Abstract
The ability of a eukaryotic cell to resist deformation, to transport intracellular cargo and to change shape during movement depends on the cytoskeleton, an interconnected network of filamentous polymers and regulatory proteins. Recent work has demonstrated that both internal and external physical forces can act through the cytoskeleton to affect local mechanical properties and cellular behaviour. Attention is now focused on how cytoskeletal networks generate, transmit and respond to mechanical signals over both short and long timescales. An important insight emerging from this work is that long-lived cytoskeletal structures may act as epigenetic determinants of cell shape, function and fate.
In a 1960 lecture, cell and developmental biologist Paul A. Weiss encouraged his audience to think of the cell as an integrated whole “lest our necessary and highly successful preoccupation with cell fragments and fractions obscure the fact that the cell is not just an inert playground for a few almighty masterminding molecules, but is a system, a hierarchically ordered system, of mutually interdependent species of molecules, molecular groupings, and supramolecular entities; and that life, through cell life, depends on the order of their interactions”1.
This statement may be more relevant today than it was 50 years ago. Despite tremendous progress, fundamental gaps remain between our understanding of individual molecules and our understanding of how these molecules function collectively to form living cells. The sequencing of genomes outpaces characterization of the cellular components they encode and far exceeds our ability to reassemble these components into the types of complex system that can provide mechanistic insight into cellular behaviour. An even more difficult task is to connect the behaviour of cells in culture with that of more complex living tissues and organisms.
Ever since muscle fibres were first examined under rudimentary microscopes in the seventeenth century, researchers have been motivated to understand how the process of self-organization generates dynamic, robust and elaborate structures that organize and ‘animate’ cells. The biological importance of establishing order over diverse length scales and timescales, as well as the challenges of understanding how systems of self-organizing molecules carry out cellular functions, is perhaps best illustrated by studies of the cytoskeleton.
The cytoskeleton carries out three broad functions: it spatially organizes the contents of the cell; it connects the cell physically and biochemically to the external environment; and it generates coordinated forces that enable the cell to move and change shape. To achieve these functions, the cytoskeleton integrates the activity of a multitude of cytoplasmic proteins and organelles. Despite the connotations of the word ‘skeleton’, the cytoskeleton is not a fixed structure whose function can be understood in isolation. Rather, it is a dynamic and adaptive structure whose component polymers and regulatory proteins are in constant flux.
Many basic building blocks of the cytoskeleton have been identified and characterized extensively in vitro, and researchers are now using advanced light microscopy to determine, with great spatial and temporal precision, the locations and dynamics of these cytoskeletal proteins during processes such as cell division and motility. For example, more than 150 proteins have so far been found to contain binding domains for the protein actin, which polymerizes to form one of the key cytoskeletal filaments in cells2. One set of actin regulators forms a macromolecular ensemble called the WAVE complex that promotes assembly of actinfilament networks at the leading edge of motile cells3. High-resolution light microscopy of rapidly crawling leukocytes revealed that the WAVE complex forms highly coherent travelling waves whose movement correlates with cell protrusion4.
Such observations in living cells can stimulate the formation of detailed hypotheses for how molecules collaborate to form functional cytoskeletal structures, but to test these hypotheses definitively, the components must be isolated from cells and purified. Remarkably, experiments that combine a small number of purified proteins have demonstrated that many complex cytoskeletal structures observed in cells can be reconstituted in vitro from purified components. For example, only three proteins are required to actively track and transport cargo on the growing end of microtubules, which are formed by the polymerization of subunits consisting of αβ-tubulin heterodimers and are another key cytoskeletal filament in cells5. Although the list of proteins associated with the cytoskeleton continues to grow, the ultimate goal remains — understanding how the interactions of the individual molecules of the cytoskeleton give rise to the large-scale cellular behaviours that depend on them.
In this Review, we discuss recent progress towards an integrated understanding of the cytoskeleton. In particular, we focus on the mechanics of cytoskeletal networks and the roles that mechanics have in many cell biological processes. Rather than focusing on one cellular process or cytoskeletal filament, we identify a set of basic concepts and link them to work in several cytoskeleton-related fields. We begin with a brief introduction to the major polymers that constitute the cytoskeleton and then shift focus from molecules to more complex structures, emphasizing three concepts that echo Weiss’s 1960 challenge to view cells as an integrated whole. The first concept is that long-range order arises from the regulated self-assembly of components guided by spatial cues and physical constraints. The second is that beyond simply composition, it is the architecture of the cytoskeleton that controls the physical properties of the cell. And the third is that cytoskeletal links to the external microenvironment can mediate both short and long timescale changes in cellular behaviour. We finish by discussing the intriguing and under-appreciated question of whether long-lived cytoskeletal structures can function as a cellular ‘memory’ that integrates past interactions with the mechanical microenvironment and influences future cellular behaviour.
Cytoskeletal building blocks
The proteins that make up the cytoskeleton have many similarities to LEGO, the popular children’s toy. Both consist of many copies of a few key pieces that fit together to form larger objects. Both can be assembled into a wide range of structures with diverse properties that depend on how the pieces are assembled. And both can be disassembled and reassembled into different shapes according to changing needs. But only the cytoskeleton fulfils all of these functions through self-assembly.
There are three main types of cytoskeletal polymer: actin filaments, microtubules and a group of polymers known collectively as intermediate filaments. Together, these polymers control the shape and mechanics of eukaryotic cells (Fig. 1). All three are organized into networks that resist deformation but can reorganize in response to externally applied forces, and they have important roles in arranging and maintaining the integrity of intracellular compartments. The polymerization and depolymerization of actin filaments and microtubules generate directed forces that drive changes in cell shape and, together with molecular motors that move along the actin filaments and microtubules, guide the organization of cellular components. The architecture of the networks that are formed by cytoskeletal polymers is controlled by several classes of regulatory protein: nucleation-promoting factors, which initiate filament formation; capping proteins, which terminate filament growth; polymerases, which promote faster or more sustained filament growth; depolymerizing factors and severing factors, which disassemble filaments; and crosslinkers and stabilizing proteins, which organize and reinforce higher-order network structures. Mechanical forces from inside or outside the cell can affect the activity of these regulatory factors and, in turn, the local organization of filaments in the networks. The most important differences between the three main cytoskeletal polymers — the differences that distinguish the architecture and function of the networks they form — are their mechanical stiffness, the dynamics of their assembly, their polarity, and the type of molecular motors with which they associate.
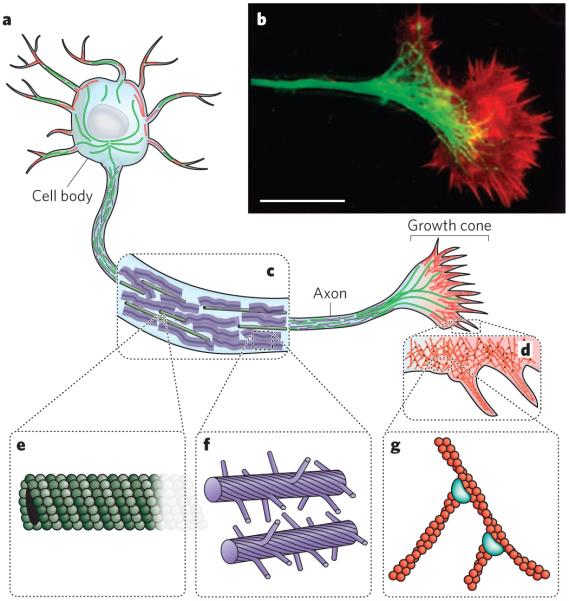
Figure 1. Elements of the cytoskeleton.
The cytoskeleton of eukaryotic cells provides structure and organization, resists and transmits stresses, and drives shape change and movement. a, Neurons are specialized eukaryotic cells that extend long processes to form connections in the nervous system. Like other eukaryotic cells, neurons have a cytoskeleton that consists of three main polymers: microtubules (green), intermediate filaments (purple) and actin filaments (red). b, A fluorescence micrograph of the neuronal growth cone, which migrates in response to chemical cues during the development of the nervous system, is shown. Microtubules (green) emanate from the axon, and actin-filament networks (red) form sheet-like structures and filopodial protrusions at the leading edge. Scale bar, 20 μm. (Image reproduced, with permission, from ref. 82.) c, The neuronal axon is a long membrane-bounded extension, in which neurofilaments (a class of intermediate filament in neurons) form a structural matrix that embeds microtubules, which transport materials from the cell body to the axon terminals at the synapse. d, The growth cone contains dendritic actinfilament networks and parallel actin-filament filopodia. e, Microtubules consist of 13 protofilaments of tubulin dimers arranged in a hollow tube. f, Neurofilaments have flexible polymer arms that repel neighbouring neurofilaments and determine the radius of the axon. g, Actin filaments are arranged into networks. These networks can have many architectures, including the branched structures depicted here, which are formed by the Arp2/3 complex (blue). The diameters of microtubules, intermediate filaments and actin filaments are within a factor of three of each other; the diagrams in e, f and g are drawn approximately to scale. But the relative flexibilities of these polymers differ markedly, as indicated by their persistence lengths: from least to most flexible, microtubules (5,000 μm), actin filaments (13.5 μm) and intermediate filaments (0.5 μm).
Microtubules are the stiffest of the three polymers and have the most complex assembly and disassembly dynamics. The persistence length of microtubules, a measure of filament flexibility that increases with stiffness, is so large (~5 mm) that single microtubules can form tracks that are almost linear and span the length of a typical animal cell, although microtubules are known to buckle under the compressive loads in cells6. During interphase, the part of the cell cycle during which cells prepare for division, many cells take advantage of this stiffness by assembling radial arrays of microtubules that function as central hubs and ‘highways’ for intracellular traffic. During mitosis, the part of the cell cycle during which cells separate chromosomes into two identical sets, the microtubule cytoskeleton rearranges itself into a high-fidelity DNA-segregating machine called the mitotic spindle. The ability of the mitotic spindle to find and align chromosomes depends, in part, on the complex assembly dynamics of individual microtubules7. A microtubule can switch between two states: stably growing and rapidly shrinking8. This ‘dynamic instability’ enables the microtubule cytoskeleton to reorganize rapidly and allows individual microtubules to search the cellular space quickly9, up to 1,000-fold faster than a polymer that is sensitive only to changes in the cellular concentration of its constituent subunits or to the actions of regulatory proteins.
Actin filaments are much less rigid than microtubules. But the presence of high concentrations of crosslinkers that bind to actin filaments promotes the assembly of highly organized, stiff structures, including isotropic networks, bundled networks and branched networks. Bundles of aligned filaments support filopodial protrusions, which are involved in chemotaxis (directed movement along a chemical gradient) and cell–cell communication. By contrast, networks of highly branched filaments support the leading edge of most motile cells and generate the forces involved in changes in cell shape such as those that occur during phagocytosis. Unlike microtubules, actin filaments do not switch between discrete states of polymerization and depolymerization; instead, they elongate steadily in the presence of nucleotide-bound monomers. This steady elongation is well suited to producing the sustained forces that are required to advance the leading edge of a migrating cell10. Also unlike the microtubule cytoskeleton, the architecture of which is often determined by one or two central organizing centres, the actin cytoskeleton is continually assembled and disassembled in response to the local activity of signalling systems. For example, protrusive, branched actin-filament networks, such as those in crawling leukocytes, are assembled at the leading edge of the cell in response to signals downstream of cell-surface receptors that guide chemotaxis11. Similarly, the assembly of contractile actin-filament bundles known as stress fibres, such as those in adherent fibroblasts, is triggered locally when cell-surface adhesion receptors called integrins engage their ligands12. And, in the final stages of endocytosis, one of the processes by which cells take up extracellular molecules, signals from the invaginating plasma membrane trigger actin filaments to assemble locally, helping this region of the membrane to become internalized as an endocytic vesicle. In addition to the network dynamics discussed here, more complex dynamics can occur when actin filaments interact with disassembly factors such as members of the cofilin family or with polymerases such as members of the formin family.
Both actin filaments and microtubules are polarized polymers, meaning that their subunits are structurally asymmetrical at the molecular level. As a result of this structural polarity, both types of polymer function as suitable tracks for molecular motors that move preferentially in one direction. For microtubules, the motors are members of the dynein or kinesin families, whereas for actin filaments, they are members of the large family of myosin proteins. These molecular motors have essential roles in organizing the microtubule and actin cytoskeletons. Microtubule-associated motors are crucial for the assembly of the microtubule array, in interphase, and the mitotic spindle. These motors also carry cargo between intracellular compartments along microtubule tracks. Some actin networks, such as the branched networks that underlie the leading edge of motile cells, seem to assemble without the aid of motor proteins, whereas others, including the contractile array at the rear of a motile cell, require myosin motor activity for their formation and function. Myosin motors also act on the bundles of aligned actin filaments in stress fibres, enabling the cells to contract, and sense, their external environment.
Intermediate filaments are the least stiff of the three types of cytoskeletal polymer, and they resist tensile forces much more effectively than compressive forces. They can be crosslinked to each other, as well as to actin filaments and microtubules, by proteins called plectins13, and some intermediate-filament structures may be organized mainly through interactions with microtubules or actin filaments. Many cell types assemble intermediate filaments in response to mechanical stresses, for example airway epithelial cells, in which keratin intermediate filaments form a network that helps cells to resist shear stress14. One class of widely expressed intermediate filament, consisting of polymerized nuclear lamins, contributes to the mechanical integrity of the eukaryotic nucleus, and phosphorylation of nuclear lamins by cyclin-dependent kinases helps trigger nuclear-envelope breakdown at the beginning of mitosis15. Unlike microtubules and actin filaments, intermediate filaments are not polarized and cannot support directional movement of molecular motors.
Long-range order from short-range interactions
The cytoskeleton establishes long-range order in the cytoplasm, helping to turn seemingly chaotic collections of molecules into highly organized living cells. Spatial and temporal information from signalling systems, as well as pre-existing cellular ‘landmarks’ such as the ‘bud scar’ left after division of budding yeast, can affect the assembly and function of cytoskeletal structures, but much of the architecture of these structures emerges from simple short-range interactions between cytoskeletal proteins. The long-range order that is generated by the cytoskeleton typically refers to cellular dimensions (tens of micrometres), which are large compared with molecular dimensions (a few nanometres).
The way that cytoskeletal structures form is studied in vivo by genetically eliminating, reducing or increasing the expression of a protein through knockout, knockdown or overexpression experiments, respectively, and is demonstrated in vitro by reconstituting cytoskeletal filament networks from purified proteins. Radially symmetrical arrays of microtubules similar to those found in interphase cells, for example, can spontaneously assemble from mixtures of microtubules and motors16. The mitotic spindle, which is more complex, has yet to be reconstituted from purified cellular components, but Heald and colleagues found that extracts from Xenopus laevis ova undergoing meiosis can robustly assemble bipolar spindles around micrometre-sized polystyrene particles coated with plasmid DNA17. The formation of such structures shows that spindles can self-assemble in vitro in the absence of both centrosomes (the microtubule-organizing centre in animal cells) and kinetochores (the site on chromosomes to which spindle microtubules attach to pull the chromosomes apart).
Long-range order of actin-filament networks is created by the activity of actin-binding proteins and nucleation-promoting factors. One example of how a set of simple rules can result in an extended structure is the formation of branched actin networks (Fig. 2). The Arp2/3 complex (which consists of seven proteins, including actin-related protein 2 (Arp2) and Arp3) binds to actin and initiates the formation of new actin filaments from the sides of pre-existing filaments, thereby generating highly branched actin filaments that form entangled ‘dendritic’ networks18. Nucleation-promoting factors activate this Arp2/3-complex-mediated branching. These factors are typically only found associated with membranes, and they specify the front (or leading edge) of a cell, ensuring that the nucleation of new filaments in a dendritic actin-filament network occurs only from filaments growing towards the membrane19,20. The growth of all filaments is eventually stopped by a capping protein, which prevents the addition of more actin monomers21. Taken together, the repeated steps of growth, branching and capping lead to the formation of micrometre-scale, protrusive, branched actin-filament networks, which are important for crawling motility. A process of disassembly then disrupts the network and recycles the actin subunits for subsequent use22.
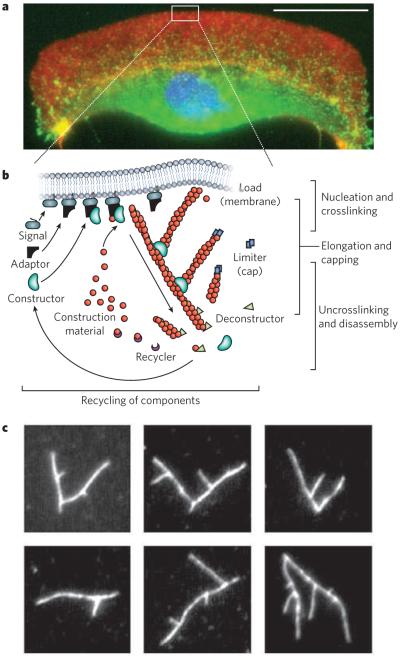
Figure 2. Building cytoskeletal structures.
Long-range order of the cytoskeleton is generated by simple rules for network assembly and disassembly. a, A fluorescence micrograph of a fish keratocyte is shown (with the nucleus in blue). Motile cells such as these form branched actin-filament networks (red) at their leading edge, and these branched networks generate protrusions. Together with coordinated adhesions to a surface (indicated by vinculin, green) and myosin-driven retraction, the protrusions lead to directed movement. Scale bar, 15 μm. (Image courtesy of M. van Duijn, Univ. California, Berkeley.) b, There are three basic steps involved in the assembly of protrusive, branched actin-filament networks: filament elongation; nucleation and crosslinking of new filaments from filaments close to the membrane; and capping of filaments. Disassembly of the network involves a separate set of proteins that severs the filaments and recycles the subunits. c, The branching of actin filaments can be reconstituted in vitro with soluble proteins, generating various branched structures such as those in these fluorescence micrographs of labelled actin (white). (Images courtesy of O. Akin, Univ. California, San Francisco.)
Crosslinkers also affect the structural organization of cytoskeletal networks, particularly as a result of their geometry and binding kinetics. For example, the crosslinker fascin preferentially stabilizes parallel bundles of filaments such as those in filopodia, owing to the rigid coupling between the filament-binding sites on fascin. By contrast, actin crosslinkers such as α-actinin, in which the filament-binding sites rotate much more freely, can stabilize either orthogonal gels (such as those found in non-aligned actin-filament networks, which support the plasma membrane of cells) or parallel bundles. In this case, the architecture of the network is determined by the kinetics of the interaction. If the dissociation rate of the crosslinker from the actin filaments is high, then filaments are aligned into bundles. If the dissociation rate is low, then filaments are stabilized in a more randomly ordered state23.
Some cytoskeletal structures can span distances much larger than that of the typical cell. ‘Cytonemes’ and ‘membrane nanotubes’, which contain actin filaments and are essentially specialized filopodia, can grow to lengths of millimetres and have been shown to mediate cell–cell signalling across sea-urchin blastulae and Drosophila melanogaster wing imaginal discs24. The mechanisms that enable filopodium-like structures to grow to such extreme lengths may involve cooperative interactions between the elastic properties of the actin filaments and the plasma membranes, stabilizing the protrusions against buckling after a membrane tube has formed25.
Network architecture and mechanics
Although distinct in their properties and the types of network they form, the polymers of the cytoskeleton are intricately linked together. The organization of these links and the resultant architecture of the cytoskeletal networks has a central role in transmitting compressive and tensile stresses and in sensing the mechanical microenvironment26. Structures formed from microtubules, actin filaments or intermediate filaments interact with each other and other cellular structures either nonspecifically (through steric interactions and entanglement) or specifically (through proteins that link one filament type to another). For example, the actin nucleation-promoting factor WHAMM binds not only to actin but also to microtubules and membranes27, and the GTPase Rac1 is activated by the growth of microtubules, which in turn stimulates the polymerization of actin in lamellipodial protrusions28. This interconnectivity creates continuous mechanical coupling through the cytoskeleton, providing a means for internal or external forces and fluctuations to be distributed throughout the cell. But, despite their interconnectivity, cytoskeletal networks have typically been investigated by studying the component polymers individually to understand their contributions to cell mechanics.
The mechanical response of gels formed from purified cytoskeletal filaments that have been reconstituted in vitro provides insight into the properties of these polymers in cells. Actin-filament networks have been of considerable interest because of the variety of structures they form and because they function as model semi-flexible polymers29. As such, their elasticity can arise from two sources: entropic elasticity, which results from a reduction in configurations available to thermally fluctuating filaments, such as when they are stretched; and enthalpic elasticity, which is due to changes in spacing of the molecules that make up the filaments, such as when they are bent, even in the absence of thermal fluctuations. The importance of these two elastic contributions seems to depend on the architecture of the network.
When shear stresses are applied to actin-filament networks, as well as to networks of intermediate filaments or extracellular-matrix filaments such as collagen and fibrin, the networks stiffen and resist additional deformation, as a result of filament entanglement (in which the displacement of one filament is impeded by another filament) and the entropic elasticity of individual filaments30. When a rigid crosslinker such as scruin is added to randomly organized actin filaments and shear stress is applied, the magnitude of the elastic modulus (a measure of the resistance of the network to deformation) increases significantly, and the network retains the stress-stiffening behaviour attributed to the entropic elasticity of individual filaments31,32. When the more flexible crosslinker filamin A is added to randomly organized actin filaments together with the molecular motor myosin, the rigidity of the network increases to more than that of an entangled filament network, and the network stiffens nonlinearly as though it were subject to external stress33. These studies demonstrate the importance of the entropic elasticity of filaments in the mechanical properties of networks without specific filament orientation.
By contrast, in networks with highly organized architectures, the bending of actin filaments, rather than their entropic stretching, can dominate the elastic properties. When branched actin-filament networks, such as those growing at the leading edge of crawling cells, are exposed to compressive forces, the network shows nonlinear stress stiffening, followed by stress softening at high stresses34. Interestingly, the softening behaviour is completely reversible, as would be expected from the buckling of actin filaments oriented towards the load, suggesting that filaments bearing a compressive load can be important contributors to network elasticity. Although the bending and stretching of filaments has been the focus of many actin-filament network studies, the mechanical properties of crosslinked networks must also depend on the properties of the crosslinkers themselves. In one study of crosslinker length, variation in the spacing between actin-binding domains significantly affected both the elastic modulus and the network architecture, with short crosslinker lengths resulting in high stiffness and bundled filament arrangements35. In general, the highly nonlinear behaviour of filament stretching and bending, together with the organizational constraints imposed by crosslinkers and nucleation-promoting factors, indicates the importance of network architecture in determining the mechanical behaviour of the cytoskeleton.
In whole cells, the actin cytoskeleton has a wide variety of architectures that are associated with specific functional structures (Fig. 3). By using in vitro reconstitutions such as those described above, researchers are beginning to identify the key molecular determinants of filament order, but how these actin-filament structures are linked to other cellular systems remains an important area of investigation. In particular, the deformation of cytoskeletal networks in response to mechanical load is coupled to changes in plasma-membrane tension and displacement of fluid in the cytoplasm. For example, Herant and colleagues observed significant increases in plasma-membrane tension as neutrophils engulfed antibody-coated beads by using phagocytosis, which is an actin-driven process36. Furthermore, the resistance to flow through dense cytoskeletal networks, known as poroelasticity, can slow the flow of fluid to the point at which it takes several seconds for stress to propagate across an individual cell37, in contrast to times of the order of microseconds for direct mechanical coupling through tensed filaments38.
Figure 3. Form meets function.
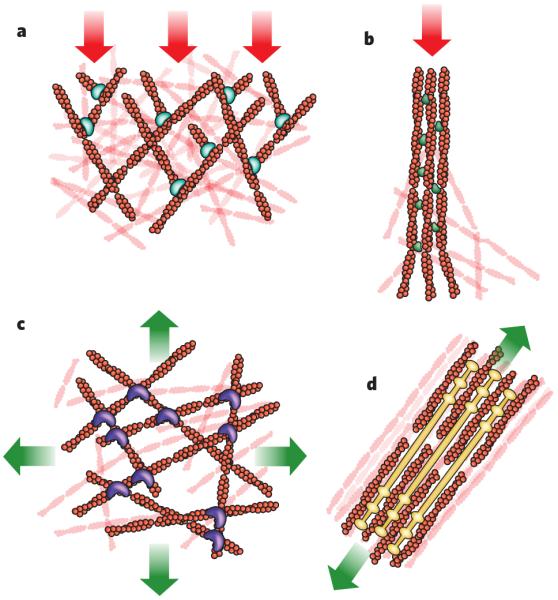
The cytoskeleton forms structures that have a wide variety of architectures and are associated with different types of cellular force. Shown are four structures generated by actin filaments and the stresses typically encountered by these structures (red arrows, compression; green arrows, tension). a, Branched actin-filament networks push against the plasma membrane and external barriers as they generate protrusions, thereby encountering an inward force of compression. b, Filaments bundled into filopodia also generate protrusive forces as they extend from the cell body, encountering similar compressive force. In this case, the linker molecule is the bundling protein fascin. c, Cortical networks (that is, non-aligned networks), such as this one involving filamin as a crosslinker, form below the plasma membrane and carry tension loads in multiple directions. d, Stress fibres form from bundled actin filaments, shown here associated with filaments of myosin, and generate tension against cell adhesions to the extracellular matrix. The illustrations are based on micrographs from refs 83-86 (a–d, respectively).
The broad range of cytoskeletal architectures and mechanisms for stress transmission has presented considerable challenges to the development of a complete model of the cytoskeleton. Accordingly, different theoretical frameworks have been developed to capture various aspects of the collective behaviour of filament networks and the cytoplasm. For example, filament hydrodynamics and molecular motors are included in a theory of active polar gels39. Some models have also attempted to describe the complex viscosity of cytoskeletal networks, which, at the molecular scale, partly arises from the binding kinetics of crosslinkers. The range of relaxation timescales that has been observed for cells at temporal frequencies (measured in hertz) has led to the suggestion that the cytoskeleton behaves as a glassy material that transitions between several kinetically trapped states40. What is needed now are more process-level models that connect the cytoskeleton with the plasma membrane or other physical boundary conditions and provide an understanding of cellular behaviours that depend on membrane–cytoskeleton interactions, as in a recent study describing shape variations in motile cells41.
What is the value of determining the cytoskeletal architecture and mechanical properties of a cell? In short, this information can provide insight into where forces are acting. Stress applied to a cell is distributed broadly by the cytoskeleton, but the magnitude of transmitted stress to a particular location depends on network mechanics and architecture and can have marked effects on cellular processes, from individual filament polymerization up to entire network reorganization. Growing microtubules that encounter resistance decrease their growth rate exponentially as the force increases42 and have a greater likelihood of complete disassembly of the polymer. The polymerization of a small number of actin filaments is similarly limited by force43, but filaments growing as part of a dendritic actin-filament network behave differently from predictions based on the single-filament model of force dependence. When reconstituted on the end of an atomic-force-microscope cantilever, dendritic networks that experience increasing loads grow at a constant velocity over a wide range of forces, suggesting that the network adapts to the increasing load by increasing local filament density through a force-sensing element in the network44. This constant-velocity behaviour of actin-filament networks under increasing load was also observed in measurements of lamellipodial protrusions in crawling cells45. The complexity of network growth, in contrast to that of single filaments, highlights the importance of thinking about the collective behaviour of cytoskeletal structures rather than just individual filaments. This challenge becomes even greater when considering how the cytoskeleton interacts with external signals.
Sensing the mechanical microenvironment
Cells are intricately connected to the external environment through their cytoskeleton. Whether in direct contact with neighbouring cells or with a dense meshwork of polymers known as the extracellular matrix, cells receive external signals that guide complex behaviours such as motility and, in some cases, differentiation (for example from stem cells into cells of a specific lineage). Whereas the contribution of chemical signals has long been understood, physical signals have only recently been widely recognized to be pervasive and powerful. The observation that physical properties of the microenvironment can affect cell shape and behaviour dates to the 1920s, when studies showed that mesenchymal cells embedded in clots of various stiffnesses had different shapes (see ref. 46 for a review). More recent studies have shown that the tension generated by a contracting cytoskeleton can be used to sense the mechanical properties of the extracellular matrix, which in turn have been shown to affect cytoskeletal organization and cell behaviour47, although whether stiffness or force is the most important signal remains a subject of debate48.
Of particular interest is how cell–extracellular matrix and cell–cell interactions can lead to long-lived changes in cellular organization in tissues and cell behaviour. Several studies have highlighted the importance of physical cues in the organization of tissues during development. For example, Thery and colleagues found that the orientation of the mitotic spindle in dividing cells, and hence the location of the division plane and the spatial arrangement of the daughter cells, is affected by the spatial distribution of extracellular matrix proteins49. Using microcontact printing to define patterns of extracellular matrix proteins to which cells adhere, they found that the cells divided with predictable orientations controlled by cortical contacts with the extracellular matrix. In addition to the pattern of adhesion sites, the mechanical properties of cells themselves also contribute to tissue organization. As one example, in gastrulating zebrafish embryos, actin- and myosin-dependent plasma-membrane tension and differential adhesion among cells drives the sorting of germ-layer progenitor cells50. Cell proliferation can even be affected when external forces are applied to tissue (Fig. 4). When a tumour spheroid, formed from murine mammary carcinoma cells grown in a cluster within an agarose gel, is exposed to compressive loads during growth, the cells proliferate more slowly at regions of high stress and undergo programmed cell death when exposed to sufficiently high stress51.
Figure 4. Force and shape.
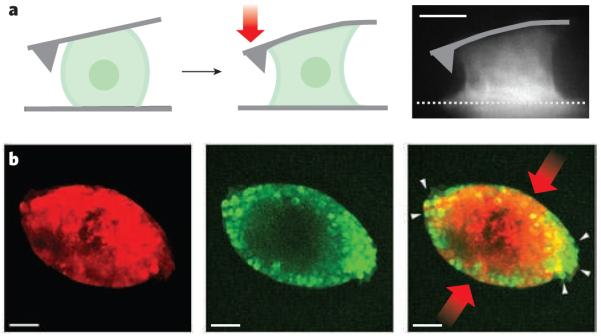
a, Single cells can exert large contractile forces that affect their shape. An osteosarcoma cell attached between the cantilever of an atomic force microscope and a surface can exert contractile forces (red arrow) of more than 100 nN (depicted diagramatically from left panel to centre panel and in a fluorescence micrograph, right panel). Actin-filament structures (white), including contractile stress fibres spanning the upper and lower surfaces, are generated in the contracting osteosarcoma cell (right). Scale bar, 10 μm. (Image reproduced, with permission, from ref. 87.) b, Mechanical stress on cancer cells in three-dimensional tumour spheroids changes their growth. There are fewer proliferating cells (green) in tumour spheroids (red) at the regions of highest compressive stress (red arrows) than at the regions of low stress (white arrows). The image on the right is an overlay of the centre and left images. Scale bar, 50 μm. (Images reproduced, with permission, from ref. 51.)
When the ‘normal’ mechanical properties of tissue are disrupted, the effects can be considerable. In epithelial cell layers, altered stiffness of the supporting tissue disrupts morphogenesis and drives the epithelial cells towards a malignant phenotype52. In fact, the stiffness of the substrate seems to be important for stem cells to differentiate properly. A substrate with a stiffness that emulates normal tissue can function as a developmental cue that directs stem cells to differentiate into cells of specific lineages, including mesenchymal stem cells53 and neural stem cells54. How substrate stiffness, as well as growth factors and matrix properties, affect stem-cell differentiation is reviewed in more detail in ref. 55.
Emulating the native properties of a cell’s environment is an important consideration that is often overlooked when studying cells ex vivo. For example, the traditional method of culturing cells on stiff substrates, which has been used for decades, can itself drive changes in the mechanical properties and gene-expression profiles of the cells. Primary human foreskin epithelial cells cultured in plastic dishes were found to increase in stiffness with passaging, with cells being twofold to fourfold stiffer after eight passages than were cells passaged fewer than three times56. Similarly, human epithelial breast carcinoma (MCF7) cells were found to stiffen with increasing passage number when cultured on glass coverslips57 and endometrial adenocarcinoma cells cultured in plastic dishes expressed more α-actin as a function of passage number, as they moved towards a stromal phenotype58. In each of these cases, the cells were grown on substrates with markedly different mechanical properties from native tissue, and long-lived changes in cytoskeletal properties and cytoskeletal organization were observed.
As more examples of cells responding to mechanical cues through the cytoskeleton are found, the questions of how, what and where physical inputs are sensed is becoming central. There is substantial evidence implicating stress-induced changes in focal adhesions and adherens junctions26, and several molecules have been identified as specific mediators of mechanical inputs. For example, mesenchymal stem cells differentiate into cells of various lineages depending on the substrate stiffness, and this elasticity-sensitive lineage-specific differentiation is blocked by inhibiting the protein non-muscle myosin53. A force-induced conformational change in p130Cas (also known as BCAR1), a scaffolding protein that is involved in focal adhesions, causes it to be more easily phosphorylated by Src59. And sensitivity to the elasticity of the extracellular matrix during angiogenesis is mediated by the Rho inhibitor p190RhoGAP (also known as GRLF1), through its effect on two antagonistic transcription factors60.
Although changes in gene expression may be the end point of mechanosensing, the process can take days for cells in culture, and it is unclear how information about physical interactions with the mechanical micro environment is stored. Are heritable changes in gene expression in mechanically perturbed cells due only to changes in chromatin structure and organization or other familiar epigenetic mechanisms? The heritability of changes that arise from mechanical interactions — and that are mediated by the cytoskeleton — raises the question of whether the organization and reorganization of long-lived cytoskeletal structures themselves might have a role in recording the cell’s mechanical ‘history’.
Cytoskeletal epigenetics
The idea that cellular structures can be passed on and influence the behaviour of subsequent generations of cells is not new. In the 1930s, embryologists recognized that regional differences in the molecular composition of ova caused daughter cells to inherit different surface molecules61. In the 1960s, Paramecium aurelia cells that were genetically identical to wild-type cells were found to pass on alterations in the orientation of their cilia for hundreds of generations62,63. Internal structures of the cytoskeleton can also persist after cells divide. Daughter 3T3 fibroblast cells have similar actin-filament stress-fibre organization and motile behaviour64,65, and adjacent epidermal ‘siamese twin’ cells in Calpodes ethlius caterpillars have the same number of actin-filament bundles66. The emergence of primary cilia from sister cells was recently found to depend on centriole age, with sister cells that inherit the old centriole growing a primary cilium before sister cells that inherit the new centriole, which was formed before cell division67.
As a look at any dish of cultured cells will confirm, genetically identical cells can have markedly different cytoskeletal structures, presumably as a result of random events as well as slight differences in external conditions. Endothelial cells grown in vitro under similar conditions, for example, contain stress fibres that are oriented randomly. But, if those cells are exposed to shear stress from fluid flowing above them, they respond by elongating and orienting their stress fibres in the direction of the flow. If the shear stress is removed, then the variability in stress-fibre orientation returns, but it does so slowly. Interestingly, if the elongated cells are detached from the surface, then the elongated shape persists68. In this way, the cytoskeleton can be a record of a cell’s past mechanical interactions. Given the interconnectedness of the cytoskeleton and its role in the transduction of mechanical signals from the external microenvironment, as well as its role as a scaffold for many reactions69, the ability of cytoskeletal structures to record the past may result in the cytoskeleton profoundly affecting the cell’s future and even the future of the cell’s progeny70.
Given that cytoskeletal structures are often highly dynamic, with specific factors that promote disassembly and recycling of the cytoskeletal building blocks competing with factors that assemble and stabilize them, is it possible for mechanical inputs to be recorded? During endocytosis, actin-filament networks can assemble around clathrin-coated pits and displace the invaginated membrane in less than 15 seconds71. But, when the timescales for assembling and stabilizing a cytoskeletal structure are longer than those for disassembling and recycling it, the result can be a persistent structure that affects the behaviour of a cell over a longer timescale than the initial signal. In essence, the system shows hysteresis, a common feature of magnetic, electrical and elastic properties of materials, in which there is a lag between application or removal of a stimulus, such as a force, and its effect. In biological systems, this hysteresis seems to involve active, energy consuming processes. In one example, a growing actin-filament network that had been reconstituted in vitro was exposed to a weak compressive force during growth. When the compressive force on the network was gradually increased and then rapidly reduced to the previous level of force, the velocity of network growth increased and persisted at a rate that was significantly higher than the original velocity at that force44. It is likely that this increase in velocity resulted from an increase in actin-filament density caused by the transient increase in load, with the system showing hysteresis. In contrast to molecular motors, for which the relationship between force and velocity is immediately reversible, the observation that there is more than one growth velocity for a given force suggests that actin-filament network growth depends on history. The cytoskeletal structure and the process by which it is built can record mechanical interactions, whereas a single filament could not.
If mechanical interactions with the external environment can alter the cytoskeleton in a lasting way, what are the implications? To the extent that the cytoskeleton is intricately involved both mechanically and biochemically in cellular processes such as cell division and motility, long-lived cytoskeletal structures could create variability in cell behaviour and may guide variation towards certain phenotypes. This behaviour could be as transient as the mirror-image movements of sister cells65 or as permanent as alterations in cell fate53. Although it is plausible, the hypothesis that specific cytoskeletal structures are necessary and sufficient determinants of cell behaviour has only just begun to be explored in detail. Further research at the level of tissues, cells and reconstituted cytoskeletal structures is needed to understand when and how the cytoskeletal history can markedly influence a cell’s future.
Future research on the cytoskeleton
With the experimental techniques now available for studying the cytoskeleton, ranging from super-resolution imaging of molecular organization to direct physical manipulation of cellular structures and processes, there are many opportunities to probe the link between physical force and cell behaviour. To guide experiments, new models are needed to search for the mechanisms and molecules that link cell mechanics and the cytoskeleton to cellular decision-making. Computational simulations will become increasingly important as a way of testing the properties of hierarchical ordered systems. If successful, this effort to follow forces, whether internally generated or externally imposed, from the mechanical input to the phenotypic output could have a profound impact on our understanding of how normal and diseased cells behave.
Because of the central role of the cytoskeleton in cell structure and intracellular organization, perturbations in the architecture of any of the three main cytoskeletal networks can result in marked pathologies. For example, if the mitotic spindle does not function properly, dividing cells cannot partition the genetic material equally between the daughter cells. Chromosomal instability is associated either with a loss of the checkpoint that ensures that all chromosomes are correctly attached to microtubules in the mitotic spindle or with the presence of too many microtubule-organizing centres at the time of cell division72. And mutations in the genes encoding intermediate filament proteins are associated with many diseases in humans (see ref. 73 for a review), including a predisposition to liver disease in the case of some keratins, amyotrophic lateral sclerosis (also known as Lou Gehrig’s disease) in the case of a neuronal class of intermediate filament called neurofilaments, and progeria (a hereditary form of premature ageing) in the case of improperly assembled nuclear lamins.
The in vitro reconstitution of purified proteins will continue to be a powerful tool for identifying the conditions that are necessary and sufficient for a cytoskeletal process and could guide the search for therapeutic drug targets and candidate drug molecules. For example, a decrease in the activity of cardiac-muscle myosin proteins is associated with heart failure, and myosin motors are now important targets for cardiovascular drug therapy. Reconstitution experiments that involve only a few proteins have the potential to uncover complex physical behaviour and long-range structural organization (Fig. 5). Like cells, reconstituted cytoskeletal polymers are sensitive to the physical boundary conditions imposed by their environment. Confining cytoskeletal polymers within lipid vesicles affects both their dynamics and their organization, as has been shown for microtubules74 and actin filaments75, and this can be used to test the effect of forces on the architecture and function of the cytoskeleton. As experiments with purified proteins increase in complexity and in fidelity to cellular processes, new methods will be needed so that not only cytoskeletal structures but also integral membrane proteins and metabolic processes can be reconstituted76.
Figure 5. Learning by building.
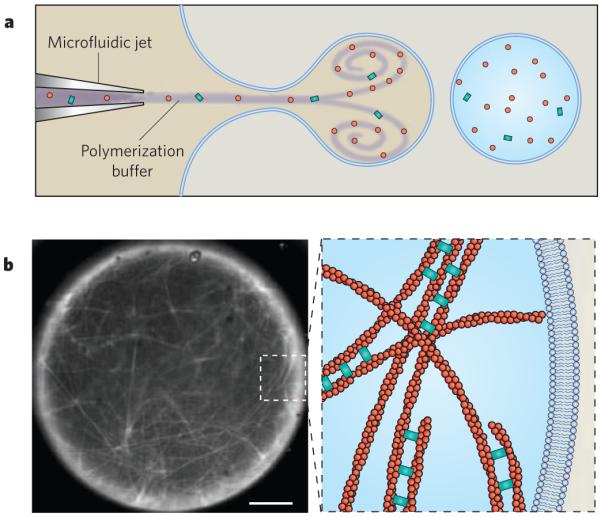
The reconstitution of cytoskeletal structures is a key method for understanding how functional behaviour emerges from discrete components. In this example, actin filaments were nucleated from beads coated with the nucleation-promoting factor ActA (not shown) and were then crosslinked by fascin inside a unilamellar lipid vesicle. a, Purified proteins were loaded into a vesicle by a microfluidic encapsulation technique that allows the dynamics of filament assembly to be observed immediately after encapsulation88. b, The micrograph (left) shows fluorescently labelled actin filaments (white) that have polymerized inside the vesicle and have assembled into a fascin-crosslinked network. Scale bar, 5 μm. The diagram (right) is a schematic depiction of the actin-filament network present in the inset box of the micrograph. (Image courtesy of D. Richmond, S. Hansen and M. Zanic, Physiology Course, Marine Biological Laboratory, Woods Hole, Massachusetts.)
Until not long ago, eukaryotic cells were thought to be distinguished from bacteria and archaea by the presence of a cytoskeleton. But the discovery of cytoskeletal polymers even in comparatively simple cells of small size and genome are revealing the central importance of internal organization for cell function. Now, clues to the origin of the eukaryotic cytoskeleton and the polymers that constitute it are emerging from the study of bacteria. Filamentous proteins with homology to actin filaments, microtubules and intermediate filaments have been identified and shown to have a role in organizing the bacterial cytoplasm. Rod-shaped bacteria such as Escherichia coli require an actin-like polymer formed from MreB to be assembled in order to define their shape77. To grow into its curved shape, Caulobacter crescentus (also known as Caulobacter vibrioides) requires an additional cytoskeletal component, an intermediate-filament-like protein called crescentin78. Other polymers have a role in organizing the DNA in bacteria. ParM, for example, is an actin-like protein that forms polymers required for segregating type II plasmids during cell division79. Reconstituting ParM-dependent DNA segregation in vitro has revealed a mechanism by which large, slowly diffusing cargo can be moved rapidly through the bacterial cytoplasm80. More than 35 actin-like proteins have been identified in bacteria, but most remain to be characterized81.
In the lecture quoted at the beginning of this Review, Weiss also stated: “Life is a dynamic process. Logically, the elements of a process can be only elementary processes, and not elementary particles or any other static units. Cell life, accordingly, can never be defined in terms of a static inventory of compounds, however detailed, but only in terms of their interactions”1. The cytoskeleton is a manifestation of those elementary interactions, exemplifying the rich behaviour that can emerge in hierarchically organized systems. The progress over the past 50 years indicates that the collective properties of the cytoskeleton, including the architecture of the networks and their mechanical history, are essential for understanding the push and pull of cellular behaviour.
Acknowledgements
We thank O. Chaudhuri, D. Richmond, V. Risca and other members of the Fletcher laboratory for discussion and assistance with this Review. We also benefited from interactions with the researchers and students in the 2009 Physiology course at the Marine Biological Laboratory, Woods Hole, Massachusetts. Work in our laboratories is supported by R01 grants from the National Institutes of Health (NIH) and by the Cell Propulsion Lab, an NIH Nanomedicine Development Center. We apologize to those colleagues whose work could not be cited because of space constraints.
Footnotes
The authors declare no competing financial interests.
References
- 1.Weiss PA. In: The Molecular Control of Cellular Activity. Allen JM, editor. McGraw-Hill; 1961. pp. 1–72. [Google Scholar]
- 2.dos Remedios CG, et al. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 3.Machesky LM, et al. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl Acad. Sci. USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 6.Brangwynne CP, et al. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J. Cell Biol. 2006;173:733–741. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nature Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 8.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 9.Holy TE, Leibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc. Natl Acad. Sci. USA. 1994;91:5682–5685. doi: 10.1073/pnas.91.12.5682. This paper showed that microtubule dynamics have a central role in spatial organization within cells.
- 10.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 11.Parent CA. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr. Opin. Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Naumanen P, Lappalainen P, Hotulainen P. Mechanisms of actin stress fibre assembly. J. Microsc. 2008;231:446–454. doi: 10.1111/j.1365-2818.2008.02057.x. [DOI] [PubMed] [Google Scholar]
- 13.Wiche G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998;111:2477–2486. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- 14.Flitney EW, Kuczmarski ER, Adam SA, Goldman RD. Insights into the mechanical properties of epithelial cells: the effects of shear stress on the assembly and remodeling of keratin intermediate filaments. FASEB J. 2009;23:2110–2119. doi: 10.1096/fj.08-124453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai MY, et al. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- 16.Nedelec FJ, Surrey T, Maggs AC, Leibler S. Self-organization of microtubules and motors. Nature. 1997;389:305–308. doi: 10.1038/38532. [DOI] [PubMed] [Google Scholar]
- 17.Heald R, et al. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. The reconstitution of spindles in a cell extract, as reported in this paper, was a remarkable demonstration of the self-assembling properties of the cytoskeleton.
- 18.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl Acad. Sci. USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. This paper presented the dendritic nucleation model for the assembly of branched actin networks.
- 19.Bailly M, et al. Relationship between Arp2/3 complex and the barbed ends of actin filaments at the leading edge of carcinoma cells after epidermal growth factor stimulation. J. Cell Biol. 1999;145:331–345. doi: 10.1083/jcb.145.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell. Mol. Biol. 2008;267:183–206. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlier MF, et al. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wachsstock DH, Schwarz WH, Pollard TD. Cross-linker dynamics determine the mechanical properties of actin gels. Biophys. J. 1994;66:801–809. doi: 10.1016/s0006-3495(94)80856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiung F, Ramirez-Weber FA, Iwaki DD, Kornberg TB. Dependence of Drosophila wing imaginal disc cytonemes on Decapentaplegic. Nature. 2005;437:560–563. doi: 10.1038/nature03951. [DOI] [PubMed] [Google Scholar]
- 25.Liu AP, et al. Membrane-induced bundling of actin filaments. Nature Phys. 2008;4:789–793. doi: 10.1038/nphys1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janmey PA, McCulloch CA. Cell mechanics: integrating cell responses to mechanical stimuli. Annu. Rev. Biomed. Eng. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- 27.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nature Cell Biol. 1999;1:45–50. doi: 10.1038/9018. [DOI] [PubMed] [Google Scholar]
- 29.MacKintosh FC, Kas J, Janmey PA. Elasticity of semiflexible biopolymer networks. Phys. Rev. Lett. 1995;75:4425–4428. doi: 10.1103/PhysRevLett.75.4425. [DOI] [PubMed] [Google Scholar]
- 30.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–194. doi: 10.1038/nature03521. In this study, the role of entropic elasticity was shown experimentally and modelled for a broad set of cytoskeletal polymers.
- 31.Gardel ML, et al. Elastic behavior of cross-linked and bundled actin networks. Science. 2004;304:1301–1305. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 32.Tharmann R, Claessens MM, Bausch AR. Viscoelasticity of isotropically cross-linked actin networks. Phys. Rev. Lett. 2007;98:088103. doi: 10.1103/PhysRevLett.98.088103. [DOI] [PubMed] [Google Scholar]
- 33.Koenderink GH, et al. An active biopolymer network controlled by molecular motors. Proc. Natl Acad. Sci. USA. 2009;106:15192–15197. doi: 10.1073/pnas.0903974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhuri O, Parekh SH, Fletcher DA. Reversible stress softening of actin networks. Nature. 2007;445:295–298. doi: 10.1038/nature05459. This paper showed that the architecture of actin-filament networks affects the relative importance of entropic and enthalpic elasticity.
- 35.Wagner B, Tharmann R, Haase I, Fischer M, Bausch AR. Cytoskeletal polymer networks: the molecular structure of cross-linkers determines macroscopic properties. Proc. Natl Acad. Sci. USA. 2006;103:13974–13978. doi: 10.1073/pnas.0510190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herant M, Heinrich V, Dembo M. Mechanics of neutrophil phagocytosis: behavior of the cortical tension. J. Cell Sci. 2005;118:1789–1797. doi: 10.1242/jcs.02275. [DOI] [PubMed] [Google Scholar]
- 37.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 2005;435:365–369. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nature Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 39.Basu A, Joanny JF, Julicher F, Prost J. Thermal and non-thermal fluctuations in active polar gels. Eur. Phys. J. E. 2008;27:149–160. doi: 10.1140/epje/i2008-10364-9. [DOI] [PubMed] [Google Scholar]
- 40.Bursac P, et al. Cytoskeletal remodelling and slow dynamics in the living cell. Nature Mater. 2005;4:557–561. doi: 10.1038/nmat1404. [DOI] [PubMed] [Google Scholar]
- 41.Keren K, et al. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 43.Footer MJ, Kerssemakers JW, Theriot JA, Dogterom M. Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl Acad. Sci. USA. 2007;104:2181–2186. doi: 10.1073/pnas.0607052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parekh SH, Chaudhuri O, Theriot JA, Fletcher DA. Loading history determines the velocity of actin-network growth. Nature Cell Biol. 2005;7:1219–1223. doi: 10.1038/ncb1336. [DOI] [PubMed] [Google Scholar]
- 45.Prass M, Jacobson K, Mogilner A, Radmacher M. Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol. 2006;174:767–772. doi: 10.1083/jcb.200601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janmey PA, Winer JP, Murray ME, Wen Q. The hard life of soft cells. Cell. Motil. Cytoskeleton. 2009;66:597–605. doi: 10.1002/cm.20382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 48.Chen CS. Mechanotransduction — a field pulling together? J. Cell Sci. 2008;121:3285–3292. doi: 10.1242/jcs.023507. [DOI] [PubMed] [Google Scholar]
- 49.Thery M, et al. The extracellular matrix guides the orientation of the cell division axis. Nature Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 50.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafish. Nature Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 51.Cheng G, Tse J, Jain RK, Munn LL. Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells. PLoS ONE. 2009;4:e4632. doi: 10.1371/journal.pone.0004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paszek MJ, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. This paper showed that substrate elasticity can control the differentiation of mesenchymal stem cells.
- 54.Saha K, et al. Substrate modulus directs neural stem cell behavior. Biophys. J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berdyyeva TK, Woodworth CD, Sokolov I. Human epithelial cells increase their rigidity with ageing in vitro: direct measurements. Phys. Med. Biol. 2005;50:81–92. doi: 10.1088/0031-9155/50/1/007. [DOI] [PubMed] [Google Scholar]
- 57.Burns JM, Cuschieri A, Campbell PA. Optimisation of fixation period on biological cells via time-lapse elasticity mapping. Jpn. J. Appl. Phys. 2006;45:2341–2344. [Google Scholar]
- 58.Kato S, et al. Characterization and phenotypic variation with passage number of cultured human endometrial adenocarcinoma cells. Tissue Cell. 2008;40:95–102. doi: 10.1016/j.tice.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 59.Sawada Y, et al. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127:1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mammoto A, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weiss PA. Principles of Development; A Text in Experimental Embryology. H. Holt; 1939. [Google Scholar]
- 62.Sonneborn TM. The differentiation of cells. Proc. Natl Acad. Sci. USA. 1964;51:915–929. doi: 10.1073/pnas.51.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beisson J, Sonneborn TM. Cytoplasmic inheritance of organization of cell cortex in Paramecium aurelia. Proc. Natl Acad. Sci. USA. 1965;53:275–282. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albrecht-Buehler G. Phagokinetic tracks of 3T3 cells: parallels between the orientation of track segments and of cellular structures which contain actin or tubulin. Cell. 1977;12:333–339. doi: 10.1016/0092-8674(77)90109-x. [DOI] [PubMed] [Google Scholar]
- 65.Albrecht-Buehler G. Daughter 3T3 cells. Are they mirror images of each other? J. Cell Biol. 1977;72:595–603. doi: 10.1083/jcb.72.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delhanty P, Leung H, Locke M. Paired cytoskeletal patterns in an epithelium of siamese twin cells. Eur. J. Cell Biol. 1991;56:443–450. [PubMed] [Google Scholar]
- 67.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 2009;19:1498–1502. doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato M, Levesque MJ, Nerem RM. Micropipette aspiration of cultured bovine aortic endothelial cells exposed to shear stress. Arteriosclerosis. 1987;7:276–286. doi: 10.1161/01.atv.7.3.276. [DOI] [PubMed] [Google Scholar]
- 69.Janmey PA. The cytoskeleton and cell signaling: component localization and mechanical coupling. Physiol. Rev. 1998;78:763–781. doi: 10.1152/physrev.1998.78.3.763. [DOI] [PubMed] [Google Scholar]
- 70.Locke M. Is there somatic inheritance of intracellular patterns? J. Cell Sci. 1990;96:563–567. This paper summarized early examples of ‘cytoskeletal epigenetics’.
- 71.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 72.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Omary MB, Coulombe PA, McLean WH. Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 2004;351:2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- 74.Fygenson DK, Elbaum M, Shraiman B, Libchaber A. Microtubules and vesicles under controlled tension. Phys. Rev. E. 1997;55:850–859. [Google Scholar]
- 75.Pontani LL, et al. Reconstitution of an actin cortex inside a liposome. Biophys. J. 2009;96:192–198. doi: 10.1016/j.bpj.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu AP, Fletcher DA. Biology under construction: in vitro reconstitution of cellular function. Nature Rev. Mol. Cell Biol. 2009;10:644–650. doi: 10.1038/nrm2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 78.Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- 79.Garner EC, Campbell CS, Mullins RD. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science. 2004;306:1021–1025. doi: 10.1126/science.1101313. [DOI] [PubMed] [Google Scholar]
- 80.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Derman AI, et al. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol. Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rochlin MW, Dailey ME, Bridgman PC. Polymerizing microtubules activate site-directed F-actin assembly in nerve growth cones. Mol. Biol. Cell. 1999;10:2309–2327. doi: 10.1091/mbc.10.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henson JH, et al. Two components of actin-based retrograde flow in sea urchin coelomocytes. Mol. Biol. Cell. 1999;10:4075–4090. doi: 10.1091/mbc.10.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Svitkina TM, et al. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nature Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 86.Svitkina TM, Verkhovsky AB, Borisy GG. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J. Struct. Biol. 1995;115:290–303. doi: 10.1006/jsbi.1995.1054. [DOI] [PubMed] [Google Scholar]
- 87.Chaudhuri O, Parekh SH, Lam WA, Fletcher DA. Combined atomic force microscopy and side-view optical imaging for mechanical studies of cells. Nature Methods. 2009;6:383–387. doi: 10.1038/nmeth.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stachowiak JC, et al. Unilamellar vesicle formation and encapsulation by microfluidic jetting. Proc. Natl Acad. Sci. USA. 2008;105:4697–4702. doi: 10.1073/pnas.0710875105. [DOI] [PMC free article] [PubMed] [Google Scholar]