Summary
The characteristic bipolar shape of the mitotic spindle is produced by the focusing of the minus ends of microtubules at the spindle poles. The focus is maintained by the centrosome, a microtubule-nucleating organelle, as well as by proteins that are capable of focusing kinetochore fibers (K fibers) even in the absence of a centrosome. Here, we have performed a small-scale RNA interference (RNAi) screen of known or suspected pole-related proteins in Drosophila S2 cells. An unexpected outcome of this screen was the finding that one of the four Drosophila Mob proteins (a family of kinase regulators) plays a role in spindle pole organization. Time-lapse microscopy of mitotic cells depleted of Drosophila Mob4 by RNAi revealed that the K fibers splay apart and do not maintain their focus either in the presence or absence of functional centrosomes. The Mob4 RNAi phenotype most closely resembles that observed after depletion of the protein encoded by abnormal spindle (Asp), although Asp localization is not substantially affected by Mob4 RNAi. Expression of a Drosophila Mob4-GFP fusion protein revealed its localization to the nucleus in interphase and to spindle poles and kinetochores during mitosis. We propose that Mob4 in Drosophila controls a mitotic kinase that in turn regulates downstream target proteins involved in K fiber focusing at the poles.
Keywords: RNAi, Centrosome, Microtubule, Kinetochore, γ-tubulin
Introduction
During mitosis, the microtubule network reorganizes into a bipolar spindle that ensures proper chromosome segregation by aligning the chromosomes along the metaphase plate before partitioning them equally into the two daughter cells. Because the fidelity of chromosome segregation depends on proper spindle function, the mechanisms by which microtubules self-organize into a bipolar spindle have been a topic of considerable interest (Karsenti and Vernos, 2001; Maiato et al., 2004; Nedelec et al., 2003). The centrosome is the primary microtubule organizing center (MTOC) in somatic animal cells, and it plays a fundamental role in spindle formation. The centrosome number determines both the location and number of spindle poles, as illustrated by the formation of monopolar spindles in cells with unseparated or unduplicated centrosomes (Heald et al., 1997) or of multipolar spindles in cells with multiple centrosomes (Nigg, 2002). However, functional spindles are capable of forming in the absence of bona fide centrosomes, as occurs in meiotic cells or in cells with disrupted centrosomes (Heald et al., 1996; Hinchcliffe et al., 2001; Khodjakov et al., 2000; Mahoney et al., 2006; Megraw et al., 2001).
The process of spindle formation in somatic cells begins typically with the duplication and separation of centrosomes in prophase. This separation is facilitated by bipolar kinesin motors (kinesin-5) (Lawrence et al., 2004), which establish spindle bipolarity by crossbridging and pushing apart antiparallel microtubules from the two centrosomes (Sharp et al., 2000). Following breakdown of the nuclear envelope, centrosome-nucleated microtubules invade the nuclear region, grow and shrink by dynamic instability and become selectively stabilized after connecting with kinetochores (Desai and Mitchison, 1997). Chromosomes also mediate microtubule nucleation, including polymerization at the kinetochore (Khodjakov et al., 2003; Mahoney et al., 2006; Maiato et al., 2004). Microtubules originating from both sources contribute to the formation of kinetochore fibers (K fibers), thick microtubule bundles connected directly to the kinetochore. K fibers become focused at the spindle poles adjacent to the two centrosomes through the action of microtubule crosslinking proteins [e.g. abnormal spindle protein (Asp) and NuMA] and motors (Ncd and dynein) that crossbridge the minus (distal) ends of the K fibers and transport them towards the centrosomes along astral microtubules (Goshima et al., 2005; Khodjakov et al., 2003; Maiato et al., 2004; Merdes et al., 2000; Morales-Mulia and Scholey, 2005). The centrosome, therefore, has a fundamental role not only in the nucleation of spindle microtubules but also in the collection and organization of K fibers into two focused poles before chromosome segregation in anaphase. However, some degree of pole focusing can be achieved in acentrosomal spindles through the actions of minus-end-directed motors and microtubule crosslinking proteins alone (Nedelec et al., 2003).
Here, through the use of RNA interference (RNAi), we have depleted 24 proteins that have been implicated in spindle pole organization in Drosophila S2 cells, a well-characterized cell line for RNAi and mitosis (Goshima et al., 2007; Bettencourt-Dias et al., 2005; Goshima and Vale, 2003; Morales-Mulia and Scholey, 2005; Rogers et al., 2002). One of the ‘hits’ from this mini-RNAi screen was Drosophila Mob4, an uncharacterized member of the highly conserved Mob protein family (Lai et al., 2005). Our experiments reveal that Drosophila Mob4 is required for both centrosome separation and focusing of K fibers. These findings represent the first demonstration of a Mob family protein serving a role in the formation of the mitotic spindle.
Results
Screen of centrosome-related proteins for mitotic defects in S2 cells
We examined RNAi phenotypes for 24 Drosophila proteins that have been implicated in spindle pole organization in Drosophila or other organisms. While this list probably represents only a subset of the proteins involved in Drosophila spindle pole formation, our intention was to uncover new functions for known or suspected pole proteins. We examined the mitotic index, mitotic spindles and γ-tubulin localization to the spindle poles in Drosophila S2 cells after a 4-day RNAi treatment of candidate spindle pole proteins (the screen was repeated with 7-day RNAi and produced identical results). The efficacy of RNAi was examined for seven proteins for which we could obtain antibodies (supplementary material Fig. S1). In these cases, substantial (>80%) protein depletion was observed. We suspect that protein depletion occurred for all of the RNAi treatments, as these results are consistent with our prior (Goshima and Vale, 2003; Rogers et al., 2003) and our unpublished observations, where comparable RNAi-induced depletion was observed for approximately 40 proteins tested by immunoblot analysis.
The results from the screen are shown in Table 1 and more details are described in supplementary material Table S2 and supplementary material Figs S1 and S2. While this screen was being performed, similar work described phenotypes for γ-tubulin and γ-tubulin-associated subunits (Goshima et al., 2007; Vérollet et al., 2006), which largely agreed with the results from this screen. Several RNAi treatments did not alter the mitotic index or spindle morphology. In some cases (e.g. Dgrip79, Dgrip223, centrin2, Niki and germ-line specific γ-tubulin37C), the gene expression was low or at background levels, as determined by DNA microarray analysis (Table 1). Recent work described a phenotype of centrosomal antigen dispersion for Nek2 depletion in S2 cells (Prigent et al., 2005); however, the reported penetrance of this phenotype was low (~ 30% of cells, which was only slightly above wild-type background levels) and most likely was not sufficiently robust to be scored by our criteria (see Materials and Methods). For other genes, we used double RNAi treatment to explore whether the lack of an RNAi phenotype might be due to redundancy with another gene product. For example, the fly genome contains multiple splice variants of pericentrin (Zimmerman et al., 2004), centrin (Beisson and Wright, 2003) and two NEK kinase-like proteins, Nek2 and Niki (Prigent et al., 2005). We performed double RNAi of these related proteins and still failed to observe any significant effects on mitotic index or spindle morphology (data not shown). Thus, these proteins either might not have a significant function in S2 cell mitosis or they might not be sufficiently depleted by RNAi.
Table 1.
Spindle phenotype and γ-tubulin localization analysis after RNAi
| Targeted gene product |
Relative mitotic index |
Spindle phenotype |
% abnormal spindles (in %), (n) |
Spindles with localized γ-tubulin (in %), (n) |
Knockdown confirmation |
|---|---|---|---|---|---|
| None | 1 | 24±9 (496)† | 100±0 (114) | ||
| γ-tubulin (23C) | 3.3±0.7* | Monopolar/anastral | 98±2 (279) | 3±2 (118) | |
| Dgrip84 | 2.8±0.9* | Monopolar/anastral | 98±3 (303) | 14±5 (146) | + |
| Dgrip91 | 2.9±0.6* | Monopolar/anastral | 95±3 (371) | 11±5 (133) | + |
| Dgrip128 | 2.6±0.3* | Monopolar/anastral | 84±4 (285) | 94±3 (125) | − |
| Dgrip163 | 2.0±0.7* | Monopolar/anastral | 76±5 (301) | 89±7 (166) | − |
| Dgrip71 | 2.5±0.3* | Monopolar/anastral | 85±7 (385) | 96±1 (82) | − |
| Dgrip75 | 2.3±0.3* | Monopolar/anastral | 82±1 (331) | 91±5 (129) | − |
| Msps | 3.2±0.4* | Multiple defects | 97±2 (292) | 99±1 (135) | + |
| Msps and Asp | 2.0±0.6* | Bipolar/anastral spindles | 88±5 (134) | 95±4 (111) | − |
| Centrosomin | 1.0±0.2 | Anastral | 93±4 (246) | 10±1 (108) | + |
| Asp | 1.2±0.7 | Detached/unfocused spindles poles | 91±6 (222) | 98±3 (109) | − |
| Dmob4 | 1.1±0.4 | Monopolar/monastral bipolar | 73±7 (342) | 96±6 (83) | − |
| Mps1 | 0.5±0.2* | Monastral bipolar/other | 51±2 (122) | 99±2 (63) | − |
| γ-tubulin (37C) | 0.9±0.5 | Normal | 33±11 (235) | 99±2 (85) | − |
| Dgrip223○ | 1.2±0.1 | Normal | 26±8 (152) | 100 (39) | ○ |
| Dgrip79○ | 0.7±0.6 | Normal | 29±4 (176) | 100±0 (84) | ○ |
| D-TACC | 1.2±0.7 | Normal | 25±7 (241) | 100±0 (101) | + |
| Centrin 1 | 0.6±0.1 | Normal | 15±6 (190) | 99±2 (105) | − |
| Centrin 2○ | 0.6±0.1 | Normal | 26±1 (174) | 100±0 (77) | ○ |
| Niki/Nek2○ | 1.1±0.1 | Normal | 30±5 (168) | 100±0 (89) | ○ |
| CP60 | 0.9±0.3 | Normal | 36±3 (260) | 94±4 (116) | + |
| CP190 | 0.9±0.2 | Normal | 24±6 (241) | 99±2 (77) | + |
| Dmob1 | 0.8±2.4 | Normal | 25±2 (247) | 100± (93) | − |
| Dmob2 | 1.0±0.2 | Normal | 24±10 (286) | 100±0 (19) | − |
| Dmob3 | 1.3±0.4 | Normal | 24±3 (484) | 95±1 (148) | − |
| Pericentrin | 0.8±0.3 | Normal | 24±12 (206) | 100±0 (99) | − |
For complete gene information, including CG-identification numbers and RNAi primer sequences, see supplementary material Table S1. Cells were analyzed on day 4 or 5 of RNAi treatment. Values of mean ± s.d. are from three individual experiments unless noted otherwise. n=number of spindles observed.
Dmob1, Dmob2, Dmob3, Dmob4; Drosophila Mob1 (CG13852), Mob2 (CG11711), Mob3 (CG4946), Mob4 (CG3403), respectively. They have been identified as Drosophila Mob proteins based on homology to the Saccharomyces cerevisiae Mob1p (He et al., 2005).
Msps, minispindles protein; Asp, abnormal spindle protein.
γ-tubulin (23C) and γ-tubulin (37C): two isotypes of the γ-tubulin gene are present in Drosphila. The 23C isotype is expressed ubiquitously, whereas the 37C isotype is primarily expressed in the ovaries and in embryos.
Relative mitotic index: Percentage of nuclei that stained positive for phsphorylated histone H3 (a marker of mitotic cells), primarily scored using an automated microscope (see Materials and Methods). Numbers give the average fold increase in mitotic index for at least three separate RNAi experiments (mean ± s.d.) relative to the average mitotic index of untreated control cells (1.41±0.47, derived from untreated cells in three independent experiments). The average mitotic index of untreated cells from each experiment was determined from measurements of 90 separate wells.
P<0.005 (t-test), fold increase in mitotic index is statistically different from untreated cells.
As reported previously (Goshima and Vale, 2003), S2 cells often show aberrant mitotic spindle phenotypes. Some S2 cells form monopolar spindles that subsequently can be converted to a bipolar spindle by a rescue process in which microtubules become organized to form a second, acentrosomal pole (~10%). These monastral bipolar spindles are capable of entering anaphase and segregating chromosomes.
Spindles with localized γ-tubulin: Values are the mean ± s.d. of three experiments, except that for Dgrip223, which was obtained from a single experiment. n, number of mitotic cells.
+, protein reduction after RNAi confirmed using specific antibodies; −, not tested because antibodies were unavailable;
○, expression of these genes was at background levels in the S2 cell line by Affimatrix DNA microarray analysis (J. Hollien and J.Weissman, personal communication), suggesting that they are not expressed in this cell line at significant levels.
Data for Drosophila pericentrin are from RNAi experiments targeting Computed Gene (CG) 6735; however, RNAi targeting of alternative segments of the Drosophila pericentrin gene contained in CG18648 and CG13459 produced similar results.
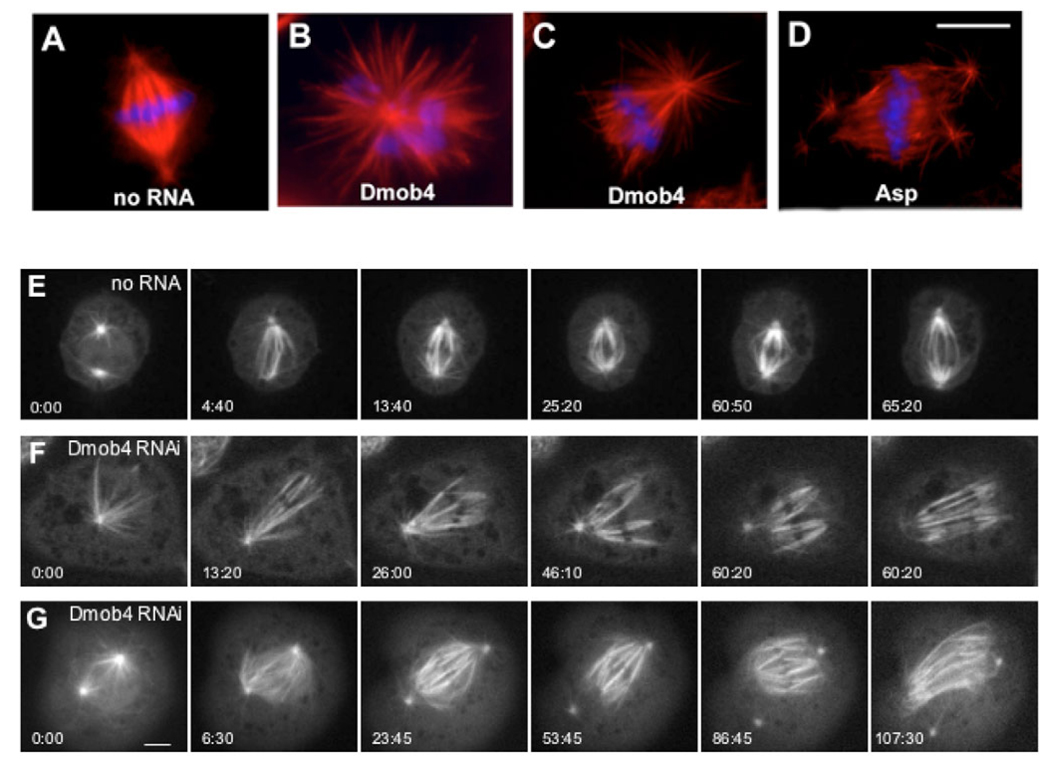
An interesting and unexpected outcome of this screen was obtained with RNAi targeting Drosophila Mob4, which has not been studied previously in Drosophila. This RNAi phenotype consisted of a high percentage of monopolar (32%; Fig. 1B) and poorly formed monastral bipolar (41%) and bipolar spindles (Fig. 1C), yet the mitotic index of the population was normal (Table 1). Mob proteins are implicated as regulators of kinases (Frenz et al., 2000). In Saccharomyces cerevisiae, Mob1p has been implicated in spindle pole body (SPB) duplication and the mitotic checkpoint through regulation of the Mps1p kinase (Fisk et al., 2004; Jones et al., 2005; Luca and Winey, 1998; Stucke et al., 2002). However, a role for a Mob protein in mitosis in higher eukaryotes has not been described. In Drosophila, there are four Mob-like proteins (25–40% similarity to S. cerevisiae Mob1p), including CG11711, the protein previously named in FlyBase as Mob1. During the course of our study, He et al. (He et al., 2005) reported similar findings, referring in their paper to CG13852 (mats or Dmob1), CG11711 (Dmob2), CG4946 (Dmob3) and CG3403 (Dmob4). Here, the Drosophila Mob proteins are referred to as Mob1, Mob2, Mob3 and Mob4. In our screen, only Drosophila Mob4 had an effect on spindle morphology, which is somewhat surprising as it is the family member most distantly related to yeast Mob1p.
Fig. 1.
Mitotic phenotypes observed after RNAi knockdown of Drosophila Mob4 (Dmob4) in S2 cells. Cells were treated with dsRNA and were fixed and stained with antibodies against tubulin (red) and with DAPI (blue) at day 4 (see Materials and Methods). (A) Untreated cells display bipolar spindles with well-defined centrosomes. (B) Mob4 RNAi cells show abnormally high numbers of monopolar spindles (32%, data not shown) as well as monastral bipolar spindles with splayed kinetochore minus ends (41%, data not shown). This latter defect resembled the frayed spindles that formed after Asp RNAi (D). Bar, 10 µm (A–D). Live imaging of GFP-tubulin revealed further details of the defects in spindle formation arising from Mob4 depletion (E–G). (E) Control cell (no RNA) with two centrosomes forming a bipolar spindle with spindle fibers organized normally at the poles. Time (in minutes and seconds) from the first frame is indicated in the lower left corner of each panel. The rightmost panel shows a cell in early anaphase (same in F and G), indicating the time of anaphase onset. (Full time-lapse movie available in supplementary material Movie 1.) (F) This Mob4-RNAi-treated cell initially formed a monopolar spindle that then converted to a monastral bipolar spindle. The acentrosomal pole on the right never becomes organized, and the left pole loses focus when the centrosome and K fibers begin to dissociate in prometaphase (4th panel). (Full time-lapse movie available in supplementary material Movie 2.) (G) Mob4-RNAi-treated cell with two centrosomes forming a bipolar spindle that becomes unorganized at both poles owing to detachment of K fibers from the centrosome and splaying during prometaphase/metaphase. (Full time-lapse movie available in supplementary material Movie 3.) Anaphase onset in Mob4-depleted cells occurs within the same time range observed in control cells. The K fiber splaying observed after Mob4 depletion resembles that described in live-cell imaging of cells depleted of the Asp protein (data not shown). Bar, 5 µm.
A particular feature of the Mob4-depleted cells was that their spindles had disorganized poles with splayed K fibers. A similar phenotype is also observed after depletion of abnormal spindle protein (Morales-Mulia and Scholey, 2005; do Carmo Avides and Glover, 1999; Saunders et al., 1997) (Fig. 1D), although the splaying of K fibers is more dramatic after RNAi of Asp. We reproduced this phenotype using dsRNA sequences corresponding to different parts of the mRNA sequence encoding Mob4 (supplementary material Fig. S3), indicating that the phenotype is specific and is not caused by an off-target RNAi effect. We thus focused our effort on characterizing the Mob4 RNAi phenotype.
Live-cell imaging of spindle formation after Mob4 RNAi reveals defects in maintenance of pole focusing
We next examined the dynamics of mitotic spindles after RNAi depletion of Mob4 using GFP-tubulin-expressing cells and time-lapse microscopy. In control cells (Fig. 1E and supplementary material Movie 1), K fibers appear rapidly and gather at the centrosomes after their minus ends are captured and transported along astral microtubules (Goshima et al., 2005; Khodjakov et al., 2003; Mahoney et al., 2006; Maiato et al., 2004). As noted in the referenced studies, wild-type cells frequently have three or more centrosomes at the beginning of mitosis, but these generally coalesce into two spindle poles during prometaphase. In Mob4-RNAi-treated cells, microtubules grew robustly from centrosomes and chromosomes following nuclear envelope breakdown (NEB), as in control cells, but we frequently observed two defects. First, many cells only had one microtubule organizing center (MTOC) present at the beginning of mitosis and initially formed a monopolar spindle, which is consistent with the results from immunofluorescence of fixed cells (Fig. 1B). However, this state was not permanent, as these spindles eventually assumed a bipolar shape through chromatin-mediated microtubule nucleation, followed by the reorganization of the microtubules into a second pole, creating a monastral bipolar spindle (Fig. 1F and supplementary material Movie 2) (see also Goshima and Vale, 2003). The second defect observed by time-lapse microscopy, again consistent with fixed-cell images, was that the spindle poles frequently became unfocused over time, as some or all K fibers detached from and splayed apart at the poles (Fig. 1G and supplementary material Movie 3). This defect in pole focusing was apparent in cells with either monastral or bipolar spindles. In some cells, the defect in pole focusing was dynamic and reversible, with K fibers rejoining the pole after a temporary detachment. But, in other cases, K fibers fully and permanently dissociated from the pole, leaving centrosomes completely unattached and free to drift away from the spindle pole (Fig. 1G). In several instances, we observed cells with centrosomes that actively nucleated microtubules but did not take part in the organization of the spindle (supplementary material Movie 4), suggesting a complete loss of crosslinking activity between K fibers and centrosomal microtubules. Regardless of the disruption in pole focusing, anaphase in Mob4-RNAi-treated cells occurred within the same time span as in control cells, and each set of separating chromatids remained in close enough proximity to form a single pronucleus in telophase. These live-cell observations are consistent with the normal mitotic index observed in our fixed-cell screen after Mob4 RNAi (Table 1).
Mob4 has a role in focusing of K fibers independent of centrosomes
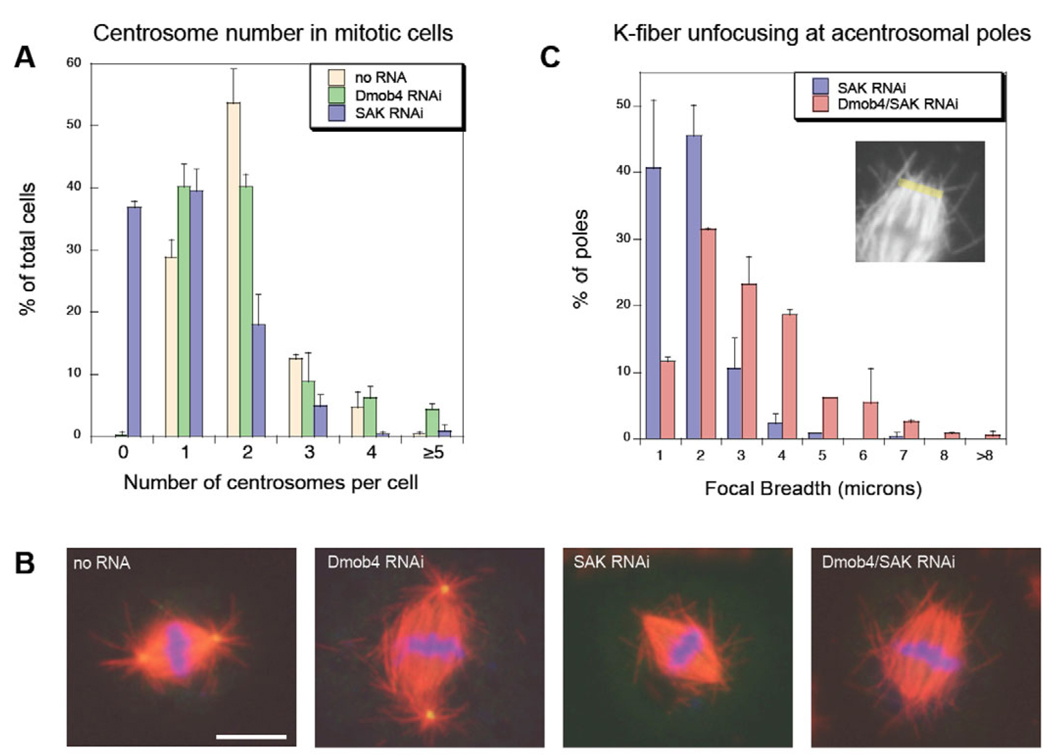
Live-cell imaging of mitotic spindle formation in cells depleted of Mob4 revealed a frequent loss of K fiber bundling at spindle poles with centrosomes or at acentrosomal poles of monastral bipolar spindles. We therefore reasoned that the primary role of Mob4 in pole focusing might be in K fiber self-assembly rather than in centrosome-mediated organization. To test this hypothesis, we analyzed K fiber unbundling in Mob4-RNAi-treated cells depleted of functional centrosomes by RNAi of SAK/PLK4, a polo-like kinase crucial for duplication of centrioles (Bettencourt-Dias et al., 2005). RNAi of SAK increased the frequency of cells with zero or 1 centrosome (Fig. 2A), as reported by Bettencourt-Dias and colleagues. In fixed-cell assays, SAK-depleted cells often had either a small, unorganized array of microtubules surrounding the condensed chromosomes, or bipolar spindles with aligned chromosomes but completely lacking astral microtubules (Fig. 2B). These spindle types were both present in cells co-depleted of Mob4 and SAK. However, the poles of the bipolar spindles were more disorganized and splayed apart than were poles after SAK RNAi treatment alone. We analyzed pole focusing quantitatively by measuring the width of the spindle fiber minus ends at these centrosome-free poles and found that SAK/Mob4 co-depletion increased the focal width by 2.5-fold compared with poles in cells depleted of SAK alone (histograms shown in Fig. 2C). Thus, Mob4 plays a role in K fiber focusing even in the absence of functional centrosomes.
Fig. 2.
Drosophila Mob4 (Dmob4) has a minor roll in the maintenance of centrosome number and functions in spindle pole organization in the absence of centrosomes. (A) Bars indicate the percentage of mitotic cells with 0, 1, 2, 3, 4 or ≥5 centrosomes (Dgrip84-stained foci that nucleate astral microtubules) in untreated cells (tan; n=401) or cells treated with RNAi to Mob4 (green; n=408) or SAK polo-like kinase (blue; n=400). (Error bars indicate s.d. of mean value of two independent experiments.) Depletion of Mob4 increased the proportion of single-centrosome cells relative to untreated cells but did not generate a significant population of acentrosomal cells, as did RNAi of SAK. (B) RNAi of SAK led to loss of centrosomes and astral microtubules from spindle poles, and co-depletion of Mob4 with SAK noticeably exacerbated spindle fiber unfocusing at acentrosomal poles (right). (α-tubulin, red; Dgrip84, green; DNA, blue; bar, 5 µm.) (C) Histogram showing the distribution of spindle pole focal breadth (yellow line; the distance between the minus ends of the outermost K fibers) at acentrosomal poles in SAK-depleted cells (blue) and in cells depleted of both Mob4 and SAK by double RNAi (red). The mean focal breadth in Mob4-SAK co-depleted cells was 5.0±2.4 µm (n=246), whereas that in cells depleted of SAK alone was 1.9±1.4 µm (n=261; P<0.0001). (Error bars indicate s.d. of mean value of two independent experiments.)
Mob4 and Asp exhibit similar RNAi phenotypes, but Mob4 is not required for Asp localization
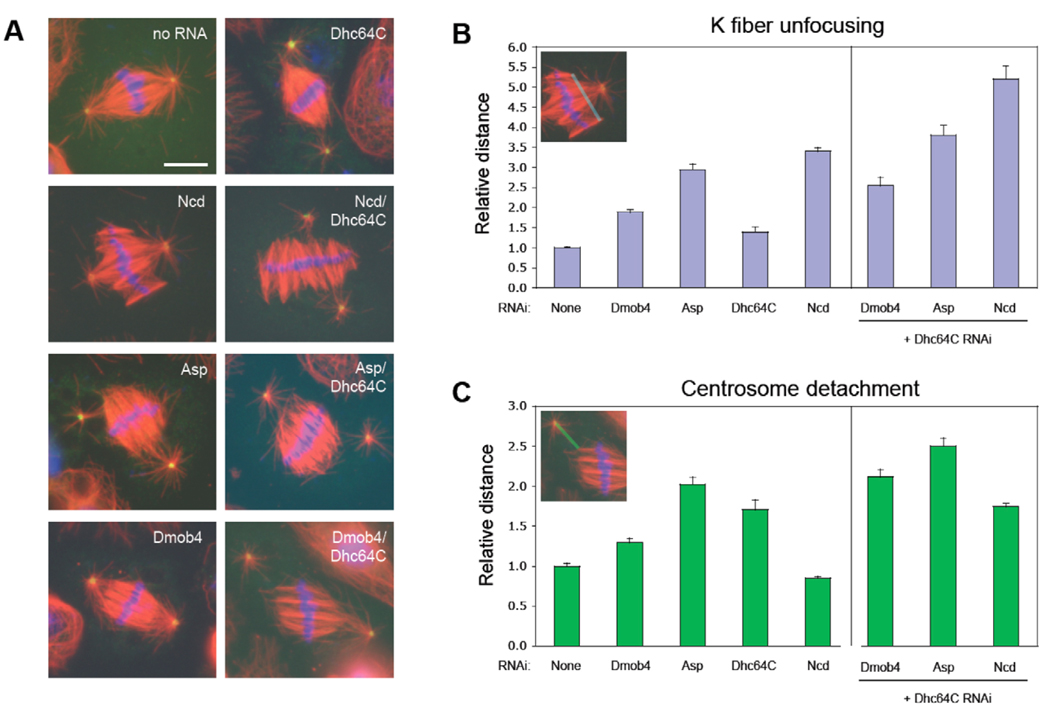
In addition to our present work on Mob4, three other proteins have been found to be required for spindle pole focusing in Drosophila cells: dynein, the kinesin-related protein Ncd (both minus-end-directed motor proteins) and Asp. To determine whether Mob4 function might be related to that of dynein, Ncd or Asp, we compared the RNAi spindle pole unfocusing phenotypes of each. As described in previous studies (Goshima et al., 2005; Maiato et al., 2004; Morales-Mulia and Scholey, 2005), RNAi of dynein heavy chain (Dhc64C) primarily disrupted centrosome attachment to K fiber minus ends, with less of an effect on K fiber focusing, whereas RNAi of Ncd generated the opposite phenotype; and depletion of Asp affected both parameters significantly (Fig. 3). The Mob4 RNAi phenotype showed an increase in both centrosome detachment and K fiber unfocusing (Fig. 3B,C). Thus, the spindle unfocusing RNAi phenotype of Mob4 more closely resembled that of Asp than of Ncd or dynein, showing both defective K fiber focusing and loss of centrosome attachment at the poles, although the magnitudes of these defects were greater for Asp than for Mob4. Double RNAi of Asp and Mob4 produced the same phenotype as depletion of Asp alone (data not shown).
Fig. 3.
Double RNAi experiments suggest that the Drosophila Mob4 (Dmob4) phenotype is more similar to Asp than dynein or Ncd. (A) Representative mitotic spindle morphology after RNAi of the genes indicated. (α-tubulin, red; Dgrip84, green; DNA, blue; bar, 5 µm.) (B) Quantitation of K fiber unfocusing in metaphase spindles. The relative mean width of K fiber minus ends – the distance between the minus ends of the outermost K fibers at each pole (blue line in inset) relative to control cells [average 2.08±0.03 µm (mean±s.d.)] – is shown after RNAi of the gene(s) indicated. RNAi of Mob4, Asp or Ncd induced significant increases in the focal width of K fibers relative to that of control cells, whereas Dhc64C had a minimal effect. Co-depletion of Dhc64C and Ncd had a strong synergistic effect, whereas the synergism in cells co-depleted of Dhc64C and either Asp or Mob4 was minimal. [Error bars indicate the s.d. of the average K fiber unfocusing distance measured in three independent experiments (n>45 spindles for each trial).] (C) Quantitation of centrosome detachment in the same spindles measured in (B). The distance of the gap between the centrosome (Dgrip84-staining foci that nucleate astral microtubules) and the minus end of the K fiber lying closest to the centrosome (green line in inset), relative to control cells [average 1.92±0.07 µm (mean± s.d)] is shown. RNAi of Mob4, Asp or Dhc64C, but not Ncd, caused an increase in centrosome detachment (statistically significant difference for Mob4 RNAi and control cells; P<0.0001). Co-depletion of Dhc64C and either Asp or Mob4 produced a synergistic increase in centrosome detachment, whereas no synergism was observed for co-depletion of Dhc64C and Ncd.
The quantitative measurements described above as well as time-lapse imaging suggests that the RNAi phenotype of Mob4 is most similar to that of Asp. Of the two minus-end-directed motors Ncd and dynein, the Mob4 RNAi phenotype was more similar to that of Ncd than dynein, as RNAi of Ncd (but not dynein) produced unfocusing of K fibers. To compare the Mob4 phenotype between Ncd and Asp more closely, we compared synergistic effects that might arise in double RNAi experiments with Dhc64C. As described earlier, Dhc64C RNAi caused centrosome detachment. This defect was magnified more by the co-depletion of Asp; in contrast, the combined effect of Ncd and Dhc64C RNAi on centrosome detachment was no greater than that of Dhc64C alone (Fig. 3C). However, the opposite effect was observed when K fiber focusing was examined. In this case, Ncd and Dhc64C RNAi synergize to produce significantly greater K fiber unfocusing than is seen for RNAi of either protein alone, while the combined effect of either Asp or Mob4 co-depletion with Dhc64C is only modestly increased relative to the unfocusing defect seen after RNAi of any of the three individually (Fig. 3B). Thus, the synergistic outcomes of double RNAi experiments with Dhc64C reveal similar trends for Asp and Mob4 compared with Ncd. Thus, the phenotypes observed for both single and double RNAi experiments suggest that the Mob4 phenotype more closely resembles that observed for Asp than Ncd.
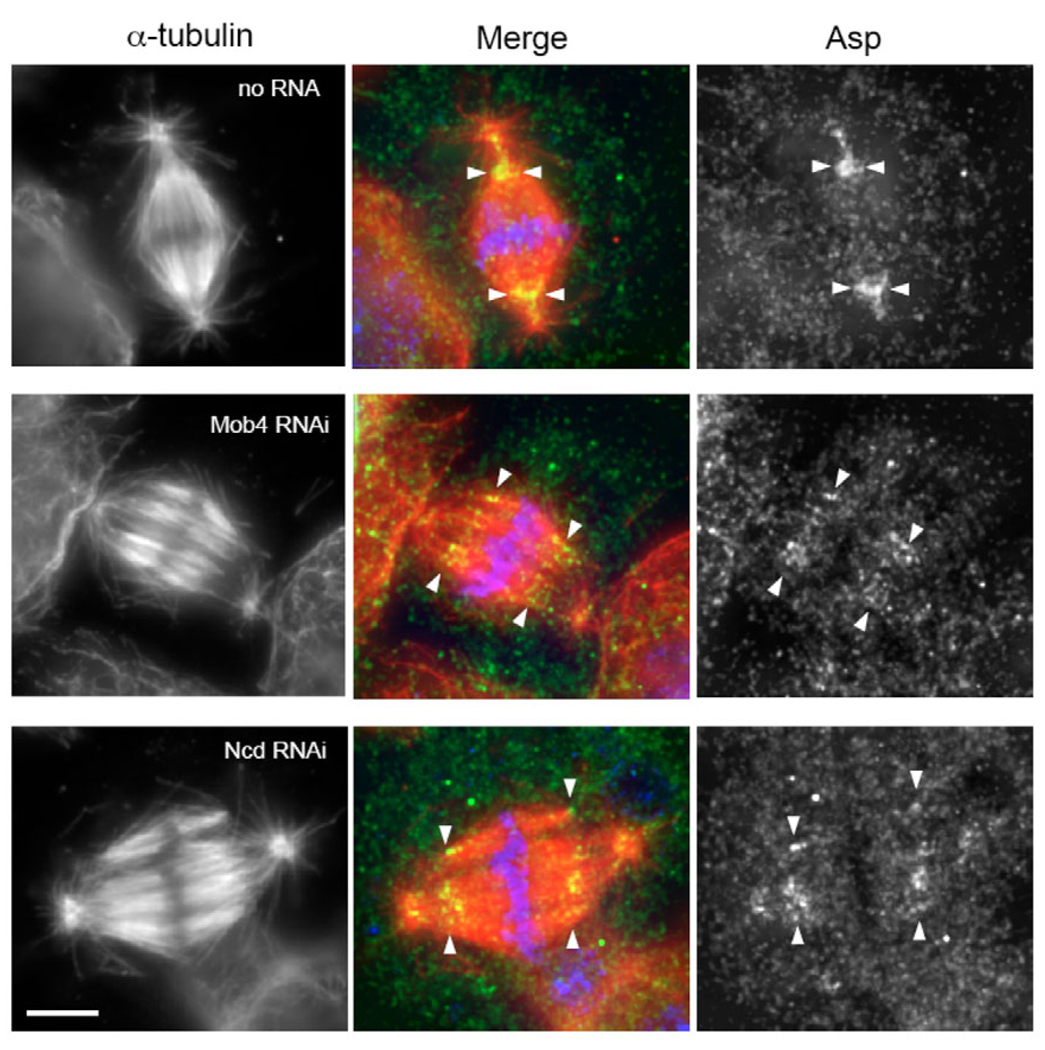
Because the RNAi phenotype of Mob4 most closely resembled that of Asp, we examined whether Mob4 is necessary for the localization of Asp to the minus ends of K fibers, the major site of microtubule crosslinking in the spindle (Morales-Mulia and Scholey, 2005; Saunders et al., 1997). After Mob4 RNAi treatment, we still observed Asp immunofluorescence at K fiber minus ends, as well as a small amount at the centrosome (Fig. 4). It was difficult, however, to compare the signal intensity with that of wild-type cells as the K fibers were unfocused after Mob4 RNAi. We therefore examined the intensity of Asp immunofluorescence after depletion of Ncd, which is required for K fiber focusing but has no known association with Asp. The Asp staining at the minus ends of K fibers in the splayed spindles of Ncd- and Mob4-RNAi-depleted cells was comparable. Therefore, although we cannot rule out a partial effect on Asp localization, Mob4 depletion does not severely disrupt the recruitment of Asp to K-fiber minus ends and the centrosome.
Fig. 4.
Drosophila Mob4 (Dmob4) is not required for proper localization of Asp. Asp (green) immunolocalizes to centrosomes (indicated by microtubule asters at the poles) and K fiber minus ends (stretching between arrowheads) in controls cells with organized poles (upper panels) as well as in cells depleted of Mob4 (middle panels) or Ncd (lower panels) by RNAi treatment. (α-tubulin, red; DNA, blue; bar, 5 µm.)
Mob4-GFP localizes to mitotic centrosomes and kinetochores
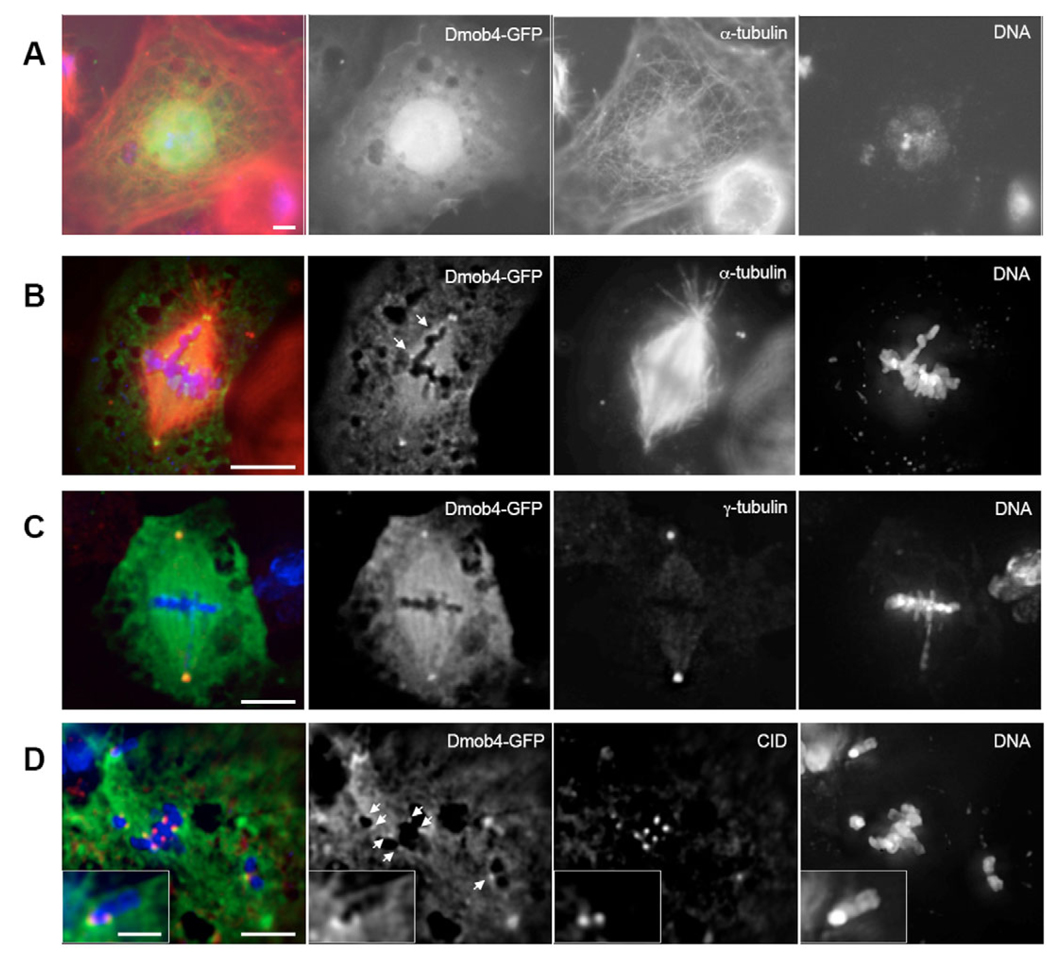
To learn more about the functions of Mob4, we made a stable S2 cell line expressing Mob4-GFP and visualized cells showing low levels of expression so as to determine best its cellular localization. In interphase cells, the GFP signal was diffuse through the cytoplasm but showed a clear enrichment in the nucleus (Fig. 5A). At prophase before NEB, a GFP signal could be observed at the centrosomes. After NEB, a bright GFP signal was visible at the spindle poles (Fig. 5B), where it colocalized with γ-tubulin (Fig. 5C), and was also visible above the background throughout the spindle. This localization differs from that of Asp, which is at the minus-end of K fibers and does not colocalize with γ-tubulin at the centrosome. We also observed several GFP foci at condensed chromosomes (Fig. 5B), which closely aligned with the centromere/inner kinetochore marker CID (the fly CENP-A homolog) (Fig. 5D). The Mob4-GFP signal was slightly offset away from CID, suggesting that Mob4 accumulates at the outer kinetochore.
Fig. 5.
Drosophila Mob4 fused to GFP (Dmob4-GFP) accumulates at mitotic spindle poles and kinetochores. (A) In interphase cells, Mob4-GFP (green in A–D) localizes primarily to the nucleus. (α-tubulin, red; DNA, blue for A and B.) (B) Mob4-GFP is diffuse throughout the cytoplasm in mitotic cells but accumulates both at spindle poles and at punctate spots immediately adjacent to condensed chromosomes (arrows in Mob4-GFP panel). (C) Mob4-GFP colocalizes with γ-tubulin staining (red) at the centrosome. (DNA, blue.) (D) Mob4-GFP foci adjacent to condensed chromosomes (arrows in Mob4-GFP panel) coincide with immunostaining for the fly CENP-A homolog CID (red). The spindle is oriented diagonally from upper left to lower right. The inset shows a magnified image of the chromosome pair at the upper left in the main panels, taken in an alternative Z section. Bar, 5 µm (2 µm for inset in D). (One CID pair does not appear to have a matching GFP-Mob4 pair because the fluorescent intensity of Rhodamine-stained CID was substantially larger than the GFP signal and bled onto focal planes in which the GFP signal was not visible.)
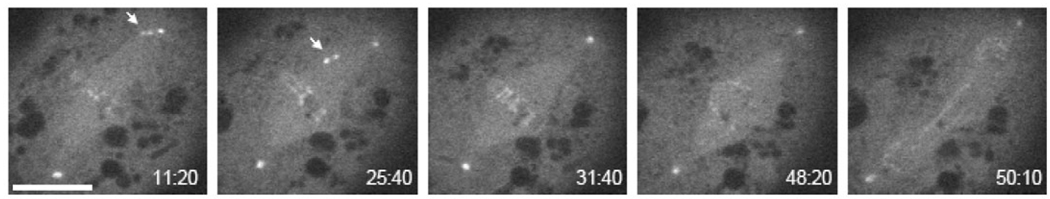
To observe Mob4 localization through mitosis, we performed time-lapse imaging of live cells expressing Mob4-GFP (Fig. 6 and supplementary material Movie 5). GFP fluorescence at centrosomes and the low level of signal throughout the spindle remained constant from prometaphase to anaphase. Kinetochore-localized Mob4-GFP, visible as bright pairs of spots adjacent to the silhouetted, condensed chromosomes, was observed before, during and after transport of the chromosomes to the metaphase plate, as exemplified by the mono-oriented chromosome visible in the left two panels of Fig. 6. This indicated that Mob4 is recruited to the kinetochore independent of whether the chromosome is attached to one or both poles. The kinetochore pair on the mono-oriented chromosome in Fig. 6 appeared to have brighter GFP fluorescence before alignment to the metaphase plate than after, suggesting that Mob4 accumulation at the kinetochore might diminish when the spindle checkpoint is satisfied. However, this effect was small and only observed in three of the five cells that were imaged from early prometaphase through anaphase. Thus, Mob4-GFP is found at several locations in the mitotic spindle and does not undergo pronounced changes in its localization at different stages of mitosis.
Fig. 6.
Time-lapse imaging of mitosis in cells expressing Drosophila Mob4 fused to GFP (Dmob4-GFP). Pairs of GFP foci indicating kinetochores are visible on both mono-oriented chromosomes (left two panels) and on chromosomes aligned at the metaphase plate. In some cases (such as the cell shown), kinetochore fluorescence of mono-oriented chromosomes was stronger before alignment at the metaphase plate than after, but otherwise GFP fluorescence at spindle poles and kinetochores is present throughout mitosis. The time in minutes and seconds from beginning of image acquisition is indicated at the lower right of each panel. Bar, 5 µm. The full time-lapse movie is shown in supplementary material Movie 5.
Discussion
Our mini RNAi screen of centrosomal proteins has uncovered an unreported phenotype associated with depletion of Mob4, consisting of an increased frequency of monopolar spindles and unorganized bipolar spindles that lack proper pole focusing. The other three Mob proteins did not produce a mitotic RNAi phenotype. The founding member of the Mob family was Mob1p from budding yeast, which interacts with Mps1p, a mitotic kinase required for SPB duplication and spindle checkpoint regulation (Luca and Winey, 1998), and Dbf2p, a multifunctional kinase that localizes to the spindle poles and is part of the mitotic exit network (Frenz et al., 2000). Phylogenetic analysis of the four Drosophila Mob proteins reveals that they belong to separate subfamilies of the Mob superfamily. Each Drosophila Mob protein is more similar to homologs in other species than to each other (supplementary material Fig. S4A). Mob4 shares nearly 80% sequence identity with human phocein (Hmob1, accession no. CAE45270) and an uncharacterized mouse protein (supplementary material Fig. S4B) and is very distantly related to S. cerevisiae Mob1p (18% identity; Mob1 and Mob3 are more closely related, with 44 and 33% identity to Mob1p, respectively). Phocein appears to function in peripheral ganglia and dendritic spines of many types of neurons, and, in unpolarized HeLa cells, it localizes to the Golgi (Baillat et al., 2002; Baillat et al., 2001; Moreno et al., 2001). The Mob4 branch of the Mob family is otherwise poorly characterized in the literature. Given the high degree of homology between Mob4 and phocein, it is striking that Mob4 does not appear to localize to the Golgi in Drosophila.
Mps1p kinase, regulated in budding yeast by Mob1p, is required for duplication of the SPB in yeast and of the centrosome in human cells (Fisk et al., 2003; Luca and Winey, 1998; Winey et al., 1991). The increased incidence of monopolar spindles after Mob4 RNAi might suggest it has a similar function to that of Mob1p, but we observed a normal number of centrosomes in prophase after Mob4 RNAi. Moreover, we could often discern two (or more) centrosomes at the center of a single monopolar aster in Mob4-depleted cells. Thus, our results indicate that Mob4 is involved in centrosome separation, but not duplication. Our results also indicate that Mob4 has a role in K fiber focusing that is separate from its role in centrosome separation. However, it is possible that our depletion of Mob4 protein is incomplete and that other centrosome defects might emerge with greater knockdown. In our fixed-cell screen, we observed an abundance of disorganized bipolar spindles, and time-lapse imaging of live cells revealed that this disorganization was the result of extensive K fiber detachment from the spindle poles. RNAi of Mob4 in cells depleted of functional centrosomes by SAK RNAi also produced a defect in K fiber focusing at acentrosomal poles. Thus, the Mob4 phenotype differs considerably from the only other known mitotic phenotype described for a Mob protein (spindle pole duplication defects associated with Mob1p mutations in yeast).
We also found that Mob4 localizes to kinetochores. However, Mob4-depleted cells did not display a change in mitotic index, normally indicative of a role in the spindle checkpoint or kinetochore assembly. Moreover, Mob4 depletion did not alter the kinetochore localization of either Rod or dynein (data not shown), two proteins that play roles in the spindle checkpoint (Karess, 2005). Dynein depletion also did not change the levels of Mob4-GFP on the kinetochores, which typically occurs with other checkpoint proteins. Therefore, the function of Mob4 at the kinetochore remains unknown.
Because other Mob family members regulate protein kinases, we postulate that Mob4 might similarly regulate one or more kinases that control the activities of spindle-organizing proteins. A possible candidate Mob4 target is Drosophila Mps1 kinase, the target of Mob1p in yeast (Castillo et al., 2002; Fischer et al., 2004; Fisk et al., 2004), but the RNAi phenotype that we observed for Mps1 is unlike that of Mob4 (Table 1). Mob1 and Mob2 also can bind to and activate NDR-family kinases in budding and fission yeast (Frenz et al., 2000; Hou et al., 2004; Komarnitsky et al., 1998; Mah et al., 2001; Weiss et al., 2002), human (hMob1 and hMob2) (Devroe et al., 2004) and flies (Mob1 and Mob2) (Giot et al., 2003; He et al., 2005; Lai et al., 2005). Interactions between Mob2 and the NDR kinases Tricornered (Trc) and Warts (Wts) have been reported, and the kinase-interacting residues in Mob2 are conserved in Mob4 (He et al., 2005), suggesting the possibility of a similar binding interaction. However, cells treated with dsRNA to either Trc or Wts did not show a noticeable spindle phenotype (data not shown). Thus, the target of Mob4 remains unknown and constitutes an important direction for future work. It is also possible that the phenotype is complex and might involve subtle interactions with multiple kinases or other target proteins that are not easily phenocopied by single RNAi knockdowns.
Our experiments suggest that the microtubule organizer Asp could be a downstream target of Mob4. However, the evidence is somewhat circumstantial, being based primarily on similarities in the RNAi phenotypes of Mob4 and Asp. Nevertheless, the Asp phenotype is rather unique even when the entire genome was explored in a large-scale RNAi screen (Goshima et al., 2007). The only other gene product with an RNAi phenotype virtually indistinguishable to that of Asp is calmodulin (Goshima et al., 2007), which probably binds to the multiple IQ motifs in Asp and is essential for Asp function. The human Mob4 homolog phocein also binds to striatin, a calmodulin-binding protein (Baillat et al., 2001), which again raises the possibility of some connection between Mob4 and calmodulin. However, an effect of Mob4 on Asp or calmodulin might not be direct as Mob4 does not colocalize with Asp or calmodulin (Goshima et al., 2007) in the spindle. Thus, regulation of Asp or calmodulin function by Mob4 through diffusable kinases (do Carmo Avides et al., 2001) represents a potential route for future investigation. Asp or calmodulin also cannot be the sole candidate target for Mob4 as the monopolar spindle defect observed after Mob4 RNAi is not characteristic of Asp or calmodulin RNAi. Identifying binding partners, target kinases and downstream phosphorylated proteins constitutes the next goal for further understanding how Mob4 functions in mitosis. In addition, as S2 cells constitute an immortalized cell line with a high percentage of abnormal mitoses (Goshima and Vale, 2003), it will be important to investigate the role of Mob4 in an organism by preparing Drosophila mutants.
Materials and Methods
Cell culture and RNAi
Drosophila Schneider cells (S2) were cultured and RNAi was performed as described previously (Goshima and Vale, 2003; Rogers et al., 2002). Templates for in vitro transcription were generated by PCR using specific primers shown in supplementary material Table S1. DsRNA was generated using the MEGAscript® T7 transcription kit (Ambion). The concentration of dsRNA was estimated by agarose gel and dsRNA was added to cell cultures in 96- or 24-well plates (1 or 5 µg per well, respectively). Cells were examined on day 4 or 5 and day 7 after RNAi treatment. Mob4-GFP (in pMT vector, Invitrogen) was expressed in S2 cells 2–3 days after transfection using Cellfectin (Invitrogen) or after hygromycin selection of stably transfected cells by the addition of 10–50 µM CuSO4 to the culture medium 24 hours before fixation and staining and/or visualization. Cells with low levels of GFP expression were selected for analysis. For fixation and staining, cells were plated in Con-A-coated 96-well imaging plates for 2–3 hours before analysis (Rogers et al., 2002).
Immunofluorescence microscopy and mitotic cell analysis
Cells were fixed in 3–6.4% formaldehyde in HL3 buffer (70 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, 5 mM HEPES, pH 7.2), permeabilized with 0.1% Triton X-100 in PBS (PBST) and then blocked with either 5% normal goat serum in PBST or 5% BSA in PBST. For Asp staining, cells were permeabilized in 0.5% SDS in PBS before treatment with PBST. Cells were then stained with DM1α (antibody against α-tubulin, 1:1000, Sigma-Aldrich) to label tubulin, antibody against phospho-histone H3 (1:300, Upstate) to label mitotic nuclei, and either GTU-88 (antibody against γ-tubulin, 1:1000, Sigma-Aldrich) or antibody against Dgrip84 (1:1000) to label centrosomes. After washing in PBST, cells were stained with secondary antibodies linked to Rhodamine Red-X or Cy2 (Jackson ImmunoResearch), washed again and mounted in ProLong Gold with DAPI mounting media (Invitrogen) to stain DNA. Specimens were imaged using a cooled CCD camera (Cooke Sensicam) mounted on an inverted microscope (Zeiss Axioplan 200M; Carl Zeiss MicroImaging), or using IXON cooled CCD cameras (Andor) in 16-bit conventional mode coupled to a wide-field fluorescent microscope developed in the Sedat Lab at UCSF. In the latter case, images were processed with constrained iterative deconvolution (Chen et al., 1996) using experimentally determined optical transfer functions specific for the objective used. The mitotic index was determined using the Cellomics Mitotic Index kit and imaged using a Cellomics ArrayScan (Cellomics). The mitotic index of untreated S2 cells varies between experiments (Goshima and Vale, 2003); therefore we included several control samples in every RNAi experiment. K fiber unfocusing and centrosome detachment distances were measured as described previously (Goshima et al., 2005). Briefly, the distance between the minus ends of the outermost K fibers at each pole (K fiber unfocusing distance) and the distance between the centrosome (Dgrip84-positive foci that nucleate astral microtubules) and the minus end of the K fiber lying closest to it (centrosome detachment distance) were measured in image software. RNAi of either Asp or Ncd frequently induces the formation of spindles with multiple asters and severe abnormalities, whereas RNAi of Dhc64C or Mob4 increases the occurrence of monopolar spindles, but we chose cells with an overall bipolar structure (chromosomes aligned on the metaphase plate) and two centrosomes for measurement.
Live imaging of GFP-tubulin
For time-lapse experiments, image sequences of microtubule dynamics were acquired using a previously characterized GFP-tubulin cell line with a constitutively active promoter (Goshima and Vale, 2003; Rogers et al., 2002) or a stable GFP-tubulin cell line with a metallothionein promoter (Mahoney et al., 2006). Cells were adhered on ConA-treated glass-bottom culture dishes (35 mm, Mattek). The majority of movies were collected at 10-second intervals with 50–200 msecond exposure times at room temperature using a cooled CCD camera Orca-ER2 (Hamamatsu) attached to a Yokogawa spinning-disk confocal scanhead (Solamere Technologies) that was mounted on a Zeiss Axiovert inverted microscope equipped with excitation and emission filter wheels (Sutter Instruments). Camera and AOTF were controlled by Metamorph software on a PC computer (Universal Imaging, Molecular Devices). Live images were also acquired using the Sedat laboratory microscope mentioned above, using 10–50 msecond exposure times at intervals of 10 seconds. Images in this case were acquired with IXON CCD cameras in 14-bit electron-multiplier gain mode.
Supplementary Material
Acknowledgments
We gratefully acknowledge the following people for providing us with antibodies: Jordon Raff (anti-D-TACC), David Glover (anti-Asp), Hiro Ohkura (anti-Msp proteins), Thomas Kaufman (anti-Cnn antibodies) and Gary Karpen (anti-CID). We are grateful to current and former members of the Vale laboratory, especially Gohta Goshima, Nico Stuurman, Steve Rogers, Eric Griffis, Sarah Goodwin, Julia Kardon, and Ursula Wiedemann, for extensive technical support and valuable discussions, and to Pete Carlton, Sebastian Haase and John Sedat for use of the OMX microscope and generous donation of time and energy in assistance with image acquisition and processing. N.M.M. was supported by a Damon Runyon Cancer Research postdoctoral award. Work was supported by the NIH and the Howard Hughes Medical Institute.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/121/8/1284/DC1
References
- Baillat G, Moqrich A, Castets F, Baude A, Bailly Y, Benmerah A, Monneron A. Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell. 2001;12:663–673. doi: 10.1091/mbc.12.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat G, Gaillard S, Castets F, Monneron A. Interactions of phocein with nucleoside-diphosphate kinase, Eps15, and dynamin I. J. Biol. Chem. 2002;277:18961–18966. doi: 10.1074/jbc.M108818200. [DOI] [PubMed] [Google Scholar]
- Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 2003;15:96–104. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Castillo AR, Meehl JB, Morgan G, Schutz-Geschwender A, Winey M. The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J. Cell Biol. 2002;156:453–465. doi: 10.1083/jcb.200111025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Devroe E, Erdjument-Bromage H, Tempst P, Silver PA. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 2004;279:24444–24451. doi: 10.1074/jbc.M401999200. [DOI] [PubMed] [Google Scholar]
- do Carmo Avides M, Glover DM. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 1999;283:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]
- do Carmo Avides M, Tavares A, Glover DM. Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat. Cell Biol. 2001;3:421–424. doi: 10.1038/35070110. [DOI] [PubMed] [Google Scholar]
- Fischer MG, Heeger S, Hacker U, Lehner CF. The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1. Curr. Biol. 2004;14:2019–2024. doi: 10.1016/j.cub.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc. Natl. Acad. Sci. USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. A field guide to the Mps1 family of protein kinases. Cell Cycle. 2004;3:439–442. [PubMed] [Google Scholar]
- Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J. Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollmann R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Emoto K, Fang X, Ren N, Tian X, Jan YN, Adler PN. Drosophila Mob family proteins interact with the related tricornered (Trc) and warts (Wts) kinases. Mol. Biol. Cell. 2005;16:4139–4152. doi: 10.1091/mbc.E05-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;91:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol. Cell. Biol. 2004;24:3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Huneycutt BJ, Pearson CG, Zhang C, Morgan G, Shokat K, Bloom K, Winey M. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah AS, Jang J, Deshaies RJ. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Goshima G, Douglass AD, Vale RD. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 2006;16:564–569. doi: 10.1016/j.cub.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr. Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 2000;149:851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S, Scholey JM. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell. 2005;16:3176–3186. doi: 10.1091/mbc.E04-12-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno CS, Lane WS, Pallas DC. A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 2001;276:24253–24260. doi: 10.1074/jbc.M102398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec F, Surrey T, Karsenti E. Self-organisation and forces in the microtubule cytoskeleton. Curr. Opin. Cell Biol. 2003;15:118–124. doi: 10.1016/s0955-0674(02)00014-5. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Prigent C, Glover DM, Giet R. Drosophila Nek2 protein kinase knockdown leads to centrosome maturation defects while overexpression causes centrosome fragmentation and cytokinesis failure. Exp. Cell Res. 2005;303:1–13. doi: 10.1016/j.yexcr.2004.04.052. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders RD, Avides MC, Howard T, Gonzalez C, Glover DM. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Rogers GC, Scholey JM. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vérollet C, Colombie N, Daubon T, Bourbon HM, Wright M, Raynaud-Messina B. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 2006;172:517–528. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL, Kurischko C, Zhang C, Shokat K, Drubin DG, Luca FC. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 2002;158:885–900. doi: 10.1083/jcb.200203094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J. Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.