF-1R engagement. This was observed in several tested cell line models, suggesting a potentially broad role for IGF-1R signaling in drug tolerance. Notably, IGF-1R activation has been linked to drug resistance and poor prognosis in several cancer settings (reviewed in (Casa et al., 2008; Pollak, 2008). Furthermore, several published reports describing cell culture models of acquired resistance to both TKIs and conventional chemotherapy drugs have similarly demonstrated the activation of IGF-1R in drug-resistant derivatives (Buck et al., 2008; Chakravarti et al., 2002; Dallas et al., 2009; Eckstein et al., 2009).
Our findings also implicate a distinct chromatin state in the maintenance of the drug-tolerant subpopulation, and the histone demethylase KDM5A was identified as at least one chromatin-modifying enzyme required to establish this state. Notably, reduced methylation of H3K4 has been linked to poor prognosis in cancer patients (Seligson et al., 2005). It is certainly possible that additional chromatin-modifying enzymes contribute to drug tolerance in various tumor contexts. Indeed, our findings also suggest a role for decreased histone acetylation in this process. Although the regulation of histone demethylase activity is poorly understood (Lan et al., 2008), our results suggest a role for IGF-1R signaling in modifying KDM5A activity, which at least partly involves the suppression of KDM5A expression.
A transiently maintained drug-tolerant state could provide a mechanism that allows a small subpopulation of tumor cells to withstand an initial onslaught of drug or other stressful stimuli to enable their survival for a period of time until more permanent resistance mechanisms can be established. This is highly reminiscent of the properties of antibiotic-tolerant bacterial subpopulations, also called “persisters”, which similarly exhibit a transient ability to endure potentially lethal stresses (Balaban et al., 2004; Dhar and McKinney, 2007). These are slower growing cells, whose survival within a more rapidly proliferating cell population is ensured by the fact that they can readily revert to a non-persister state via epigenetic mechanisms. Consequently, the “burden” of protecting the population from eradication is shared equally among all members of the population (Lewis, 2007). Our findings suggest that a subpopulation of drug-tolerant cancer cells may behave similarly, and that all of the tumor cells in a population potentially have the ability to stochastically acquire and relinquish this protective phenotype at a low frequency. Our collective findings support a striking analogy between bacterial and cancer cell-derived persisters; thus, both populations reflect dynamic phenotypic heterogeneity of a non-genetic nature, both exhibit intrinsic multi-drug tolerance that does not involve drug efflux, and upon drug withdrawal, both give rise to progeny that are as susceptible to drug exposure as their ancestors. These similarities raise the possibility that a tumor cell population invokes more “primitive” properties associated with microbial populations to ensure survival.
Our findings suggest a more complex “pathway” to stable genetically-conferred resistance to cancer drugs than is implied by the detection of specific drug resistance mutations in tumors. Such mutations are generally thought to arise spontaneously at low frequency in tumor cells prior to drug treatment and are selected during treatment. However, our observations implicate a multi-step “process” (Fig. S15) mediated by metastable drug-tolerant states associated with chromatin alterations. Importantly, the proposed model is not incompatible with “pre-existing” resistance-conferring mutations. Thus, while drug resistance mutations, such as EGFR T790M, may be present in rare tumor cells prior to EGFR TKI exposure, they might also arise from reversibly drug-tolerant cells. Significantly, accumulating evidence supports a role for stress-induced mutagenesis as an adaptive mechanism both in bacteria and in cancer cells (Galhardo et al., 2007), raising the possibility that an increased mutagenesis rate within drug-tolerant cells leads to a greater opportunity for drug resistance mutations to emerge.
The relationship between the reversibly drug-tolerant subpopulation and cancer stem cells is potentially complex. Although DTPs display markers associated with CSCs, their ability to survive lethal drug exposure does not involve drug efflux, a property attributed to at least some drug-resistant CSCs. Moreover, during the transition of DTPs to DTEPs, CSC-specific markers are lost, and yet both cell populations are equally drug-insensitive. Emerging studies of CSCs have clearly revealed their reduced sensitivity to a variety of toxic exposures (Eyler and Rich, 2008), and recent studies have demonstrated that exposure of mouse tumors to certain chemotherapy drugs can cause tumor regression yielding a population of drug-refractory cells with CSC properties (Kang and Kang, 2007). Considered with our studies, such findings point to a likely relationship between a reversibly drug-tolerant cancer cell subpopulation and the CSC subpopulation. However, this relationship appears to be complex and certainly merits further exploration.
Reversible drug tolerance may account for accumulating clinical reports demonstrating that cancer patients treated with a variety of anti-cancer drugs can be successfully re-treated with the same drug after a “drug holiday”. The detection of a distinct chromatin state in drug-tolerant cancer cells and consequent hypersensitivity to HDAC inhibitors potentially yields a therapeutic opportunity to prevent the development of stable drug resistance. To test this possibility, we have recently initiated a clinical study to examine the ability of combining a chromatin-modifying agent with erlotinib in NSCLC patients. While the trial is not yet completed, the early clinical data indicate that the inclusion of a chromatin-modifying agent can dramatically improve clinical benefit in a subset of patients demonstrating acquired TKI resistance (S.V.S. and J.S., unpublished observation). When considering that acquired drug resistance may involve multiple distinct molecular mechanisms that arise independently within the same patient, thereby complicating strategies to overcome such resistance with a single rationally-targeted agent, the potential ability to prevent the development of resistance by disrupting a drug-tolerant state is provocative. However, further studies will certainly be required to establish the in vivo significance of the cell culture findings, as well as to determine more precisely the molecular mechanisms underlying reversible drug tolerance.
EXPERIMENTAL PROCEDURES
Cell survival assays
3×104 cells were plated in each well of a 12-well cluster dish. 24 hours after plating, media was removed and replaced with media containing drugs. Fresh media was replaced every 2 days until untreated cells reached confluence. Media was then removed, cells were washed with Phosphate Buffered Saline (PBS), and then fixed for 15 min with 4% formaldehyde in PBS. Cells were then washed with PBS and stained with the fluorescent nucleic acid stain, Syto60 (1 nM in PBS; Molecular Probes) for 15 min. Dye was removed, cell monolayers were washed with PBS, and fluorescence quantitation was carried out at 700nm with an Odyssey Infrared Imager (Li-Cor Biosciences). Each experiment was performed in quadruplicate.
Generation of drug-tolerant persisters (DTPs)
Drug-sensitive cells were treated with the relevant drugs, at concentrations exceeding 100 times the established IC50 values, for three rounds, with each treatment lasting 72h. Viable cells remaining attached on the dish at the end of the third round of drug treatment were considered to be DTPs, and were collected for analysis.
Generation of DTEPs
105 cells were plated in five 10cm plates and allowed to adhere for 24h. Cells were then treated with 1μM gefitinib or 2.5μM erlotinib (for PC9), 2.5 μM cisplatin for Colo205, or 2μM AZ628 for M14. Fresh media containing drug was replaced every 3 day until clones of drug-resistant cells appeared. ~50 clones per dish appeared after 30 days of drug selection. Isolated clones were individually expanded in drug-containing media. Where indicated, cells were withdrawn from drug for analysis, but “stock cultures” of DTEPs were maintained in drug. To assess the effect of the various pharmacological agents on the generation of DTEPs following exposure to targeted agents, 106 cells were plated in triplicate under the indicated conditions. After 24h, the various indicated compounds were added to culture medium and replenished every 3 days. On day 6, untreated cells and cells treated with the individual pharmacological agents alone had reached confluence and were fixed with 4% paraformaldehyde in PBS and stained with Syto 60. Treated cells were cultured for 33 days, after which plates were fixed and stained as described above.
Cell Harvesting and Protein Analysis
Cell lysates were prepared in Laemmli sample buffer and analyzed by immunoblotting as described previously (McDermott et al., 2007).
Immunofluorescence
Immunofluorescence analysis of CD133 on cells plated on glass coverslips was performed after fixing cells in 4% paraformaldehyde in PBS, as described previously (Quinlan et al., 2008).
Mouse tumor studies
Bitransgenic CCSP-rtTA/Tet-op-EGFR del exon 19 mice with lung adenocarcinomas driven by EGFR were exposed to a doxycycline-containing diet for 8 to 12 weeks, and subjected to magnetic resonance imaging (MRI) to document tumor burden as described previously (Ji et al., 2006). Mice identified with lung tumor burden were then treated with erlotinib at 50mg/kg/day or vehicle via gavage. Two hours after the second dose, mice were sacrificed and the tumor nodules were grossly dissected. As per Dana-Farber Cancer Institute Animal Care and Use Committee approval, all mice were housed in a pathogen-free environment at the Harvard School of Public Health and were handled in accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare.
shRNA studies
Lentivirus-mediated delivery of shRNAs directed against KDM5A was performed as previously described (Rothenberg et al., 2008), using shRNA constructs from the Broad Institute TRC consortium. KDM5A expression constructs were from Addgene.
Supplementary Material
Acknowledgments
We thank the members of the Settleman laboratory, Luisa Di Stefano, Daniel Haber, and Kurt Isselbacher for helpful discussions through the course of these studies. We also thank Lisa Drew and Jeff Hanke (AstraZeneca) for providing AZ628; James Christensen (Pfizer) for providing PF-2341066; Robert Schlegel (Novartis) for providing AEW541; Jeff Engelman for providing the drug-resistant HCC827 cells and Toshi Shioda for providing shRNA plasmids and performing gene expression array analysis. This work was supported by an award from the LUNGevity Foundation and Goldman Philanthropic Partnerships to S.V.S. and NIH RO1CA115830 and NIH P20 CA090578 to J.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed S, Palermo C, Wan S, Walworth NC. A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast. Mol Cell Biol. 2004;24:3660–3669. doi: 10.1128/MCB.24.9.3660-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baege AC, Disbrow GL, Schlegel R. IGFBP-3, a marker of cellular senescence, is overexpressed in human papillomavirus-immortalized cervical cells and enhances IGF-1-induced mitogenesis. J Virol. 2004;78:5720–5727. doi: 10.1128/JVI.78.11.5720-5727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Buck E, Eyzaguirre A, Rosenfeld-Franklin M, Thomson S, Mulvihill M, Barr S, Brown E, O’Connor M, Yao Y, Pachter J, et al. Feedback mechanisms promote cooperativity for small molecule inhibitors of epidermal and insulin-like growth factor receptors. Cancer Res. 2008;68:8322–8332. doi: 10.1158/0008-5472.CAN-07-6720. [DOI] [PubMed] [Google Scholar]
- Cara S, Tannock IF. Retreatment of patients with the same chemotherapy: implications for clinical mechanisms of drug resistance. Ann Oncol. 2001;12:23–27. doi: 10.1023/a:1008389706725. [DOI] [PubMed] [Google Scholar]
- Casa AJ, Dearth RK, Litzenburger BC, Lee AV, Cui X. The type I insulin-like growth factor receptor pathway: a key player in cancer therapeutic resistance. Front Biosci. 2008;13:3273–3287. doi: 10.2741/2925. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Loeffler JS, Dyson NJ. Insulin-like growth factor receptor I mediates resistance to anti-epidermal growth factor receptor therapy in primary human glioblastoma cells through continued activation of phosphoinositide 3-kinase signaling. Cancer Res. 2002;62:200–207. [PubMed] [Google Scholar]
- Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, 2nd, Samuel S, Kim MP, Lim SJ, Ellis LM. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defeo-Jones D, Huang PS, Jones RE, Haskell KM, Vuocolo GA, Hanobik MG, Huber HE, Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Eckstein N, Servan K, Hildebrandt B, Politz A, von Jonquieres G, Wolf-Kummeth S, Napierski I, Hamacher A, Kassack MU, Budczies J, et al. Hyperactivation of the insulin-like growth factor receptor I signaling pathway is an essential event for cisplatin resistance of ovarian cancer cells. Cancer Res. 2009;69:2996–3003. doi: 10.1158/0008-5472.CAN-08-3153. [DOI] [PubMed] [Google Scholar]
- Eot-Houllier G, Fulcrand G, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase inhibitors and genomic instability. Cancer Lett. 2009;274:169–176. doi: 10.1016/j.canlet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Ernst M, Rodan GA. Increased activity of insulin-like growth factor (IGF) in osteoblastic cells in the presence of growth hormone (GH): positive correlation with the presence of the GH-induced IGF-binding protein BP-3. Endocrinology. 1990;127:807–814. doi: 10.1210/endo-127-2-807. [DOI] [PubMed] [Google Scholar]
- Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839–2845. doi: 10.1200/JCO.2007.15.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattaey AR, Helin K, Dembski MS, Dyson N, Harlow E, Vuocolo GA, Hanobik MG, Haskell KM, Oliff A, Defeo-Jones D, et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Echeverria C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, Gao J, Brueggen J, Capraro HG, Cozens R, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5:231–239. doi: 10.1016/s1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Glasspool RM, Teodoridis JM, Brown R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer. 2006;94:1087–1092. doi: 10.1038/sj.bjc.6603024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, Mahmood U, Mitchell A, Sun Y, Al-Hashem R, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Kang MK, Kang SK. Tumorigenesis of chemotherapeutic drug-resistant cancer stem-like cells in brain glioma. Stem Cells Dev. 2007;16:837–847. doi: 10.1089/scd.2007.0006. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Kurata T, Tamura K, Kaneda H, Nogami T, Uejima H, Asai Go G, Nakagawa K, Fukuoka M. Effect of re-treatment with gefitinib (‘Iressa’, ZD1839) after acquisition of resistance. Ann Oncol. 2004;15:173–174. doi: 10.1093/annonc/mdh006. [DOI] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, Archibald H, Raudales R, Tam A, Lee D, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- Quinlan MP, Quatela SE, Philips MR, Settleman J. Activated Kras, but not Hras or Nras, may initiate tumors of endodermal origin via stem cell expansion. Mol Cell Biol. 2008;28:2659–2674. doi: 10.1128/MCB.01661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond KM, Wilson TR, Johnston PG, Longley DB. Resistance mechanisms to cancer chemotherapy. Front Biosci. 2008;13:5138–5154. doi: 10.2741/3070. [DOI] [PubMed] [Google Scholar]
- Rothenberg SM, Engelman JA, Le S, Riese DJ, Haber DA, Settleman J. Modeling oncogene addiction using RNA interference. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0803217105. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Stewart MH, Bosse M, Chadwick K, Menendez P, Bendall SC, Bhatia M. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods. 2006;3:807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Wiestler OD. Mechanisms of Disease: cancer stem cells--targeting the evil twin. Nat Clin Pract Oncol. 2008;5:337–347. doi: 10.1038/ncponc1110. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S, Nakataki E, Ohtsuka S, Inayama M, Tomimoto H, Edakuni N, Kakiuchi S, Nishikubo N, Muguruma H, Sone S. Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: a report of three cases. Oncol Res. 2005;15:107–111. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.