Abstract
Molecular genetic data provide powerful tools for genealogy reconstruction to reveal mechanisms underlying disease ecology. White-tailed deer (Odocoileus virginianus) congregate in matriarchal groups; kin-related close social spacing may be a factor in the spread of infectious diseases. Spread of chronic wasting disease (CWD), a prion disorder of deer and their cervid relatives, is presumed to be associated with direct contact between individuals and by exposure to shared food and water sources contaminated with prions shed by infected deer. Key aspects of disease ecology are yet unknown. DNA tools for pedigree reconstruction were developed to fill knowledge gaps in disease dynamics in prion-infected wild animals. Kinship indices using data from microsatellite loci and sequence haplotypes of mitochondrial DNA were employed to assemble genealogies. Molecular genealogy tools will be useful for landscape-level population genetic research and monitoring, in addition to epidemiologic studies examining transmission of CWD in captive and free-ranging cervids.
Résumé
Les données de génétique moléculaire sont des outils puissants pour la reconstruction généalogique afin de révéler les mécanismes sous-jacents à l’écologie des maladies. Les cerfs de Virginie (Odocoileus virginianus) se rassemblent en groupes matriarcaux; le rapprochement social d’animaux apparentés dans un espace restreint pourrait être un facteur dans la transmission de maladies infectieuses. La transmission de la maladie débilitante chronique (CWD), une maladie à prion des cerfs et des cervidés apparentés, est présumée être associée à des contacts directs entre les individus et par exposition à des aliments partagés et des sources d’eau contaminées par des prions excrétés par des cerfs infectés. Les aspects importants de l’écologie de la maladie sont encore inconnus. Les outils génétiques pour la reconstruction des pedigrees ont été développés afin de combler les manques de connaissance dans la dynamique de l’infection chez les animaux sauvages infectés par les prions. Les indices de parenté utilisant les données des loci des microsatellites et la séquence des haplotypes de l’ADN mitochondrial ont été utilisés pour former les généalogies. Les outils moléculaires en généalogie seront utiles pour la recherche et le suivi de la génétique des populations sur le terrain, en plus de permettre les études épidémiologiques examinant la transmission de CWD chez les cervidés sauvages et en captivité.
(Traduit par Docteur Serge Messier)
Molecular genetics and computational tools now allow visualization of relationships in wildlife systems that was not previously possible. One exciting application is genealogy reconstruction to reveal patterns and mechanisms underlying disease ecology in individuals of unknown ancestry. Emerging statistical methods for kinship analysis will provide effective new tools for the study of wildlife epidemiology (1). Analyses of genotypic DNA data from highly polymorphic markers provide the means to disclose not only parent-offspring and sibling relationships, but also secondary and tertiary family relationships such as half siblings, niece-aunt, and grandparent-grand offspring (2).
Chronic wasting disease (CWD) is a transmissible spongiform encephalopathy (TSE) or prion accumulation disorder of cervid ruminants in the United States and Canada (3). The TSEs are a novel group of disorders, in which the transmissible agent is a mis-folded isoform of the host cellular prion protein (4). Although the routes of transmission under natural conditions are not known, a transmissible agent is detected in saliva of experimentally infected deer (5) and there is evidence for indirect transmission and persistent environmental contamination by the infectious agent (6). Relative disease susceptibility and/or incubation period have been correlated with prion gene (PRNP) polymorphisms encoding amino acid substitutions at residue 96 for white-tailed deer (7). Dams dying of CWD on elk farms posed an increased CWD risk factor to calves (8). Because deer congregate in matriarchal groups with dispersal of males at adolescence (9), CWD may follow transmission patterns aligning within kin lines. These relationships require clarification to explain the dynamics of CWD in natural populations.
We developed a set of molecular genealogy reconstruction tools to analyze family relationships in deer from areas of high CWD prevalence. Genotypes from 38 nuclear microsatellite loci and mitochondrial sequences from 135 white-tailed deer were analyzed using statistical genetic algorithms to reconstruct genealogical relationships. Samples included 133 deer from a previously described CWD-infected semi-free-ranging herd (7) and 2 additional deer not previously described from the herd. Kin groups inferred from DNA analyses were assessed for association of CWD pathology and relatedness.
Sequence analysis of 418 bp of the 5′-peripheral domain of the mitochondrial control region extending from the tRNAPro gene through TAS1 bordering the central conserved region (10) was performed. Sequences were confirmed using Sequencher software (Gene Codes Corporation, Ver. 4.7; Ann Arbor, Michigan, USA) then aligned. Haplotypes were characterized with TCS (Version 1.21). Variation at 48 single nucleotide sites and 1 insertion site of the mitochondrial control region revealed 19 haplotypes (GenBank accession numbers EF644627-EF644645), including 11 haplotypes each containing at least 2 individuals. There was no evidence of nuclear copies of the control region: replicate sequences from DNA yielded identical sequences. There was a single base pair insertion at position 21 in Haplotypes 2 and 16.
Thirty-eight microsatellite loci were polymerase chain reaction (PCR) amplified and included a panel of 18 loci routinely used for white-tailed deer parentage analysis (ADCYC, AGLA226, BL42, BM203, BM4107, BM4208, BM6438, BM6506, CERVID1, CERVID2, CP026, ETH152, FCB193, JP15, JP38, B9, SRCRSP1, and TGLA94) (11,12–18). These loci were augmented by 21 loci previously developed for mule deer (19), designated as A, B, C, D, E, F, G, H, J, K, L, M, N, O, P, Q, R, S, T, V, and D1 and optimized in our laboratory for white-tailed deer. Products from PCR were electrophoretically separated with a 3730 DNA Analyzer (Applied Biosystems, Foster City, California, USA), then visualized and scored. Of the 38 microsatellite loci tested, 28 amplified consistently, conformed to Hardy-Weinberg and linkage equilibrium expectations, and displayed < 10% probability of null alleles (ADCYC, AGLA226, BL42, BM4107, CERVID1, CERVID2, ETH152, FCB193, GNZ106, SRCRSP1, TGLA94, A, C, E, F, H, J, K, L, M, O, P, Q, R, S, T V, and D1). The PRNP locus did not deviate from expectations of linkage equilibrium with the microsatellite loci. Deer did not exhibit evidence of inbreeding or population substructure: HE = 0.71, FIS = −0.0007, average relatedness (standard deviation), R = 0.03 (0.06).
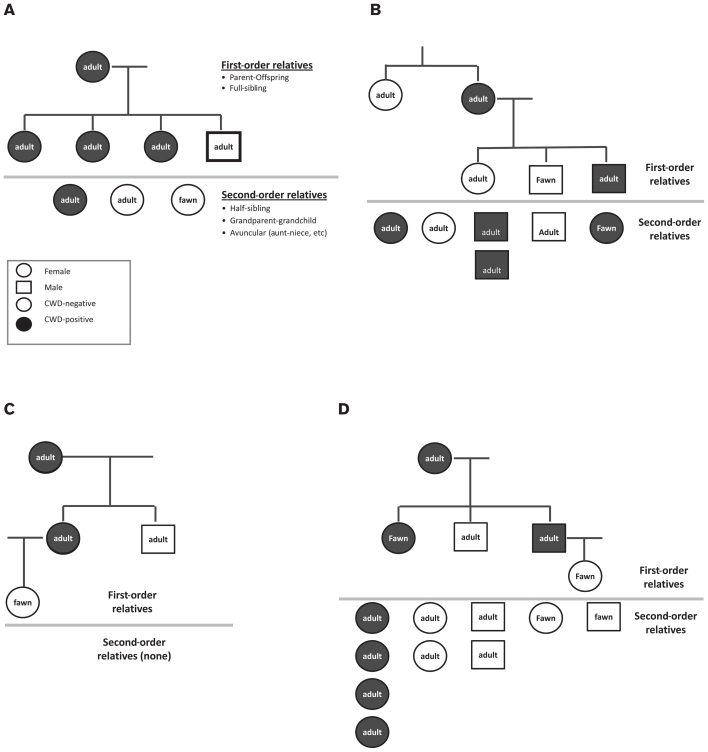
Of all possible pairs of deer, 47 dyads qualified as first order relatives using a maximum likelihood relatedness analysis of microsatellite and mitochondrial DNA data and yielded 24 parent-offspring and 23 full sib pairs (Table I). Four kin-groups containing at least 3 first-order relatives were identified and matched with qualifying second-order relatives of the same mitochondrial haplotype (Figure 1A–D; note that only a subset of the parent-offspring and full sib dyads of Table I are shown in the figure). Of the 47 dyads of first-order relatives, 14 were co-infected with CWD (9 parent-offspring dyads and 5 full sibling dyads; Table I). Four of the 19 mitochondrial haplotype groups contained at least 4 adult females; 1 of these 4 haplotype groups displayed a greater than expected proportion of CWD-positive adult females (91%; versus prevalence of 75% for female adults tested in this study). The 2 mitochondrial haplotype groups that contained at least 4 adult males had CWD proportions of 47% and 50% (as compared with 40% for all male adults in this study). No haplotypes contained at least 4 fawns.
Table I.
Dyads of deer qualified as first order relatives using microsatellite and mitochondrial DNA analyses. Dyads with both members infected in parentheses. Fawns include both sexes. Chronic wasting disease prevalence of individuals tested in this study (n = 135) was 0.75 for adult females, 0.40 for adult males, and 0.13 for fawns
| Relationship | Total | Dyads of adult female adult female | Dyads of adult female adult male | Dyads of adult male adult male | Dyads of adult female fawn | Dyads of adult male fawn | Dyads of fawn fawn |
|---|---|---|---|---|---|---|---|
| Parent-offspring | 24 (9) | 5 (4) | 6 (2) | 0 (0) | 12 (3) | 1 (0) | 0 (0) |
| Full sibling | 23 (5) | 6 (4) | 6 (0) | 2 (0) | 2 (0) | 4 (1) | 3 (0) |
| First order relative | 47 (14) | 11 (8) | 12 (2) | 2 (0) | 14 (3) | 5 (1) | 3 (0) |
Figure 1.
A, B, C, and D. Four reconstructed pedigrees with at least 3 first-order relatives (parent-offspring or full siblings). Individuals above the gray line represent first-order relatives. Parent-offspring relationships depicted by vertical lines. Full siblings are represented by individuals connected by horizontal lines. Individuals below the gray line correspond to matrilineal second-order relatives (half siblings, aunt-niece, etc.) as determined by shared mitochondrial haplotypes and kinship analysis of genotypes from 28 microsatellites. Filled symbols represent individuals testing CWD positive; empty symbols are CWD-negative individuals. Circles are females and squares are males. Note that not all dyads listed in Table I are represented in the figure.
Spatial pattern of kinship, likely a key factor influencing CWD dynamics in free-ranging deer populations, is critically important to understanding the ecology of CWD. These molecular tools will aid in establishing control programs to reduce risk of CWD transmission. Future molecular genetic work to address these factors across scales of population densities will clarify the patterns and mechanisms of chronic wasting disease in deer.
Acknowledgments
We thank D. Anderson, T. Gilliland, L. Gordon, and C. M. T. Penedo at UC Davis Veterinary Genetics Laboratory and A. Lyda, C. Durfee, and L. Hamburg at the USDA Agricultural Research Service (ARS) for technical assistance. This work was supported in part by support from the UC Davis Veterinary Genetics Laboratory and grants from University of California Genetics Resource Conservation Program and the USDA ARS (5348-32000-021-00D). Tissues were collected by the Nebraska Game & Parks (NGP) Commission and data on age and sex were provided by B. Morrison, D. Oates, and J. Boner, NGP. CWD diagnostic testing was performed by T. R. Spraker, Colorado Veterinary Diagnostic Laboratory, and E. S. Williams, Wyoming Veterinary Diagnostic Laboratory. Roger Sahara of the Nebraska Department of Agriculture provided information on the origin and management of the animals in this study.
Footnotes
Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that may be suitable. This article is the material of the US government.
References
- 1.Weir BS, Anderson AD, Hepler AB. Genetic relatedness analysis: Modern data and new challenges. Nature Rev Genet. 2006;7:771–780. doi: 10.1038/nrg1960. [DOI] [PubMed] [Google Scholar]
- 2.Blouin MS. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol Evol. 2003;18:503–511. [Google Scholar]
- 3.Williams ES. Chronic wasting disease. Vet Path. 2005;42:530–549. doi: 10.1354/vp.42-5-530. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB. Shattuck lecture — Neurodegenerative diseases and prions. N Engl J Med. 2001;344:1516–1526. doi: 10.1056/NEJM200105173442006. [DOI] [PubMed] [Google Scholar]
- 5.Mathiason CK, Powers JG, Dahmes SJ, et al. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science. 2006;314:133–136. doi: 10.1126/science.1132661. [DOI] [PubMed] [Google Scholar]
- 6.Mathiason CK, Hays SA, Powers J, et al. Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS One. 2009;4:e5916. doi: 10.1371/journal.pone.0005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Rourke KI, Spraker TR, Hamburg LK, et al. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004;85:1339–1346. doi: 10.1099/vir.0.79785-0. [DOI] [PubMed] [Google Scholar]
- 8.Argue CK, Ribble C, Lees VW, et al. Epidemiology of an outbreak of chronic wasting disease on elk farms in Saskatchewan. Can Vet J. 2007;48:1241–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Kie JG, Bowyer RT. Sexual segregation in white-tailed deer: Density-dependent changes in use of space, habitat selection, and dietary niche. J Mamm. 1999;80:1004–1020. [Google Scholar]
- 10.Purdue JR, Oleksyk TK, Smith MH. Independent occurrences of multiple repeats in the control region of mitochondrial DNA of white-tailed deer. J Hered. 2006;97:235–243. doi: 10.1093/jhered/esj032. [DOI] [PubMed] [Google Scholar]
- 11.Georges M, Massey J. Polymorphic DNA markers in Bovidae. Geneva: World Intellectual Property Org; 1992. WO Publication. No. 92/13120. [Google Scholar]
- 12.Bishop MD, Kappes SM, Keele JW, et al. A genetic-linkage map for cattle. Genetics. 1994;136:619–639. doi: 10.1093/genetics/136.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talbot J, Haigh J, Plante Y. A parentage evaluation test in North American elk (Wapiti) using microsatellites of ovine and bovine origin. Anim Genet. 1996;27:117–119. doi: 10.1111/j.1365-2052.1996.tb00480.x. [DOI] [PubMed] [Google Scholar]
- 14.Dewoody JA, Honeycutt RL, Skow LC. Microsatellite markers in white-tailed deer. J Heredity. 1995;86:317–319. doi: 10.1093/oxfordjournals.jhered.a111593. [DOI] [PubMed] [Google Scholar]
- 15.Slate J, Coltman DW, Goodman SJ, et al. Bovine microsatellite loci are highly conserved in red deer (Cervus elaphus), sika deer (Cervus nippon) and Soay sheep (Ovis aries) Anim Genet. 1998;29:307–315. doi: 10.1046/j.1365-2052.1998.00347.x. [DOI] [PubMed] [Google Scholar]
- 16.Slate J, Marshall T, Pemberton J. A retrospective assessment of the accuracy of the paternity inference program CERVUS. Mol Ecol. 2000;9:801–808. doi: 10.1046/j.1365-294x.2000.00930.x. [DOI] [PubMed] [Google Scholar]
- 17.Marshall TC, Slate J, Kruuk LEB, et al. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. [DOI] [PubMed] [Google Scholar]
- 18.Crawford AM, Dodds KG, Ede AJ, et al. An autosomal genetic-linkage map of the sheep genome. Genetics. 1995;140:703–724. doi: 10.1093/genetics/140.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones KC, Levine KF, Banks JD. DNA-based genetic markers in black-tailed and mule deer for forensic applications. Calif Fish Game. 2000;86:115–126. [Google Scholar]