Abstract
Background
Insect odorant receptors (ORs) are heteromers comprised of highly variable odorant-binding subunits associated with one conserved co-receptor. They are potential molecular targets for the development of novel mosquito attractants and repellents. ORs have been identified in the malaria mosquito, Anopheles gambiae, and in the yellow fever mosquito, Aedes aegypti. However, they are still unknown in the Southern house mosquito, Culex quinquefasciatus, which transmits pathogens that cause human diseases throughout the world, including West Nile Virus in the United States.
Methodology
We have employed a combination of bioinformatics, molecular cloning and electrophysiology approaches to identify and characterize the response profile of an OR in Cx. quinquefasciatus. First, we have unveiled a large multigenic family of one-hundred-fifty-eight putative ORs in this species, including a subgroup of conserved ORs in three mosquito species. Using the Xenopus oocytes expression system, we have determined the response profile of CquiOR2, an antennae-specific OR, which shares high identity with putative orthologs in Anopheles gambiae (AgamOR2) and Aedes aegypti (AaegOR2).
Conclusion
We show that CquiOR2 is highly sensitive to indole, an oviposition attractant for Cx. quinquefasciatus. The response profile of CquiOR2 expressed in Xenopus oocytes resembles that of an olfactory receptor neuron housed in the antennal short blunt-tipped sensilla (A2) of Cx. quinquefasciatus, which are natural detectors for oviposition attractants. This first Culex OR de-orphanized is, therefore, a potential molecular target for screening oviposition attractants.
Introduction
Insect odorant receptors (ORs), members of a highly divergent multigenic family [1], [2], are expressed in olfactory receptor neurons (ORNs) and housed in olfactory sensilla. Initially, insects ORs were hypothesized to be G-protein coupled receptors (GPCRs), but they have recently been shown to function as heteromeric ligand gated ion channels [3], [4], [5] comprised of at least one copy of a variable odorant-binding OR subunit along with at least one copy of an OR83-like co-receptor [6]. The release of the genome sequences of several species has paved the way for the identification of large families of ORs in different taxa. In mosquitoes, seventy-nine and one-hundred-thirty-one putative OR genes have been identified in Anopheles gambiae [7] and Aedes aegypti [8], respectively. Recently, a repertoire of fifty ORs from the malaria mosquito A. gambiae has been functionally characterized using both the “empty neuron” system of Drosophila melanogaster [9] and the Xenopus oocyte system [10] providing significant insight into the sense of smell in the malaria mosquito [11]. Interestingly, the authors identified ORs which responded strongly to human derived odorants and may be involved in host recognition.
The complete mapping of olfactory sensilla on the antennae [12], [13] and maxillary palps [14] of the Southern house mosquito Culex pipiens quinquefasciatus ( = Cx. quinquefasciatus) led to the identification of multiple functional classes of sensilla. Some of these sensilla harbor ORNs highly sensitive to nonanal [13], DEET detectors [15] and ORNs specialized for reception of oviposition attractants [16]. However, the characterization of odorant receptor proteins from the Southern house mosquito remains terra incognita. Hitherto, only one OR subunit, the OR83b-like co-receptor CquiOR7 has been characterized at the molecular level [17], but putative odorant-binding subunits are unknown.
In A. gambiae, AgamOR2 and AgamOR10 have been shown to respond to a narrow set of chemicals when expressed in heterologous systems [9], [10], [18], [19], including indole and 3-methylindole, which are oviposition attractants for Culex mosquitoes [16], [20], [21], [22], [23]. We have mined the genome of Cx. quinquefasciatus (The genome sequence of Culex pipiens quinquefasciatus; Culex Genome Consortium) in attempt to identify ORs likely to be involved in the detection of oviposition attractants. Using bioinformatics approaches we have now identified one-hundred-fifty-eight putative OR genes in the Cx. quinquefasciatus genome. Interestingly, two Culex genes, CquiOR2 and CquiOR10, are highly related to their putative orthologs in A. gambiae (AgamOR2, AgamOR10) and A. aegypti (AaegOR2, AaegOR10). We then hypothesized that these ORs are involved in the detection of oviposition attractants in Cx. quinquefasciatus. Here, we show that CquiOR2 expressed in Xenopus oocytes responds to indole and other oviposition attractants, similar to the response profile of a specific neuron housed in the blunt-tipped trichoid sensilla in Cx. quinquefasciatus antennae.
Results and Discussion
Identification of putative OR genes in Cx. quinquefasciatus
To explore the diversity of the OR family in the genome of Cx. quinquefasciatus, we have used the previously identified OR sequences from other dipteran species as probes to look for structurally similar proteins by Blast search [24]. Candidate sequences which displayed significant similarity have been manually screened for characteristic features of the OR family. All resulting proteins exhibited the presence of predicted multiple transmembrane domains, the main hallmark of the OR family. Additionally most candidates, when blasted in NCBI conserved domain database (CDD), exhibited the presence of characteristic motifs (pfam02949, pfam08395) conserved in the insect OR family. Finally, multiple alignments revealed the presence in most candidates of a conserved region near the C-terminus (Ser-Tyr-Ser or Ser-Tyr-Thr) also found in ORs from other dipteran species [8].
Homology searches combined with bioinformatics analysis allowed the identification of one-hundred-fifty-eight putative OR genes including previously identified CquiOR7 [17]. The number of Culex OR genes identified here is comparable to the previously characterized OR family from another Culicidae species, A. aegypti, which encompasses one-hundred-thirty-one putative OR genes [8]. We have confirmed full-length sequence annotations by cDNA cloning for two genes, CquiOR2 (XM_001864509/CPIJ014392) and CquiOR10 (XM_001844036/CPIJ002479) [17]. Accession numbers and structural features of one-hundred-fifty-eight putative OR genes identified in this study, including those originated from VectorBase automated annotations, are described in Table S1.
Comparative analysis of mosquito ORs
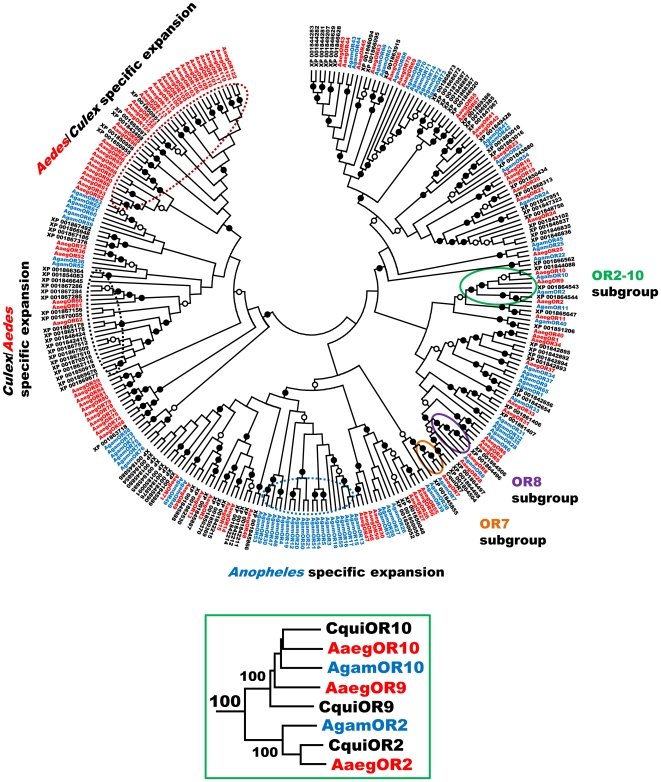
To obtain a better understanding of the relationships among mosquito ORs, we have carried out a phylogenetic analysis using putative amino acid sequences of ORs from three mosquito species, A. gambiae [7], [25], A. aegypti [8] and Cx. quinquefasciatus (this work). A sequence comparison tree reveals the existence of several species-specific lineages as well as different subgroups of conserved ORs within two or three mosquito species (Fig. 1). Focusing in this study on the most conserved OR genes in mosquitoes, we have identified four members of the OR2-OR10 clade in Cx. quinquefasciatus which share high identity with related ORs in both Anopheles and Aedes species (Fig. 1, inset). Three genes (XM_001864507/CPIJ014390; XM_001864508/CPIJ014391; XM_001864509/CPIJ014392) are found at close range on supercontig 3.258, as it has been observed with AaegOR2, AaegOR9 and AaegOR10 genes, which are also clustered together [8]. These findings suggest that these Culex and Aedes OR genes might be orthologs. Interestingly, another highly related gene (XM_001844036, CPIJ002479) was found on another genomic location (supercontig 3.32), suggesting a recent Culex specific duplication event. Of notice, XM_001864507 and XM_001844036 are both related to OR10 but only the latter was included for phylogenetic analysis as XM_001864507 includes at least three gaps in its predicted sequence.
Figure 1. Phylogenetic relationships of mosquito ORs.
Culex ORs are in black, Anopheles ORs are in blue and Aedes ORs are in red. Filled circles and empty circles represent 94–100% and 79–93% bootstrap support, respectively. The green box represents relationships in the conserved OR2-OR10 subgroup. CquiOR2 corresponds to (XM_001864509/XP_001864544), CquiOR9 to (XM_001864508/XP_001864543) and CquiOR10 to (XM_001844036/XP_001844088). Major species-specific expansions and conserved OR7 and OR8 subgroups are indicated.
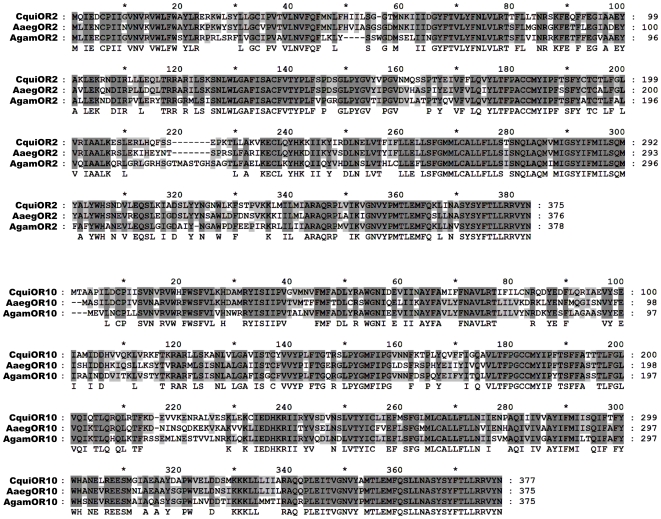
Among these four genes, XM_001864509 and XM_001844036 displayed the highest identity to AgamOR2/AaegOR2 and AgamOR10/AaegOR10, respectively, and were, therefore, named CquiOR2 and CquiOR10. Full-length sequences of these two genes were obtained by cDNA cloning and confirmed the predicted annotations, with only minor differences (four amino acid differences in CquiOR2 and a slightly different junction between exons 4 and 5 in CquiOR10). Comparative analysis revealed that, except for OR7 orthologs, OR2 and OR10 are the most conserved within identified ORs in three mosquito species. CquiOR2 shares 81% amino acid identity (91% a. a. similarity) and 70% (83%) with AaegOR2 and AgamOR2, respectively. Likewise, CquiOR10 shares 72% (87%) and 70% (84%) with AaegOR10 and AgamOR10, respectively. Alignment of CquiOR2 and CquiOR10 amino acid sequences with related proteins in the other two mosquito species is shown in Figure 2. Such high sequence conservation for ORs from different species is an interesting feature considering that in general ORs display a high level of divergence [8].
Figure 2. Alignment of mosquito OR2 and OR10 amino acid sequences.
Dark grey and light grey shading indicate residues conserved among three species and between two of the three species, respectively.
Several other subgroups of related ORs were identified based on sequence similarity and subsequent grouping in the tree (Fig. 1) but only one, the OR8 clade, displayed a similar level of conservation between species. Interestingly, we have identified two putative OR8 orthologs in Cx. quinquefasciatus (XM_001864471/CPIJ013954 and XM_001864461/CPIJ013944) which share around 65% and 75% identity with AgamOR8 and AaegOR8, respectively, based on predicted annotations. Recently, functional analysis showed that AaegOR8 acts as an enantioselective detector for (R)-(–)-1-octen-3-ol [26] and AgamOR8 was also shown to strongly respond to 1-octen-3-ol [9], [10], [19]. These findings suggest that putative OR8 orthologs in Cx. quinquefasciatus may be sensitive to 1-octen-3-ol, a compound which is detected with high sensitivity by sensilla involved in the reception of plant-derived compounds in the maxillary palps of Cx. quinquefasciatus [14].
CquiOR2 and 10 are exclusively expressed in olfactory tissues
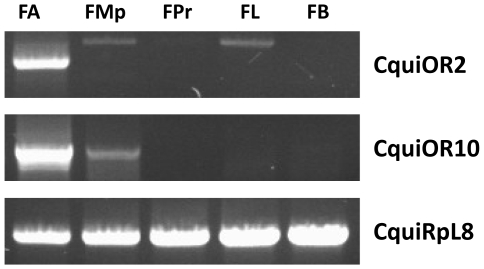
Expression patterns of CquiOR2 and CquiOR10 have been studied using RT-PCR and cDNA templates prepared from olfactory (antennae, maxillary palps and proboscis) and non-olfactory tissues (legs and bodies) of adult females. CquiOR2 was detected only in antennae, which is involved in the reception of oviposition attractants [13], [16]. On the other hand, CquiOR10 was detected in both antennae and maxillary palps, but not in non-olfactory tissues (Fig. 3). Corresponding PCR products were further cloned and sequenced to confirm CquiOR2 and CquiOR10 identities. Given that maxillary palps are involved in the reception of plant-volatile compounds and carbon dioxide [14], we reasoned that CquiOR2 is more likely to be involved specifically in the reception of oviposition attractants. Therefore, functional studies were focused on CquiOR2.
Figure 3. Expression profiles of two ORs in Cx. quinquefasciatus female tissues by RT-PCR.
Olfactory tissues: antennae (FA); maxillary palps (FMp); proboscis (FPr). Non olfactory tissues: legs (FL); bodies (FB). CquiRpL8 was used as control gene.
Functional expression of CquiOR2
Previously, AgamOR2 has been shown to strongly respond to indole, 3-methylindole, and 2-methylphenol [9], [10], [18], [19], whereas AgamOR10 has been shown to strongly respond to indole, 3-methylindole and 4-methylphenol [9], [10], [19]. Indoles and cresols have been demonstrated to function as oviposition attractants for Culex mosquitoes [16], [20], [21], [22], [23]. Displaying an expression profile restricted to antennae, CquiOR2 was selected for functional characterization in Xenopus oocytes to decipher its ligand specificity. Full-length coding sequence of CquiOR2, as well as CquiOR7 [17], the necessary OR83b-like co-receptor, were cloned into pGEMHE [27] for in vitro expression in Xenopus laevis oocytes.
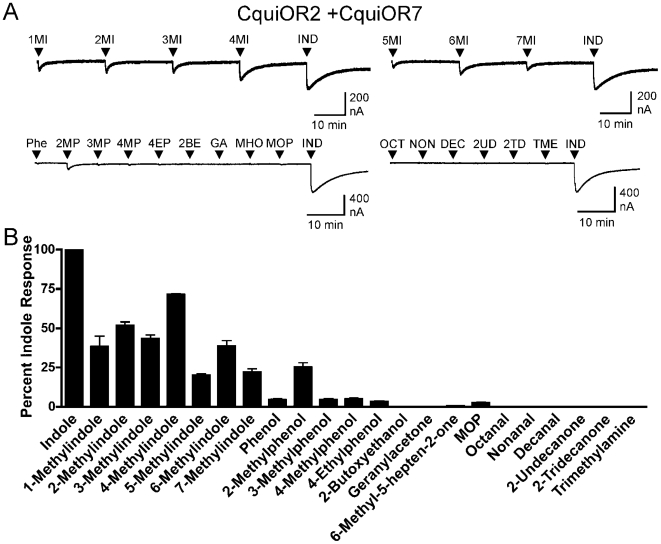
Oocytes expressing CquiOR2 + CquiOR7 were screened with a panel of compounds, each applied for 20 s at a concentration of 10 µM (Fig. 4). Indole elicited the largest current responses, but the receptor also responded well to each of the methylindoles and 2-methylphenol. To provide more detail about the sensitivity of the CquiOR2 + CquiOR7 receptor, we performed concentration response analysis for indole and each of the compounds that yielded responses that were at least 20% of the response to indole. Indole was the most potent of the tested compounds, activating the CquiOR2 + CquiOR7 receptor with an EC50 of 280 nM (Fig. 5, Table 1). The receptor was also activated by the oviposition attractants 3-methylindole and 2-methylphenol, with EC50 values of 20 µM and 7.3 µM, respectively (Fig. 5, Table 1). In addition, each of the other methylindoles were able to activate the receptor with EC50's ranging from 3.0 µM for 6-methylindole to 20 µM for 1-methylindole (Fig. S1, Table 1). Interestingly, while several of the compounds (1-methylindole, 2-methylindole, 3-methylindole) displayed relative efficacies (maximal responses) similar to that of indole, the other compounds (4-methylindole, 5-methylindole, 6-methylindole, 7-methylindole, 2-methylphenol) had lower relative efficacies than indole (Table 1).
Figure 4. CquiOR2 + CquiOR7 responds to indole, various methylindoles and 2-methylphenol.
Xenopus oocytes expressing CquiOR2 + CquiOR7 were challenged with a panel of odorant compounds, each applied for 20 s at 10 µM. A) Upper left trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with 1-methylindole (1MI), 2-methylindole (2MI), 3-methylindole (3MI), 4-methylindole (4MI) and indole (IND). Upper right trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with 5-methylindole (5MI), 6-methylindole (6MI), 7-methylindole (7MI) and indole (IND). Lower left trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with phenol (Phe), 2-methylphenol (2MP), 3-methylphenol (3MP), 4-methylphenol (4MP), 4-ethylphenol (4EP), 2-butoxyethanol (2BE), geranylacetone (GA), 6-methyl-5-hepten-2-one (MHO), mosquito oviposition pheromone (MOP) and indole (IND). Lower right trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with octanal (OCT), nonanal (NON), decanal (DEC), 2-undecanone (2UD), 2-tridecanone (2TD), trimethylamine (TME) and indole (IND). B) Quantification of current responses of CquiOR2 + CquiOR7 expressing receptors. All responses are normalized to the response of the same oocyte to 10 µM indole (mean ± SEM, n = 3–4).
Figure 5. CquiOR2 + CquiOR7 is highly responsive to indole.
A) Upper trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with 20 s applications of a range of concentrations of indole. Middle trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with 20 s applications of a range of concentrations of 3-methylindole. Lower trace, an oocyte expressing CquiOR2 + CquiOR7 is challenged with 20 s applications of a range of concentrations of 2-methylphenol. B) Concentration-response relationships for CquiOR2 + CquiOR7 expressing oocytes when activated with a range of indole, 3-methylindole and 2-methylphenol concentrations. All data are normalized to the response of each oocyte to 300 nM indole and the curves were fit as described in Materials and Methods (means ± SEM; n = 5–16). EC50 and nH values are provided in Table 1.
Table 1. Functional potencies and relative efficacies for activation of CquiOR2 + CquiOR7.
| Compound | EC50 (µM) | nH | Relative Efficacy |
| Indole | 0.28±0.12 | 0.8±0.2 | 100 |
| 1-Methylindole | 20±6 | 1.3±0.5 | 84±9 |
| 2-Methylindole | 7.5±4.1 | 0.9±0.4 | 106±16 |
| 3-Methylindole | 20±6 | 0.9±0.2 | 92±8 |
| 4-Methylindole | 3.6±0.8 | 1.0±0.2 | 60±4 |
| 5-Methylindole | 8.6±2.0 | 1.1±0.2 | 59±4 |
| 6-Methylindole | 3.0±0.6 | 0.9±0.2 | 69±4 |
| 7-Methylindole | 18±3 | 1.1±0.2 | 67±4 |
| 2-Methylphenol | 7.3±1.5 | 1.0±0.2 | 70±9 |
EC50 and Hill coefficient (nH) values were derived by fitting the data in Figure 5 and Figure S1 to a Hill equation (see Materials and Methods). Relative efficacies are the maximum values derived from fitting the data and are expressed as a percentage of the maximum value for indole. Values are the mean ± SEM derived from concentration-response data from 5–16 separate oocytes.
with response profiles recorded from Culex antennae
Previously, we have shown by direct electrophysiological recordings from Cx quinquefasciatus female antennae that indole elicits strong excitatory responses from a type of trichoid sensillum characterized by short length and blunt tip (See Fig. S4, in [13]). This sensillum, classified as type A2 after McIver [28], houses two ORNs as indicated by the spontaneous firing of two distinct amplitudes [13]. The neuron with a larger spike amplitude (ORN-A) was demonstrated to be highly sensitive to nonanal [13], an attractant for host-seeking females [13], which also elicits egg deposition by gravid females [16]. The neuron sensitive to indole, ORN-B, is characterized by spikes with smaller amplitude (See Fig. 4B,C in [13]). Of all the indole-related compounds tested individually, indole elicited the strongest dose-dependent responses (Fig. S4 in [13]). The response threshold was at least three orders of magnitude higher when the same sensilla were challenged with methyl derivatives of indole [13]. The indole-sensitive ORN-B also responded, albeit with lower sensitivity, to phenolic compounds, with 2-methylphenol (o-cresol) eliciting the strongest response. The response thresholds for 3- and 4-methylphenol (m- and p-phenol, respectively) were three orders of magnitude higher than that observed for 2-methylphenol (Fig. S4 in [13]).
Taken together these findings suggest that ORN-B in the blunt-tipped trichoid sensilla A2 in Cx. quinquefasciatus female antennae may express CquiOR2–an odorant receptor sensitive to Culex oviposition attractants. The electrophysiological responses elicited by stimulating CquiOR2-expressing Xenopus oocytes with indole were similar to the profiles obtained by single sensillum recordings from A2 [13]. Both responded to indole in a dose-dependent manner and with high sensitivity (low threshold). Additionally, 2-methylphenol elicited the best responses for phenolic compounds in the mosquito antennae and Xenopus oocytes. The natural and heterologous systems did differ in the degree of the selectivity for indole over the methylindoles. CquiOR2 expressed in the Xenopus oocyte system is 10- to 70-fold selective for indole over the various methylindoles, while single sensillum recordings from A2 show a three orders of magnitude selectivity for indole [13]. A possible explanation for this discrepancy is that odorant-binding proteins (OBPs) [29] may contribute to and enhance the selectivity of the mosquito olfactory system. Interestingly, we have recently demonstrated that knockdown of an OBP from Cx. quinquefasciatus, CquiOBP1, by RNA interference generated a phenotype with reduced electroantennographic (EAG) responses to indole and other oviposition attractants, but no significant changes in responses to nonanal [30]. These experiments with OBP gene silencing and heterologous expression of an OR sensitive to oviposition attractants suggest that OBPs may contribute to both selectivity and sensitivity of insect's olfactory system. Similarly, pheromone-binding proteins (PBPs) have been shown to contribute to the sensitivity [31] and selectivity [32] of moth's reception of sex pheromones. Thus, it is likely that both odorant receptors and odorant-binding proteins contribute to the remarkable selectivity and sensitivity of the insect's olfactory system.
Comparison with Anopheles gambiae ORs
Indole and 3-methylindole, demonstrated here to stimulate CquiOR2, were among the most narrowly tuned odorants in An. gambiae ([9], odorants #5 and #1, respectively). Furthermore, AgamOR2 [9], [10] was among the most narrowly tuned ORs, indicating it detects important chemical cues with high specificity. In brief, one of the most narrowly tuned ORs in An. gambiae, AgamOR2, activates in response to one of the most narrowly tuned odorants, indole. Therefore, it is conceivable that indole plays a significant role in Anopheles chemical ecology. Culex eggs are deposited in rafts of 200–250 eggs confined to small areas, offering an opportunity for Culex population control at this point of their life cycle. Consequently, oviposition behavior has been more thoroughly studied in Culex mosquitoes. Indeed, indole and 3-methylindole have been demonstrated in indoor bioassays and field tests to be attractants for gravid Culex mosquitoes [16], [20], [21], [22], [23]. An exciting area for future research will be to investigate the response profiles for the related receptor in Aedes aegypti, AaegOR2.
Conclusion
We have identified one-hundred-fifty-eight putative ORs in the genome of Cx. quinquefasciatus. Large scale annotations and/or cDNA cloning should now be performed to confirm the functionality of these putative ORs. Here we de-orphanized CquiOR2, which is sensitive to oviposition attractants. These findings open up new avenues for reverse chemical ecology-based approaches [33] aimed at the development of better oviposition attractants by using OBPs [16] as well as ORs as molecular targets.
Materials and Methods
Identification of putative OR sequences from Cx. quinquefasciatus genome
A predicted peptide sequences database of the whole genome of Cx. quinquefasciatus (CpipJ1.2 geneset) available at VectorBase (http://cpipiens.vectorbase.org/index.php) was entered into BioEdit v7.0.9.0 [34] to perform homology searches using Blastp algorithm [24]. Available OR sequences of two mosquito species, A. gambiae (seventy-nine sequences) [7] and A. aegypti (one-hundred-thirty-one sequences) [8] were used as queries in Blast searches. Candidates were further blasted in NCBI conserved domain database (CDD) to identify motifs conserved of the insect OR family (pfam02949: 7tm Odorant receptor and pfam08395: 7tm Chemosensory receptor). Presence of multiple transmembrane domains was predicted using TMHMM server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Multiple alignments and calculation of sequence identities and similarities were made using GeneDoc software (http://www.nrbsc.org/gfx/genedoc/ebinet.htm).
Phylogenetic analysis of mosquito ORs
Amino acid sequences of putative ORs from three mosquito species were combined to create an entry file for phylogenetic analysis in MEGA 4.0.2 [35]. An unrooted consensus neighbor joining tree was generated based on 1000 bootstrap replicates with pairwise gap deletions. Seventy-nine A. gambiae ORs, one-hundred-and-one A. aegypti ORs and one-hundred-and-three Cx. quinquefasciatus ORs were used in this study. Twenty-one pseudogenes (P) and nine incomplete sequences of A. aegypti were omitted (AaegOR12, 18, 22P, 29P, 32P, 35, 38P, 39, 51P, 53, 54P, 57P, 64P, 68P, 73P, 77P, 82P, 83P, 86, 108P, 112P, 116P, 118P, 120P, 126, 127, 128P, 129P, 130, 131P). Fifty-five putative Cx. quinquefasciatus ORs were omitted (see Table S1) as they were likely incomplete and/or looked to be partially wrongly annotated sequences when subjected to multiple alignments comparison. Nomenclature of A. gambiae and A. aegypti ORs used in this study follow the nomenclature established in [7] and [8], respectively.
Expression patterns and cloning of full-length CquiOR2 and CquiOR10
Cx. quinquefasciatus mosquitoes, tissues dissection, RNA extraction and cDNA synthesis were performed as described in [29] with minor modifications. Gene specific primers were designed based on gene annotations to amplify full-length coding sequences of CquiOR2 (XM_001864509/CPIJ014392) and CquiOR10 (XM_001844036/CPIJ002479). Antennae, maxillary palps, proboscis, legs and bodies (thorax and abdomen without head/legs) cDNAs from female adult mosquitoes (one-to-seven-day-old) were used as templates for tissue-specificity study. PCR reactions were carried out using equivalent amount of cDNA and one unit of GoTaq DNA polymerase (Promega, Madison, WI) in a final volume of 25 µl. Integrity of each cDNA template was confirmed by the amplification of a ribosomal L8 protein encoding gene (CquiRpL8, XM_001841875). For cloning, full-length sequences of CquiOR2 and CquiOR10, as well as the necessary co-receptor CquiOR7 (ABB29301) [17], were amplified from female antennal cDNA using Pfu Ultra II polymerase (Stratagene, La Jolla, CA). PCR products were purified using QIAquick Gel Extraction kit (Qiagen, Valencia, CA) and ligated into pBlueScript SK (+) (Stratagene). Ligation products were used to transform One Shot OmniMAX competent cells (Invitrogen, Carlsbad, CA) and positive clones were grown in LB medium containing ampicilline. Plasmids were purified using QIAprep Spin Miniprep kit (Qiagen) and sent for sequencing (Davis Sequencing Inc, Davis, CA). CquiOR2 and CquiOR10 cDNA sequences were deposited into GenBank. Accession numbers are GU945396 and GU945397 for CquiOR2 and CquiOR10, respectively.
flCquiOR2up: 5′-ATGCAGATCGAAGACTGCCCCAT-3′
flCquiOR2do: 5′-TTAGTTGTAAACACGACGCAGCA-3′
flCquiOR10up: 5′-ATGACCGCGGCACCCATTTTGGACT-3′
flCquiOR10do: 5′-TCAATTATAAACGCGTCTCAGCAGGG-3′
flCquiOR7up: 5′-ATGAACGTCCAGCCGACCAAGTAC-3′
flCquiOR7do: 5′-TTACTTCAGCTGCACCAACACCAT-3′
CquiRpL8: 5′-AGTCGTGAAGCACATCATCCACG-3′
CquiRpL8: 5′-GCCTTACCGATGTGCTGATGGTT-3
Expression of ORs in Xenopus Oocytes
Oocytes were surgically removed from mature Xenopus laevis frogs (Nasco). The care and use of Xenopus laevis frogs in this study were approved by the University of Miami Animal Research Committee and meet the guidelines of the National Institutes of Health. Follicle cells were removed by treatment with Collagenase B (Boehringer Mannhem) for 2 h at room temperature. CquiOR2 and CquiOR7 were transferred into pGEMHE [27]. Capped cRNA encoding each OR subunit was generated using mMessage mMachine kits (Ambion). 25 ng of cRNA encoding each OR subunit was injected into Stage V-VI Xenopus oocytes. Oocytes were incubated at 18°C in Barth's saline (in mM: 88 NaCl, 1 KCl, 2.4 NaHCO3, 0.3 CaNO3, 0.41 CaCl2, 0.82 MgSO4, 15 HEPES, pH 7.6, and 100 µg/ml amikacin) for 2–5 days prior to electrophysiological recording.
Electrophysiology and Data Analysis
Odorant-induced currents were recorded under two-electrode voltage clamp from oocytes expressing ORs, using an automated parallel electrophysiology system (OpusXpress 6000A; Molecular Devices). Oocytes were perfused with ND96 (in mM: 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, 5 HEPES, pH 7.5). Odorants were diluted in ND96 and applied for 20 s at a flow rate of 1.65 ml/min with extensive washing in ND96 (7–20 min at 4.6 ml/min) between applications. Current responses approached a plateau during the 20 sec application (Figure S2). Micropipettes were filled with 3 M KCl and had resistances of 0.2–2.0 MΩ. The holding potential was −70 mV. Current responses were filtered (4-pole, Bessel, low pass) at 20 Hz (-3 db), sampled at 100 Hz and were captured and stored using OpusXpress 1.1 software (Molecular Devices). Initial analysis of electrophysiological data was done using Clampfit 9.1 software (Molecular Devices). Curve fitting of concentration-response data was done using Prism 4 (Graphpad). Concentration-response data were fit to the equation: I = Imax/(1+(EC50/X)n) where I represents the current response at a given concentration of odorant, X; Imax is the maximal response; EC50 is the concentration of odorant yielding a half maximal response; n is the apparent Hill coefficient.
Supporting Information
Concentration-response analysis for methylindoles. The concentration-response relationships for CquiOR2 + CquiOR7 expressing oocytes when activated with a range of methylindole concentrations are shown. All data are normalized to the response of each oocyte to 300 nM indole and the curves were fit as described in Materials and Methods (means ± sem; n = 6–9). The data for indole is from Figure 5 and is shown for comparison. EC50 and nH values are provided in Table 1.
(1.60 MB TIF)
Kinetics of the response of CquiOR2 + CquiOR7 to 10 µM indole. An oocyte expressing CquiOR2 + CquiOR7 is challenged with a 20 s application of 10 µM indole (IND). Note that the response to indole approaches a plateau during the 20 second application. The response diminishes very slowly during washout, suggesting that the receptor is supersaturated and that indole is likely to be highly potent. This is borne out by the concentration-response data in Figure 5.
(1.07 MB TIF)
(0.22 MB DOC)
Acknowledgments
We thank lab members, particularly Dr. Zain Syed and Wei Xu, for enlightening discussions and Dr. Anthon Cornel for providing Culex quinquefasciatus mosquitoes for RNA extraction and cDNA synthesis. We thank Ana Castro for help with oocyte preparation.
Footnotes
Competing Interests: Dr. Walter Leal is an Editorial Board member.
Funding: This work was supported in part by National Science Foundation (0918177 to W.S.L), National Institutes of Health (DC008119 to C.W.L.), and a Cooperative Agreement with Bedoukian Research, Inc., but the funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, et al. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 2.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 3.Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452:1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- 4.Smart R, Kiely A, Beale M, Vargas E, Carraher C, et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol. 2008;38:770–780. doi: 10.1016/j.ibmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008;452:1007–1011. doi: 10.1038/nature06861. [DOI] [PubMed] [Google Scholar]
- 6.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298:176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 8.Bohbot J, Pitts RJ, Kwon HW, Rutzler M, Robertson HM, et al. Molecular characterization of the Aedes aegypti odorant receptor gene family. Insect Mol Biol. 2007;16:525–537. doi: 10.1111/j.1365-2583.2007.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey A, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odourant reception in the malaria mosquito Anopheles gambiae. Nature. 2010 doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Carey A, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci U S A in press. 2010 doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leal WS. The treacherous scent of a human. Nature. 2010;464:37–38. doi: 10.1038/464037a. [DOI] [PubMed] [Google Scholar]
- 12.Hill SR, Hansson BS, Ignell R. Characterization of antennal trichoid sensilla from female Southern house mosquito, Culex quinquefasciatus Say. Chem Senses. 2009;34:231–252. doi: 10.1093/chemse/bjn080. [DOI] [PubMed] [Google Scholar]
- 13.Syed Z, Leal WS. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proc Natl Acad Sci USA. 2009;106:18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed Z, Leal WS. Maxillary palps are broad spectrum odorant detectors in Culex quinquefasciatus. Chem Senses. 2007;32:727–738. doi: 10.1093/chemse/bjm040. [DOI] [PubMed] [Google Scholar]
- 15.Syed Z, Leal WS. Mosquitoes smell and avoid the insect repellent DEET. Proc Natl Acad Sci U S A. 2008;105:13598–13603. doi: 10.1073/pnas.0805312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leal WS, Barbosa RM, Xu W, Ishida Y, Syed Z, et al. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS ONE. 2008;3:e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Y, Zwiebel LJ. Identification and characterization of an odorant receptor from the West Nile virus mosquito, Culex quinquefasciatus. Insect Biochem Mol Biol. 2006;36:169–176. doi: 10.1016/j.ibmb.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallem EA, Nicole Fox A, Zwiebel LJ, Carlson JR. Olfaction: mosquito receptor for human-sweat odorant. Nature. 2004;427:212–213. doi: 10.1038/427212a. [DOI] [PubMed] [Google Scholar]
- 19.Xia Y, Wang G, Buscariollo D, Pitts RJ, Wenger H, et al. The molecular and cellular basis of olfactory-driven behavior in Anopheles gambiae larvae. Proc Natl Acad Sci U S A. 2008;105:6433–6438. doi: 10.1073/pnas.0801007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du YJ, Millar JG. Electroantennogram and oviposition bioassay responses of Culex quinquefasciatus and Culex tarsalis (Diptera: Culicidae) to chemicals in odors from Bermuda grass infusions. J Med Entomol. 1999;36:158–166. doi: 10.1093/jmedent/36.2.158. [DOI] [PubMed] [Google Scholar]
- 21.Millar JG, Chaney JD, Mulla MS. Identification of oviposition attractants for Culex quinquefasciatus from fermented Bermuda grass infusions. J Am Mosq Control Assoc. 1992;8:11–17. [PubMed] [Google Scholar]
- 22.Mordue AJ, Blcakwell A, Hansson BS, Wadhams LJ, Pickett JA. Behavioural and electrophysiological evaluation of oviposition attractants for Culex quinquefasciatus Say (Diptera: Culicidae). Experientia. 1992;48:1109–1111. [Google Scholar]
- 23.Olagbemiro TO, Birkett MA, Mordue Luntz AJ, Pickett JA. Laboratory and field responses of the mosquito, Culex quinquefasciatus, to plant-derived Culex spp. oviposition pheromone and the oviposition cue skatole. J Chem Ecol. 2004;30:965–976. doi: 10.1023/b:joec.0000028461.86243.19. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci U S A. 2001;98:14693–14697. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohbot JD, Dickens JC. Characterization of an enantioselective odorant receptor in the yellow fever mosquito Aedes aegypti. PLoS ONE. 2009;4:e7032. doi: 10.1371/journal.pone.0007032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 28.McIver S, Charlton C. Studies on the sense organs on the palps of selected culicine mosquitoes. Can J Zool. 1970;48:293–295. doi: 10.1139/z70-048. [DOI] [PubMed] [Google Scholar]
- 29.Pelletier J, Leal WS. Genome analysis and expression patterns of odorant-binding proteins from the Southern House mosquito Culex pipiens quinquefasciatus. PLoS One. 2009;4:e6237. doi: 10.1371/journal.pone.0006237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J Chem Ecol. 2010;36:245–248. doi: 10.1007/s10886-010-9762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Syed Z, Ishida Y, Taylor K, Kimbrell DA, Leal WS. Pheromone reception in fruit flies expressing a moth's odorant receptor. Proc Natl Acad Sci U S A. 2006;103:16538–16543. doi: 10.1073/pnas.0607874103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forstner M, Breer H, Krieger J. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int J Biol Sci. 2009;5:745–757. doi: 10.7150/ijbs.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leal WS. Pheromone reception. Top Curr Chem. 2005;240:1–36. [Google Scholar]
- 34.Hall TA. BioEdit:a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 35.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentration-response analysis for methylindoles. The concentration-response relationships for CquiOR2 + CquiOR7 expressing oocytes when activated with a range of methylindole concentrations are shown. All data are normalized to the response of each oocyte to 300 nM indole and the curves were fit as described in Materials and Methods (means ± sem; n = 6–9). The data for indole is from Figure 5 and is shown for comparison. EC50 and nH values are provided in Table 1.
(1.60 MB TIF)
Kinetics of the response of CquiOR2 + CquiOR7 to 10 µM indole. An oocyte expressing CquiOR2 + CquiOR7 is challenged with a 20 s application of 10 µM indole (IND). Note that the response to indole approaches a plateau during the 20 second application. The response diminishes very slowly during washout, suggesting that the receptor is supersaturated and that indole is likely to be highly potent. This is borne out by the concentration-response data in Figure 5.
(1.07 MB TIF)
(0.22 MB DOC)