Abstract
The Escherichia coli 30S ribosomal subunit self-assembles in vitro in a hierarchical manner, with the RNA binding by proteins enabled by the prior binding of others under equilibrium conditions. Early 16S rRNA binding proteins also bind faster than late-binding proteins, but the specific causes for the slow binding of late proteins remain unclear. Previously, a pulse-chase monitored by quantitative mass spectrometry method was developed for monitoring 30S subunit assembly kinetics, and here a modified experimental scheme was used to probe kinetic cooperativity by including a step where subsets of ribosomal proteins bind and initiate assembly prior to the pulse-chase kinetics. In this work, 30S ribosomal subunit kinetic reconstitution experiments revealed that thermodynamic dependency does not always correlate with kinetic cooperativity. Some folding transitions that cause subsequent protein binding to be more energetically favorable do not result in faster protein binding. Although 3′ domain primary protein S7 is required for RNA binding by both proteins S9 and S19, prior binding of S7 accelerates the binding of S9, but not S19, indicating there is an additional mechanistic step required for S19 to bind. Such data on kinetic cooperativity and the presence of multiphasic assembly kinetics reveal complexity in the assembly landscape that was previously hidden.
Keywords: mass spectrometry, RNA folding, RNA–protein interactions
Ribosome biogenesis is a central cellular program that accounts for a significant fraction of the energy budget for rapidly growing bacteria, and is an essential process in all living cells. In eukaryotes, ribosome biogenesis in the nucleus requires hundreds of proteins (1, 2), whereas in bacteria the cytoplasmic assembly of ribosomes is facilitated by approximately 20 cofactors (3). Remarkably, the Escherichia coli 30S (4) and 50S (5) ribosomal subunits can be reconstituted in vitro, which has facilitated mechanistic studies on ribosome assembly. The 30S ribosomal subunit is a 900 kDa complex composed of 20 ribosomal proteins (r-proteins; S2, S3, ... S21) and a 1500-nucleotide rRNA (16S RNA). The 30S subunit is a well-characterized model system for studying macromolecular self-assembly in vitro (4), where protein binding occurs in a defined hierarchy and in a cooperative manner (6). The protein binding hierarchy was determined by a series of equilibrium reconstitution experiments that are summarized in the Nomura assembly map (6) (Fig. 1A). Primary proteins bind directly and independently to the 16S RNA, whereas secondary and tertiary proteins require prior binding of one or more proteins, respectively. The thermodynamic order of protein binding is generally consistent with kinetic binding data that show primary proteins binding fastest and tertiary proteins binding slowest (7, 8). The assembly mechanism is also organized according to three structural domains, the 5′ domain, the central domain, and the 3′ domain (9), that can be reconstituted independently in vitro (10–12). Kinetic reconstitution experiments (7, 8) have demonstrated that 30S assembly proceeds in a 5′ to 3′ direction, which is consistent with the cotranscriptional assembly that likely occurs in vivo, where proteins may bind as soon as their binding site becomes available (13–15). In this model, the 5′ domain of the rRNA is already folded and protein-bound by the time the 3′ domain is transcribed.
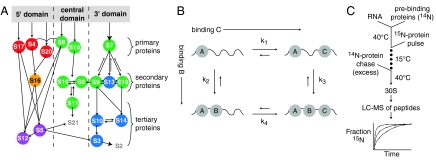
Fig. 1.
(A) Nomura equilibrium assembly map (6, 23). Protein binding dependencies at equilibrium are shown as arrows. Colored circles correspond to protein-binding rates at 15 °C as measured by PC/QMS: 7.8 - 3.1 min-1 (Red), 0.7 min-1 (Orange), 0.042 - 0.015 min-1 (Green), 0.01 - 0.005 min-1 (Blue), and 0.003 - 0.0008 min-1 (Purple). The rates of S2 and S21 are not shown because those proteins bind transiently and hence kinetics cannot be observed using pulse-chase (16). (B) Kinetic cycle with three proteins binding an RNA, showing two potential intermediates. (C) Prebinding PC/QMS experimental scheme. Prebinding proteins are incubated with 16S rRNA at 40 °C before the pulse-chase.
The pulse-chase monitored by quantitative mass spectrometry (PC/QMS) method was previously developed in our laboratory to monitor the in vitro assembly kinetics of 30S subunits. Using this method, the binding kinetics of 18 of the 20 r-proteins can be measured simultaneously. The tertiary proteins S2 and S21 bind transiently with exchange, therefore their binding kinetics cannot be monitored. Initial PC/QMS experiments were performed using MALDI of whole proteins (8), and more recently were implemented with electrospray ionization with time-of-flight MS (ESI-TOF) of tryptic peptides (16). The MALDI studies showed that r-proteins bind over a range of rates at multiple temperatures, and that there is no single populated intermediate or single rate-limiting step during reconstitution (8). Those results also suggested that assembly occurs through multiple parallel assembly pathways, and led to the idea of an assembly landscape as a metaphor for in vitro 30S subunit reconstitution.
A recent time-resolved hydroxyl radical footprinting study has shown that r-proteins associate very rapidly with their binding sites, forming initial encounter complexes that are short-lived and lack many interactions involved in stable thermodynamic association (17). Assembly was nucleated concurrently at multiple sites along the rRNA, and the protections for most nucleotides exhibited multiphasic kinetics (17). These results provide further evidence that assembly proceeds along multiple parallel assembly pathways. The data also suggested that protein binding occurs in stages and concurrently with RNA folding, with loosely associated proteins likely assisting the rRNA in assuming a conformation that allows them to bind more stably (17). The slow refolding observed for some rRNA regions suggests that some particles were caught in kinetic traps stabilized by nonnative RNA conformations. Such slow refolding from kinetic traps has frequently been observed in studies of other large RNAs (18–20). Three lines of evidence led to the hypothesis that the 3′ domain has a stronger tendency to fall into kinetic traps than the two other domains: The 3′ domain assembles slowest at all temperatures (8), and many 3′ domain proteins are clustered around the mRNA decoding site (9), which undergoes conformational changes during the late stages of assembly (21). In addition, chemical modification data from various stages of 30S reconstitution (22) have shown reactivity enhancements to occur in the 3′ domain in the late stages of assembly, which suggests that nonnative interactions within the 3′ domain are unmade.
The classic Nomura assembly map provides a thermodynamic framework for understanding 30S subunit assembly in vitro (6). A significant increase in affinity for a late (i.e., secondary or tertiary) r-protein is observed when an early (i.e., primary or secondary) r-protein is already bound. There is evidence that this cooperative and hierarchical assembly involves stabilization of RNA tertiary structure by the early r-proteins, enabling late r-proteins to bind (12, 24, 25). Given this thermodynamic cooperativity, it is plausible that the rate and the activation energy of late protein binding would also change following binding of an early protein, resulting in kinetic cooperativity that parallels the observed thermodynamic dependence. In contrast, the lack of correlation between thermodynamic and kinetic cooperativity would reveal additional energy barriers during protein binding. Whereas the kinetics of assembly are expected to roughly parallel the Nomura assembly map, the identities and populations of intermediates and the fluxes through the assembly landscape remain to be understood. The phrase “kinetic cooperativity” has been used to describe certain protein folding behaviors (26), and is used in enzymology to describe a cooperative relationship with no thermodynamic component (27). Here, kinetic cooperativity describes a protein-dependent change in binding rate that may or may not correspond to a previously observed thermodynamic relationship. However, given the cooperative nature of assembly, it is unlikely that r-proteins act as simple catalysts for binding, but rather exert a combination of stabilizing effects that alter both the kinetics and thermodynamics of binding.
The Nomura map displays information about thermodynamic dependencies, but does not contain mechanistic information about intermediates that are populated during assembly. A more mechanistic depiction of the binding of three proteins to an RNA is shown in Fig. 1B as a kinetic cycle. Binding of proteins B and C occurs to an intermediate where A is already bound. There are two paths to the final A/B/C-RNA complex, through two distinct intermediates, where either B or C binds first. If B binds faster to the A-RNA complex when C is prebound, that is, if k3 > k2, then C is kinetically cooperative with B. Alternatively, binding of the two proteins could be kinetically independent. Unlike cooperativity in a thermodynamic cycle (28), there is no path-independent constraint on the values of the rates. However, the relative values of the rates do determine the fluxes through possible parallel assembly routes, and therefore affect the populations of possible intermediates.
In the context of ribosome assembly, where protein-RNA contacts outnumber protein–protein contacts, both kinetic and thermodynamic dependencies are likely mediated through RNA folding events (29–31). If S9 is kinetically dependent on S19, the binding of protein S19 may promote a folding event that is required for the binding of S9. In this way, defining the relationships between the kinetics of the proteins can reveal rate-limiting steps in the assembly landscape. Here, the PC/QMS assay is used to investigate kinetic cooperativity between proteins during assembly by looking for changes in protein binding kinetics when defined subsets of r-proteins are prebound.
Results
In a typical PC/QMS experiment, binding kinetics are monitored using a pulse of 15N labeled r-proteins that bind to the rRNA for varying amounts of time before a chase of excess 14N protein is added. The rates of r-proteins binding in a control experiment carried out at 15 °C are depicted on the Nomura assembly map in Fig. 1A. All of the proteins bind slower at low temperature (0.001–8 min-1) than they do under optimal conditions at 40 °C (0.2–30 min-1), but the general order of binding is maintained over this temperature range (8, 16). In the modified experimental scheme (Fig. 1B), a subset of r-proteins is prebound before the pulse-chase, and the kinetics of the remaining proteins are monitored by PC/QMS to look for specific changes in protein binding kinetics and to determine the extent of kinetic cooperativity.
Prebinding of Primary, Secondary, and the Combination of Primary and Secondary Ribosomal Proteins.
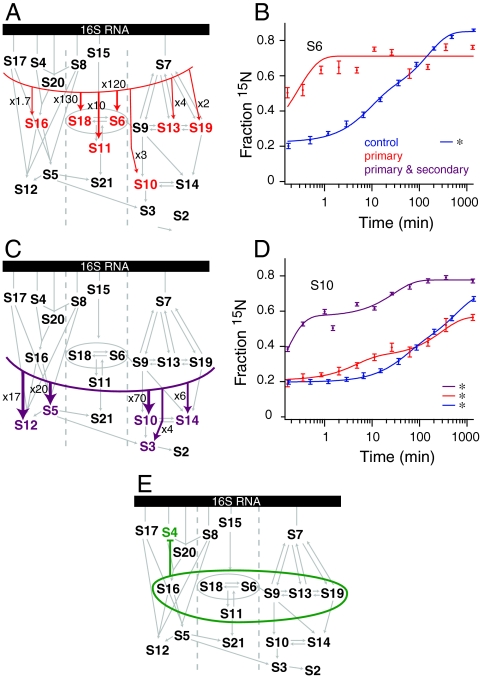
The primary proteins S17, S4, S20, S8, S15, and S7 were prebound as a group to test whether kinetic dependencies of the secondary and tertiary proteins would mirror the observed thermodynamic dependencies. Results were similar for groups of proteins within domains, with the 3′ and 5′ domain proteins showing little acceleration, whereas the central domain proteins showed large accelerations (Fig. 2A). In particular, the binding of central domain proteins S18, S6, and S11 was dramatically accelerated by 130-fold, 120-fold, and 10-fold, respectively (Table 1). A significant portion of the binding of S6 is already complete by the first time point (Fig. 2B). There are likely few significant assembly barriers for the remainder of the central domain after S15 and S8 have bound, which is consistent with previous work showing that S15 stabilizes the native rRNA conformation (12, 24, 32). In the central domain, kinetic cooperativity closely tracks with the known thermodynamic dependency. This is in contrast to the results from the 5′ and 3′ domains, which showed less kinetic cooperativity (Fig. S1), suggesting there are additional kinetic barriers to assembly subsequent to primary protein binding. Half the proteins in the 3′ domain showed modest acceleration (< 5 - fold), whereas proteins in the 5′ domain showed little to no change.
Fig. 2.
(A) Kinetic assembly map from primary protein prebinding experiment. Observed kinetic cooperativity is indicated with colored arrows and colored proteins. The thickness of the arrow relates to the magnitude of the cooperativity, with large accelerations indicated with thick arrows and modest accelerations indicated with thinner arrows. Small gray arrows are from the Nomura map. (B) Protein binding progress curves for protein S6 from experiments prebinding only the primary proteins (Red) and a control experiment (Blue). All curves are single exponential fits except those marked with an asterisk, which were best fit with a double exponential. (C) Kinetic assembly map from primary and secondary protein prebinding experiment. (D) As for (B), except showing S10 progress curves and including data from an experiment prebinding both the primary and secondary proteins (Purple). (E) Kinetic assembly map from secondary proteins prebinding experiment. No kinetic cooperativity was observed, and the binding of protein S4 was inhibited (Fig. S1).
Table 1.
Significant rate increases observed with prebinding (s-1). Fold changes are are of first order fitted rate for the prebinding experiment to the control experiment. Bracketed rates are for datasets that are biphasic according to statistical F-tests (Tables S1–S10), and the resulting fold changes are approximate.
| Protein |
Experiment |
kobs, control |
kobs, prebinding |
Fold change |
| S6 | 1° | [0.030 ± 0.002] | 3.6 ± 0.4 | 120 |
| S10 | 1° | [0.014 ± 0.002] | [0.045 ± 0.008] | 3 |
| S11 | 1° | [0.032 ± 0.003] | 0.32 ± 0.04 | 10 |
| S13 | 1° | [0.014 ± 0.003] | [0.06 ± 0.01] | 4 |
| S16 | 1° | [0.81 ± 0.03] | 1.4 ± 0.1 | 1.7 |
| S18 | 1° | [0.025 ± 0.003] | 3.3 ± 0.2 | 13 |
| S19 | 1° | 0.0188 ± 7.E-04 | [0.047 ± 0.003] | 2 |
| S5 | 1° + 2° | 9.E-04 ± 6.E+04 | 0.020 ± 0.002 | 20 |
| S10 | 1° + 2° | [0.014 ± 0.002] | [0.96 ± 0.08] | 70 |
| S12 | 1° + 2° | 0.003 ± 0.001 | 0.052 ± 0.003 | 17 |
| S14 | 1° + 2° | 0.008 ± 0.001 | 0.048 ± 0.002 | 6 |
| S3 | 5′ + c + S7 | 0.007 ± 0.001 | [0.02 ± 0.002] | 3 |
| S9 | 5′ + c + S7 | [0.018 ± 0.002] | [0.12 ± 0.01] | 7 |
| S10 | 5′ + c + S7 | [0.014 ± 0.002] | [0.023 ± 0.002] | 1.6 |
| S14 | 5′ + c + S7 | 0.008 ± 0.001 | [0.013 ± 0.001] | 1.6 |
| S9 | S7 | [0.018 ± 0.002] | [0.47 ± 0.05] | 26 |
| S9 | S7 + S13 | [0.018 ± 0.002] | [0.082 ± 0.004] | 5 |
| S9 | S7 + S19 | [0.018 ± 0.002] | [4.9 ± 0.2] | 300 |
| S10 | S7 + S19 | [0.014 ± 0.002] | [1.26 ± 0.08] | 90 |
| S13 | S7 + S19 | [0.014 ± 0.003] | [3.1 ± 0.4] | 200 |
| S14 | S7 + S19 | 0.008 ± 0.001 | 0.0121 ± 8.E-04 | 1.5 |
In a similar manner, the set of primary and secondary proteins, S17, S4, S20, S16, S8, S15, S18, S6, S11, S7, S9, S13, and S19, were prebound to determine the effects on subsequent tertiary protein binding. In this experiment, the binding of all of the tertiary proteins for which kinetics can be measured by PC/QMS was accelerated (Fig. 2C and Fig. S1). The most significant acceleration was 7-fold for S10, which also showed a significant burst phase (Fig. 2D). These results confirm that known thermodynamic dependencies also generally correlate with kinetic cooperativity for the tertiary binding proteins.
To determine if secondary proteins can nucleate assembly in the absence of primary proteins, the set of secondary proteins S16, S18, S6, S11, S9, S13, and S19 was prebound in the absence of the primary proteins. In this experiment, none of the secondary proteins bound tightly without exchange during the prebinding stage, consistent with the thermodynamics of the Nomura map (Fig. S1). Prebinding of the secondary proteins alone did not have any significant positive effects on the binding rates of primary or tertiary proteins, and had a negative effect on 5′ domain primary protein S4 (Fig. 2E). These data show that secondary proteins do not have the same capacity for organizing rRNA as primary proteins. Instead, their role in assembly may be to influence the conformation of the rRNA through initially weak interactions. If the secondary binding proteins are weakly associating with 16S rRNA during the prebinding, their association does not facilitate binding of primary proteins, which is consistent with the thermodynamic hierarchy.
Prebinding of 5′ and Central Domain Proteins.
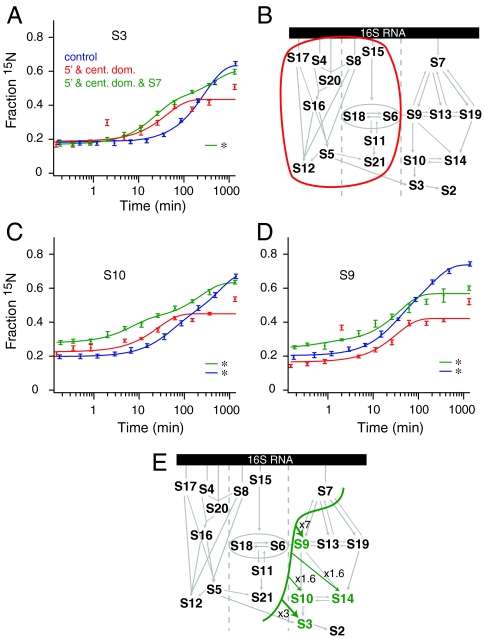
The 5′ and central domain proteins were prebound to measure the effect of interdomain kinetic cooperativity of 3′ domain assembly. A pre-30S particle with the 5′ and central domain proteins bound might correspond to a cotranscriptional assembly intermediate, and the formation of this complex might be expected to rescue efficient assembly of the 3′ domain. Surprisingly, prebinding 5′ and central domain proteins did not accelerate binding of 3′ domain proteins except for a subtle (3-fold) acceleration for binding of S3 (Fig. 3A and B). Furthermore, there was an unexpected decrease in the extent of binding, (Fig. 3C and D and Fig. S2). The lower binding extents indicate that the reconstitution was less efficient, perhaps due to nonspecific binding of 5′ and central domain proteins to the 3′ domain rRNA.
Fig. 3.
(A), Protein binding progress curves for S3 from a control experiment (Blue), the prebinding of the 5′ and central domains (Red), and the prebinding of the 5′ and central domains plus S7 (Green). All curves are single exponential fits except those marked with an asterisk, which are double exponential. (B) Kinetic assembly map showing no significant kinetic cooperativity with the 5′ and central domains prebound. (C)–(D) As in (A) except showing progress curves for (C) S10 and (D) S9. (E). Kinetic assembly map for prebinding of 5′ and central domain proteins plus S7 (Fig. S2).
Another potential cotranscriptional assembly intermediate contains the 5′ and central domain proteins and the primary protein S7. Because S7 nucleates 3′ domain assembly, prebinding this set of proteins could be expected to accelerate the binding of downstream 3′ domain proteins; however, efficient 3′ domain assembly was not observed in this experiment (Fig. 3E). The binding rates display relatively subtle (2- to 7-fold, Table 1) rate enhancements and additional kinetic phases in the binding kinetics of S9, S10, S14, and S3, that are shown in Fig. 3A–C and summarized in a kinetic map in Fig. 3E. The extent of 15N protein binding was again lower compared to the control, indicating that the efficiency of the reconstitution is lower. Taken together, these two experiments show that the assembly state of the 5′ and central domains has little effect on the assembly kinetics of the 3′ domain and that there are additional assembly barriers after S7 has bound. These data suggest the yield of low-temperature reconstitutions is sensitive to changes in the starting point, and that the 16S rRNA with the 5′ domain and central domain proteins bound with S7 is likely not an authentic in vitro assembly intermediate. Furthermore, prebinding of this extensive set of proteins does not rescue efficient assembly of the 3′ domain, indicating that the kinetic trap associated with 3′ domain folding in vitro is likely due to misfolding localized within the 3′ domain.
Prebinding 3′ Domain Proteins.
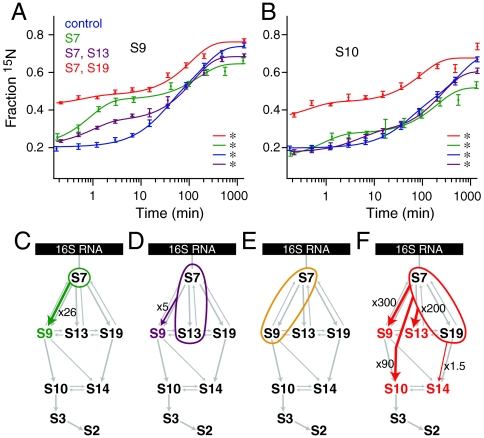
Kinetic cooperativity within the 3′ domain was investigated by prebinding different subsets of 3′ domain proteins. Prebinding S7 alone caused significant acceleration in S9 binding (Fig. 4A and Table 1) and subtle acceleration in S10 binding (Fig. 4B), with no other changes except an overall lower binding extent (Fig. 4C). The kinetic cooperativity between S7 and S9 is consistent with thermodynamic data, but the lack of acceleration of S13 and S19 (Fig. S3) is not, because all three proteins bind immediately downstream of S7 in the Nomura map. These results suggest that there are additional slow steps required for the binding of S19 and S13 that are independent of S7 binding.
Fig. 4.
(A)–(B) Protein binding progress curves for proteins (A) S9 and (B) S10 from a control experiment (Blue), prebinding of S7 (Green), prebinding of S7 and S19 (Red), and prebinding of S7 and S13 (Purple). Double exponential curves are indicated with an asterisk. (C)–(F) Kinetic assembly maps of the 3′ domain only from experiments prebinding (C) S7 alone, (D) S7 and S13, (E) S7 and S9, (F) S7 and S19 (Fig. S3).
Prebinding experiments were then conducted with S7 in combination with each of the three secondary proteins S9, S13, and S19. Prebinding S7 and S13 together only caused the acceleration of S9 binding (Fig. 4A and D). Unexpectedly, prebinding S7 and S9 together gave rise to no significant changes (Fig. 4E), even though S10, S14, and S13 are all thermodynamically dependent on S9 based on the Nomura map. Prebinding both S7 and S19 together showed significant accelerations across the 3′ domain (Fig. 4F and Table 1). The extensive kinetic changes upon prebinding of S7 and S19 were unexpected because S9 and S10 have no known thermodynamic dependency on S19. These data suggest a previously unappreciated role for S19 as a secondary organizer of the 3′ domain. It is notable that although S9 and S19 occupy similar places in the Nomura map, the trends in their binding kinetics in these experiments diverge. S9 was affected significantly and differently by each set of prebinding proteins (Fig. 4A), whereas S19 was affected only subtly by the prebinding of other 3′ domain proteins (Fig. 4D). The binding kinetics of tertiary proteins in the 3′ domain did not change significantly except for the experiment in which S7 and S19 were prebound (Fig. 4B and Fig. S3).
Discussion
In general, these results provide a significantly deeper insight into the mechanism of 30S ribosome assembly than can be inferred from the Nomura map alone. Although some of the established thermodynamic dependencies do correspond directly to observed kinetic cooperativity, some do not correspond, and new kinetic cooperativity has been observed with no thermodynamic effect as a counterpart. Prebinding of subsets of r-proteins did not promote more efficient 3′ domain assembly, and several of the prebinding experiments showed a reduced extent of reconstitution (Fig. S4C). The most efficient and complete reconstitution reactions occurred with all the proteins added to the rRNA at the same time, which is consistent with previous work showing that protein binding and rRNA folding occur concurrently (17). This is in contrast to observations of reconstitutions with a full complement of recombinant r-proteins, for which an ordered addition of proteins produces a more efficient reconstitution (33). In these prebinding PC/QMS experiments, the absence of certain proteins during the prebinding stage may have led to a less efficient reconstitution by causing a portion of the molecules to form kinetic traps. Whereas the secondary protein prebinding experiment showed that late-binding proteins do not bind stably or independently, their transient associations (17) could facilitate ordered and complete assembly by subtly directing local RNA folding.
Additional kinetic phases were a feature of the data from several prebinding experiments. Most proteins that were monophasic or had only a minor second phase in the control experiment exhibited biphasic behavior in at least one prebinding experiment (Fig. S5A). In some cases, a burst phase caused a significant portion of the binding to be complete by the first time point (Fig. S5B). Additional kinetic phases are evidence of multiple populations of particles binding proteins at different rates, that is, through multiple parallel assembly pathways (8, 16, 17). It is clear in these cases that kinetic cooperativity is a result of the prebinding step rendering some portion of the 16S molecules more competent to bind downstream proteins, presumably because some slow folding steps that are required have already been completed during the prebinding.
Comparison of Kinetic and Thermodynamic Cooperativity.
These experiments were expected to reveal kinetic cooperativity that is similar to the thermodynamic dependencies previously observed, however, only some thermodynamic dependencies translated into kinetic cooperativities, revealing additional complexity in the assembly landscape. Prebinding the primary proteins showed that kinetic cooperativity in the central domain closely mirrors thermodynamic dependency. Kinetic cooperativity is also closely associated with thermodynamic dependency for tertiary binding proteins in the 5′ and 3′ domains, as shown by the results of the experiment prebinding both primary and secondary proteins. It appears that there is a higher degree of kinetic cooperativity in the late stages of assembly as compared to the early stages, which may reflect the requirement for prior organization of much of the 16S rRNA. In the 3′ domain, S9 was accelerated to varying extents in all three 3′ domain prebinding experiments, suggesting there are several assembly pathways leading to S9 binding. Because no such acceleration was observed for S19, which occupies a similar position in the assembly map, it appears that the two assembly branches in the 3′ domain, the S9 and S19 branch, differ in their assembly mechanisms. Specifically, it is likely that an additional slow folding event is required after the binding of S7, before S19 can bind stably without exchange. In the case of S19 prebinding, kinetic cooperativity was observed where there was no thermodynamic dependency, suggesting S19 may direct assembly through channels not evident from thermodynamic data. Although this was surprising, it has been previously noted that S19 likely plays a greater role than other 3′ domain secondary proteins in 16S rRNA conformational changes during the late stages of assembly (21). In addition, S19 is known to interact with the ribosome biogenesis factor RimM (34), so those two proteins together may facilitate efficient 3′ domain assembly in vivo. In the case of S9 and S10, prebinding of S19 revealed kinetic cooperativity where no thermodynamic cooperativity had been previously observed. It is possible that S19 acts in part as a chaperone in the binding of these proteins, both lowering the energy barrier to S9 and S10 binding and also stabilizing the pre-30S conformations that are competent to bind them.
The thermodynamic data that inspired these prebinding experiments are the dependencies described by the Nomura assembly map. These dependencies were revealed by omission and addition experiments, where the extents of r-protein binding to 16S rRNA at equilibrium changed depending on the presence or absence of other r-proteins. In a two-state system, the ratio of the binding and dissociation rates corresponds to the equilibrium constant, and the apparent binding rate, kobs, corresponds to the sum of the binding and dissociation rates. PC/QMS measures the rate of formation of a kinetically stable complex, and complexes with a fast dissociation rate are not observed because the chase provides an opportunity for exchange. In contrast, such unstable initial encounter complexes were observed in a recent time-resolved hydroxyl radical footprinting study (17). Unlike a simple two-state system, ribosome assembly is a complex multistate system with many potential unseen intermediates with unknown rates of formation. Although kinetic data from prebinding experiments help describe the effects of specific proteins, it can be difficult to attribute the rates observed to specific molecular events. The complex relationship between the thermodynamic data and the prebinding kinetic data is particularly apparent in the 3′ domain, where fluxes and populated intermediates are not well predicted from the Nomura map.
Interdomain Cooperativity.
The 5′ and central domain proteins were prebound to test the hypothesis that creating a putative assembly intermediate might rescue fast assembly of the 3′ domain. However, there was little interdomain kinetic cooperativity, which suggests that the formation of interdomain long-range tertiary interactions is not necessary for stable r-protein binding within domains, consistent with the ability of individual domains to assemble independently (10–12). These experiments also provide evidence that nonnative interdomain interactions are likely not a source of kinetic traps in reconstitution, and instead, the assembly of the 3′ domain is likely limited by folding events that occur within the domain. However, given that the central domain proteins bind mainly to the 5′ half of the central domain rRNA (35), the possibility remains that there is nonnative association between the 3′ domain rRNA and the 3′ half of central domain. Recent work has shown that a circularly permutated 16S rRNA with the 3′ domain positioned 5′ to the remainder of the 16S in the genome is competent to form functional ribosomes (36), indicating that the domain order is nonessential. The native domain order may still be preferred, as the permuted 16S rRNA requires the presence of the DeaD helicase. In general, the permutation experiment provides further support to the hypothesis that interdomain interactions are not essential during assembly.
Efficiency of Prebinding.
There is evidence that initial r-protein binding is transient and kinetically unstable (17). Unstable rRNA-protein complexes were also observed in our experiments, during the prebinding phase. Proteins that prebound stably had a low fraction 15N throughout all time points, because the 14N prebound protein occupied all available binding sites (Fig. S5A and B). Other proteins, that prebound unstably, showed a higher, time-dependent fraction 15N as transiently prebound protein fell off and was partially replaced with 15N protein during the pulse (Fig. S4B). It is also possible that some 14N prebinding proteins failed entirely to associate with the rRNA, but given the encounter complex observations (17) it is more likely that these proteins were transiently associated with their binding sites. In either case, partially or transiently prebound proteins produce a heterogeneous population of pre-30S particles, which is revealed by high and time-dependent fraction 15N values. In most cases, the rate of increase of the fraction 15N of the prebound protein was similar to the binding rate observed in the control experiment. The control experiment also shows evidence of multiple populations of particles assembling through different pathways, so it seems likely that prebinding alters the pre-30S populations and that observed kinetic changes are the result of some of the populations assembling faster, whereas the rate of assembly of other populations may remain unchanged. The proteins that bound transiently or partially in prebinding experiments were S5, S7, S9, S11, S13, and S19. A similar subset of proteins was found to bind transiently in a previous pulse-chase experiment designed to test binding stability in the presence of a chase (37). Unstable binding also occurred in the experiment with secondary proteins prebound, which showed no acceleration for primary or tertiary proteins. These results suggest that although secondary proteins may bind transiently to the rRNA (17) they do not significantly drive rRNA folding in the absence of the primary proteins.
Conclusions
In E. coli, ribosome assembly is very efficient, because exponentially growing cells need to produce tens of thousands of ribosomes every generation (38). During in vivo ribosome biogenesis, it is thought that r-proteins bind as soon as their binding sites have been transcribed (14, 15), which likely avoids some kinetic traps that occur in vitro during reconstitution with intact renatured 16S rRNA. The 5′–3′ terminal stem loop flanking the mature 16S rRNA (39) may also limit the conformations that the rRNA can assume prior to processing by Rnase III. Furthermore, the most significant conformational changes that occur during ribosome biogenesis may be assisted by assembly factors such as helicases, GTPases, and other proteins (3). The rate-limiting steps that have been identified here, as locations where kinetic cooperativity is absent and thermodynamic dependency is present, may be points where assembly factors facilitate ribosome biogenesis.
This set of prebinding PC/QMS experiments has revealed additional complexity in the landscape of in vitro assembly that will inform and enable future work probing the mechanism of 30S ribosomal subunit assembly. These experiments revealed cases where kinetic and thermodynamic cooperativity aligned, cases where kinetic cooperativity was absent, and cases where kinetic cooperativity without thermodynamic precedent was observed. The kinetic cooperativity described here, such as that between S19 and other 3′ domain proteins, has significant implications for the understanding of intermediate populations and assembly pathways. Defining the rate-limiting steps during in vitro assembly provides valuable general insights into the kinetic traps for formation of ribonucleoprotein complexes, in addition to providing hints at the roles of ribosome assembly factors during biogenesis.
Methods
Stable-isotope pulse-chase experiments were performed as described previously with some modifications (8). For prebinding experiments, a prebinding step was added before the 15N pulse, in which mixtures of individually purified proteins or buffer were mixed with 180 pmol 16S rRNA and incubated at 40 °C for 40 min to 60 min in reconstitution buffer (RB, 25 mM Tris pH 7.5, 20 mM MgCl2, 330 mM KCl, 2 mM DTT). The concentration of 16S rRNA during the prebinding step was 0.33 μM, and the concentration of protein was 0.49 μM. Five minutes before the start of the pulse, the samples were transferred to a 15 °C water bath. The pulse consisted of 45 μl of 6 μM 15N TP30 in RB, and ranged in length from 10 sec to 22 h. Upon addition of a chase of 135 μl of 10 μM 14N TP30 in RB, the samples were immediately transferred to 40 °C and incubated for 40 min, after which they were placed on ice. Subsequent purification and LC-MS analysis was carried out as described previously (16, 40). The kinetic curves were fit to a double or single exponential curve, depending on the results of F-tests, using a 95% confidence interval (Fig. S6 and Tables S1–S10.)
Supplementary Material
Acknowledgments.
We thank Prof. Gary Siuzdak and Dr. Sunia Trauger of the Scripps Center for Mass Spectrometry for advice and access to instrumentation; Prof. Gloria Culver for ribosomal protein plasmids; Edit Sperling for purified ribosomal proteins; Dr. Michael T. Sykes for mass spectrometry data analysis programs; and Mr. William Ridgeway, Dr. Zahra Shajani, and Dr. Michael T. Sykes for critical comments on the manuscript. This work was supported by the National Institutes of Health Grant R37-GM-53757 (J.R.W.), and by the Skaggs Institute for Chemical Biology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912007107/DCSupplemental.
References
- 1.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Henras AK, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 4.Traub P, Nomura M. Structure and function of E. coli ribosomes v. reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc Natl Acad Sci USA. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc Natl Acad Sci USA. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Held W, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from Escherichia Coli: Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 7.Powers T, Daubresse G, Noller H. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J Mol Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 8.Talkington M, Siuzdak G, Williamson J. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuwirth B, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 10.Weitzmann C, Cunningham P, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–180. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 11.Samaha R, O’Brien B, O’Brien T, Noller H. Independent in vitro assembly of a ribonucleoprotein particle containing the 3′ domain of 16S rRNA. Proc Natl Acad Sci USA. 1994;91:7884–7888. doi: 10.1073/pnas.91.17.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Recht M, Williamson J. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol R. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Narvaez CC, Schaup HW. In vivo transcriptionally coupled assembly of Escherichia coli ribosomal subunits. J Mol Biol. 1979;134:1–22. doi: 10.1016/0022-2836(79)90411-x. [DOI] [PubMed] [Google Scholar]
- 15.Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;231:581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 16.Bunner AE, Trauger SA, Siuzdak G, Williamson JR. Quantitative ESI-TOF analysis of macromolecular assembly kinetics. Anal Chem. 2008;80:9379–9386. doi: 10.1021/ac8020505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adilakshmi T, Bellur D, Woodson S. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan J, Thirumalai D, Woodson S. Folding of RNA involves parallel pathways. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 19.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. Kinetic intermediates trapped by native interactions in RNA folding. Science. 1998;279:1943–1946. doi: 10.1126/science.279.5358.1943. [DOI] [PubMed] [Google Scholar]
- 20.Zarrinkar P, Williamson J. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat Struct Biol. 1996;3:432–438. doi: 10.1038/nsb0596-432. [DOI] [PubMed] [Google Scholar]
- 21.Holmes KL, Culver GM. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol. 2004;11:179–186. doi: 10.1038/nsmb719. [DOI] [PubMed] [Google Scholar]
- 22.Holmes KL, Culver GM. Analysis of conformational changes in 16 S rRNA during the course of 30 S subunit assembly. J Mol Biol. 2005;354:340–357. doi: 10.1016/j.jmb.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 23.Grondek J, Culver G. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–1866. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batey RT, Williamson JR. Effects of polyvalent cations on the folding of an rRNA three-way junction and binding of ribosomal protein S15. RNA. 1998;4:984–997. doi: 10.1017/s1355838298980426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. Structure of the S15,S6,S18-rRNA complex: Assembly of the 30S ribosome central domain. Science. 2000;288:107–113. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 26.Chan HS, Shimizu S, Kaya H. Cooperativity principles in protein folding. Methods Enzymol. 2004;380:350–379. doi: 10.1016/S0076-6879(04)80016-8. [DOI] [PubMed] [Google Scholar]
- 27.Neet KE. Cooperativity in enzyme function: Equilibrium and kinetic aspects. Methods Enzymol. 1980;64:139–192. doi: 10.1016/s0076-6879(80)64009-9. [DOI] [PubMed] [Google Scholar]
- 28.Williamson JR. Cooperativity in macromolecular assembly. Nat Chem Biol. 2008;4:458–465. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 29.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 30.Stern S, Powers T, Changchien LM, Noller HF. RNA–protein interactions in 30S ribosomal subunits: Folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 31.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Recht MI, Williamson JR. Central domain assembly: Thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- 33.Culver G, Noller H. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–843. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lövgren JM, et al. The PRC-barrel domain of the ribosome maturation protein RimM mediates binding to ribosomal protein S19 in the 30S ribosomal subunits. RNA. 2004;10:1798–1812. doi: 10.1261/rna.7720204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agalarov SC, Williamson JR. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA. 2000;6:402–408. doi: 10.1017/s1355838200991945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitahara K, Suzuki T. The ordered transcription of RNA domains is not essential for ribosome biogenesis in Escherichia coli. Mol Cell. 2009;34:760–766. doi: 10.1016/j.molcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–7055. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- 38.Neidhardt FC. Chemical Composition of Escherichia coli. In: Neidhardt FC, editor. Echerichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology. Vol 1. Washington, D.C.: American Society of Microbiology; 1987. pp. 3–6. [Google Scholar]
- 39.Kaczanowkska M, Rydén-Aulin M. Ribosome Biogenesis and the Translation Process in Escherichia coli. Microbiol Mol Biol R. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sperling E, Bunner AE, Sykes MT, Williamson JR. Quantitative analysis of isotope distributions in proteomic mass spectrometry using least-squares fourier transform convolution. Anal Chem. 2008;80:4906–4917. doi: 10.1021/ac800080v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.