Abstract
Protein localization mechanisms dictate the functional and structural specialization of cells. Of the four polar surface organelles featured by the dimorphic bacterium Caulobacter crescentus, the stalk, a cylindrical extension of all cell envelope layers, is the least well characterized at the molecular level. Here we apply a powerful experimental scheme that integrates genetics with high-throughput localization to discover StpX, an uncharacterized bitopic membrane protein that modulates stalk elongation and is sequestered to the stalk. In stalkless mutants StpX is dispersed. Two populations of StpX were discernible within the stalk with different mobilities: an immobile one near the stalk base and a mobile one near the stalk tip. Molecular anatomy provides evidence that (i) the StpX transmembrane domain enables access to the stalk organelle, (ii) the N-terminal periplasmic domain mediates retention in the stalk, and (iii) the C-terminal cytoplasmic domain enhances diffusion within the stalk. Moreover, the accumulation of StpX and an N-terminally truncated isoform is differentially coordinated with the cell cycle. Thus, at the submicron scale the localization and the mobility of a protein are precisely regulated in space and time and are important for the correct organization of a subcellular compartment or organelle such as the stalk.
Keywords: Caulobacter, fluorescence loss in photobleaching/fluorescence recovery after photobleaching, polar organelle, protein localization, protein mobility
Mechanisms exist in prokaryotic and eukaryotic cells to direct specialized proteins to distinct subcellular sites where they execute topologically constrained functions, for example, morphogenesis (1). Dissecting the underlying molecular mechanisms for localization is facilitated by the availability of suitable proteins that can be used as molecular probes. In the dimorphic Gram-negative bacterium Caulobacter crescentus localization probes for cellular organelles such as the medial cytokinetic apparatus, the cell-fate signaling hub at the old pole, and surface organelles positioned at the new pole (pili or the flagellum) led to the establishment of molecular localization hierarchies (2–5). C. crescentus also features another polar surface organelle, the stalk, whose molecular anatomy is not well characterized. Until recently, no molecular probes for the study of the stalk ultrastructure and biogenesis were known. Conducting a cytological survey of 75% of the predicted translation products of Caulobacter, Werner et al. uncovered candidate proteins that appear associated with the stalk (6). The stalk is a cylindrical protrusion of all envelope layers (inner membrane, periplasm, outer membrane, and S-layer) at the old cell pole and encloses cytoplasmic material that is free of chromosomal DNA, ribosomes, and most cytoplasmic proteins (7, 8). The absence of ribosomes precludes a cotranslational protein targeting mechanism within the stalk. Instead, mobile molecules might diffuse into the stalk where they are subsequently retained, i.e., immobilized, essentially resembling a “diffusion-and-capture”-based localization mechanism (9). Alternatively, proteins might be inserted into the stalk envelope near the stalk base prior to and/or during stalk outgrowth.
Stalk outgrowth occurs at a specific time during the cell division cycle (10). At division, two functionally and morphologically specialized cell types are formed: a stalkless swarmer (SW) cell that harbors a single flagellum and several pili at the old cell pole and a stalked (ST) cell that features a stalk at the old cell pole. Although the SW cell performs flagellar-mediated chemotaxis akin to a dispersal cell, it is unable to replicate DNA. By contrast, the ST cell is sessile and replication competent. Unknown signals orchestrate a developmental transition (also known as G1 → S transition) in which the SW cell loses the flagellum and pili, elaborates a stalk from the vacated pole, and acquires replication competence (11). The G1 → S transition marks the onset of stalk elongation and the stalk continues to elongate during each subsequent ST cell cycle. Thus, the ST cell is terminally differentiated, but continues to spawn fresh SW cells through its asymmetric division.
Here, we report the identification of a hitherto uncharacterized protein (stalk-specific protein X, StpX) and its use (i) to probe the molecular anatomy of the stalk, (ii) to define the localization requirements that underlie sorting into the stalk, (iii) to observe unexpected mobility properties of StpX as a function of its position in the stalk, and (iv) to show that the production of StpX and an N-terminally shortened isoform is differentially cell-cycle regulated.
Results and Discussion
Identification of StpX Using a Random Protein Localization Screen.
To uncover localized proteins in C. crescentus in an unbiased fashion, we generated libraries of strains with translational fusions to the GFP gene (SI Text) at random positions to genomic sequences on a plasmid or on the chromosome [using a mini-Tn5-GFP derivative (ref. 12); see Experimental Procedures]. Strains were grown in 96-well plate format and then imaged by epifluorescence microscopy in pools of 8 (Fig. S1A). From >55,000 strains investigated, 50 clones harboring chimeric genes that direct GFP from cytoplasmic homogeneity into a cluster(s) at various subcellular sites were isolated. Sequencing revealed in-frame fusions of GFP to ORFs encoding polarity factors (TipN, PodJ) (4, 5, 13, 14) and developmental or chemosensory kinases (DivL, DivJ, CheA) (15, 16) that are known to localize to the pole(s) (Fig. S2A and Table S1). Additionally, fusions to a number of ORFs encoding regulatory proteins, metabolic enzymes, and other proteins with diverse predicted cellular function(s) and with unknown function were identified that can impart focal localization to GFP (Fig. S2 B and C and Table S2). The overlap between our set of localized proteins and that of Werner et al. (6) is quite low, possibly due to the difference of (i) the fusion junction (position within the ORF and the linker sequence), (ii) the fluorophore [(s)EGFP vs. mCherry], (iii) the context of expression (chromosomal, episomal), and (iv) errors in genome annotation that were unknowingly included with the design of the ORFeome library (see below).
The discovery of a strain (BR-E7) that expresses a Tn5-GFP fusion yielding an extended patch of fluorescence apparently restricted to the stalk compartment as judged by differential interference contrast (DIC) imaging (Fig. S1A) was particularly interesting. BR-E7 harbors a translational fusion of GFP to residue 553 (Fig. S1B) of the predicted 642-residue protein CC1238 that is encoded from nucleotides 1396070 to 1394142 of the CB15 genome [note that the original annotation of CC1238 is incorrect (ref. 17) and therefore CC1238 had not been localized]. The localization pattern of BR-E7 was recapitulated in a strain engineered to express a functional fusion (Fig. S3A) of sEGFP (GFP) to the C-terminal residue of CC1238 at the native chromosomal locus (Fig. 1 A and F; see Fig. 3A).
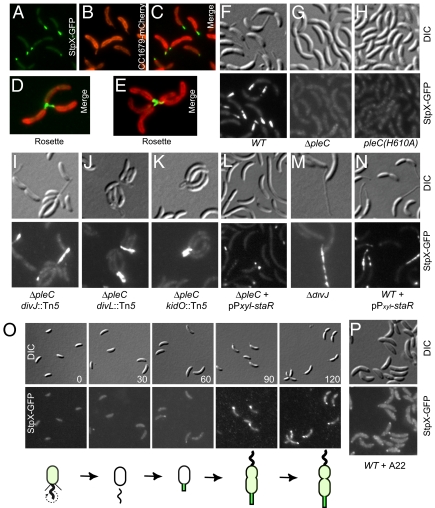
Fig. 1.
Localization of StpX-GFP in wild-type (WT) and mutant cells that are stalkless or that have elongated stalks in which WT stpX is replaced by stpX-GFP under the control of the native promoter. (A–E) Colocalization of CC1679-mCherry and StpX-GFP in NA1000 (A–C) or CB15 cells (D and E). Cells harbor stpX-GFP and pPvan-CC1679-mCherry, a low-copy plasmid encoding CC1679-mCherry under the control of the vanillate-inducible promoter (Pvan). (F–N and P) Mutants harboring stpX-GFP. (L and N) StpX-GFP cells with pPxyl-StaR, a low-copy plasmid encoding StaR under the control of the xylose-inducible promoter (Pxyl). (O) Time-course fluorescence imaging of stpX-GFP cells during cell-cycle progression. Numbers in DIC panels indicate the time in minutes samples were imaged, relative to the start of the cell cycle (SW cell stage). A summarizing schematic shown beneath the images depicts delocalized (light green) and localized (bright green) StpX-GFP in SW and ST cells, respectively. (P) Localization of StpX-GFP after exposure of stpX-GFP SW cells to A22 (12.5 μg/mL) to inhibit stalk outgrowth. Addition of the same amount of solvent (methanol) had no effect on StpX-GFP localization.
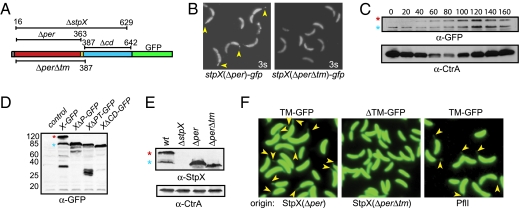
Fig. 3.
Localization determinants and accumulation of StpX isoforms. (A) Schematic of the predicted primary structure predictions of StpX, StpX-GFP, and deletion derivatives, including the signal sequence peptide (SS, gray box), the periplasmic domain (per, red box), the transmembrane (TM) domain (yellow box), the cytoplasmic domain (cd, turquoise box), and the GFP moiety (green box). The numbers refer to amino acid residues of StpX. (B) Fluorescence image (3s, 3-sec exposure) analyses of cells expressing StpX(Δper)-GFP or StpX(ΔperΔtm)-GFP from the native chromosomal locus under control of its endogenous promoter in place of WT StpX. Yellow arrowheads point to fluorescent stalks. (C) Immunoblot analysis of StpX-GFP at different stages of the cell cycle (see schematic in Fig. 10; 20-min intervals indicated by multiples of 20), using polyclonal antibodies to GFP (α-GFP). (D) Immunoblot analysis of NA1000 (control), stpX-GFP, stpX(Δcd)-GFP, and the strains in B, using a polyclonal antibody to GFP. StpX-GFP, StpX(Δper)-sEGFP, StpX(ΔperΔtm)-GFP, and StpX(Δcd)-GFP are abbreviated as X-GFP, XΔP-GFP, XΔPT-GFP, and XΔCD-GFP, respectively. (E) Immunoblots of endogenous (untagged) StpX in WT and deletion strains using polyclonal antibodies to the C-terminal domain of StpX. (C–E) Full-length (red asterisk) and truncated (blue asterisk) StpX (E) or StpX-GFP (C and D) are indicated. (C and E) Immunoblots of CtrA are shown as a control for the cell cycle and/or loading. (F) NA1000 cells expressing a short polypeptide with (Left) or without (Center) the TM domain of StpX or that from the bitopic membrane protein PflI (CC2060, Right) fused to GFP under Pvan control from plasmids. Cells were imaged after 5 h of growth in the presence of 0.5 mM vanillate. Cells harbor pCWR435, pCWR437, and p2060-TM-long-510 encoding the short polypeptide-GFP chimeras TM-GFP, ΔTM-GFP, and TM-GFP that derive from StpX(Δper), StpX(ΔperΔtm), and PflI, respectively (labeled beneath the images as “origin”).
CC1238 does not exhibit primary structure resemblance to any protein in the database with a known function. Sequence analysis using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM) predicts that CC1238 is a bitopic membrane protein (Fig. S1B; see Fig. 3A) with a large N-terminal periplasmic domain (residues 27–366), followed by a transmembrane (TM) domain (residues 367–389) and a C-terminal domain facing the cytoplasm (residues 390–642). Moreover, SignalP 3.0 analysis using Hidden Markov models (http://www.cbs.dtu.dk/services/SignalP) predicts a SecA-dependent secretion signal (SS) at the N terminus with a putative cleavage site between residues 26 and 27. On the basis of the localization data we henceforth refer to CC1238 as StpX.
StpX Is Sorted to the Stalk.
To simultaneously observe StpX and the stalk by fluorescence microscopy, we colocalized StpX-GFP in cells expressing mCherry-tagged CC1679 (CC1679-mCherry) in which the entire cell envelope, including the stalk, is red fluorescent (Fig. 1B). The peripheral distribution of CC1679, a predicted periplasmic protein harboring an N-terminal SS, but no TM domains, is consistent with the notion that the periplasm is contiguous between the cell body and the stalk (18). In support of this, fragments with the putative SS of the BlaA (CC2139) β-lactamase (residues 1–50) or StpX (residues 1–49) are sufficient to yield mCherry-derived fluorescence along the cell body and the stalk (Fig. S1C).
The overlay of the fluorescence micrographs (Fig. 1C) shows that the StpX-GFP signal overlaps with that of CC1679-mCherry in the stalk. In the wild C. crescentus isolate CB15, cells can adhere symmetrically to one another via the holdfast located at the tip of the stalk to form a multicellular structure known as a rosette. Rosettes of CB15 cells expressing StpX-GFP appear as fluorescent lines converging toward the rosette center, reflecting the arrangement of the stalks within the rosette (Fig. 1 D and E).
To obtain genetic evidence that StpX is sorted to the stalk, we localized StpX-GFP in mutant strains that are stalkless or that have elongated stalks. The bifunctional PleC phosphatase/kinase is a positive regulator of a phospho-signaling cascade that promotes stalk elongation (19, 20). PleC− (ΔpleC) cells are stalkless, presumably due to the failure to express the StaR transcriptional regulator and perhaps additional proteins (19). StpX-GFP is dispersed throughout the cell body in ΔpleC cells (Fig. 1G) and immunoblotting revealed comparable steady-state levels of StpX-GFP in wild-type (WT) and ΔpleC cells (Fig. S3B). Three lines of evidence demonstrate that the stalk-specific disposition of StpX is regulated by PleC's activity on the phospho-cascade, rather than by the mere presence of the PleC polypeptide. First, StpX-GFP is diffuse in the pleC(H610A) strain (Fig. 1H) that stably expresses the catalytically inactive form (lacking phosphatase/kinase activity) in place of WT PleC (21). Second, StpX-GFP localizes correctly to the stalk in ΔpleC cells that harbor a compensatory Tn5 mutation in the gene encoding DivJ, DivL or the recently identified KidO oxido-reductase homolog (Fig. 1 I–K) (22, 23). These mutations reenable the phospho-cascade via other entry points to allow stalk formation, presumably by restoring StaR expression and/or other proteins involved in stalk biogenesis. Third, and in support of the aforementioned idea, expression of StaR from the xlyose-inducible promoter on a low-copy plasmid (pPxyl-staR) in ΔpleC cells partly restores StpX-GFP localization to the stalk (Fig. 1L).
Conversely, we also explored the localization of StpX-GFP in strains with long stalks. DivJ− (ΔdivJ) cells (i) accumulate the staR message to high levels (19), (ii) have markedly elongated stalks, and (iii) have StpX-GFP scattered throughout the stalk (Fig. 1M). In support of the idea that stalk elongation in ΔdivJ cells is due to elevated StaR levels, overexpression of StaR in WT cells from the xylose-inducible promoter on a low-copy plasmid phenocopies the effects of the ΔdivJ mutation, yielding cells with extended stalks in which StpX-GFP fluorescence is heterogeneously distributed (Fig. 1N). The irregularity in StpX-GFP fluorescence, particularly notable in cells with extended stalks, may be related to the presence of the enigmatic “crossbands” (24), transverse linings of the stalk that are discernible by transmission electron microscopy and might provide selective barrier functions for microcompartments and/or regions in the stalk to exclude proteins like StpX.
Finally, we conducted cell-cycle localization experiments to explore if StpX-GFP localization correlates with the onset of stalk biogenesis (Fig. 1O). Whereas no distinct StpX-GFP foci were observed in SW cells, a polar focus of fluorescence formed between the 30- and 60-min time point, the time SW cells are reprogrammed into ST cells. This fluorescence signal lengthened and intensified over time and clearly enveloped the elaborating stalk by the 90-min time point. The onset of stalk outgrowth at the G1 → S transition can be inhibited with the small molecule A22 that interferes with MreB-dependent stalk formation (25). Brief exposure of SW cells to A22 (12.5 μg/mL) results in stalkless cells that appear otherwise phenotypically normal (25). The finding that StpX-GFP is mostly delocalized, occasionally with a dim polar focus in A22-treated cells (Fig. 1P), suggests that the elaboration of StpX-fluorescent stalks is also MreB dependent.
Role of StpX in Stalk Elongation Control.
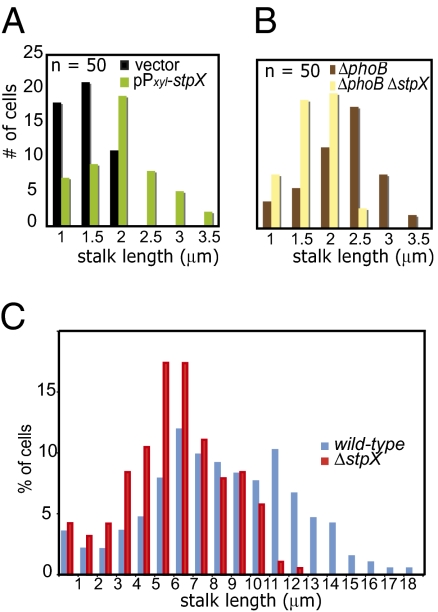
The genetic and pharmacological evidence for StpX localization to the stalk described above prompted us to test if StpX plays a role in stalk morphogenesis and/or function. In support of this, overexpression of StpX from the xylose-inducible promoter on a low-copy plasmid (pPxyl-stpX) promotes elongation of the stalk in rich peptone-yeast extract (PYE) medium. As shown in Fig. 2A, distribution of cells with long stalks (>2 μm) increased when StpX is overexpressed compared to cells with normal StpX levels. To test if the absence of StpX has the opposite effect on stalk length, we determined the distribution of stalk lengths in WT and StpX− cells harboring an in-frame deletion in stpX (ΔstpX) (Fig. 3A). Whereas no difference in stalk length was apparent between WT and ΔstpX cells grown in PYE, StpX is required for stalk elongation under phosphate-limiting conditions [Hutner base–imidazole-buffered–glucose–glutamate (HIGG) medium + 30 μM phosphate, henceforth referred to as HIGG; Fig. 2C]. The mean length of ΔstpX stalks (6.1 ± 2.6 μm, n = 190) is significantly [t(382) = 7.4, P < 0.0001] shorter than that of WT stalks (8.6 ± 3.8 μm, n = 194). Next, we tested if StpX can promote stalk elongation in PYE when cells are artificially coaxed into perceiving phosphate limitation. To this end, the ΔstpX mutation was introduced into PhoB− (ΔphoB) cells lacking the PhoB phosphate regulon response regulator that prevents stalk elongation in rich medium (26). When the distribution of stalk lengths of ΔstpX ΔphoB double-mutant cells was compared to that of ΔphoB single-mutant cells, only few stalks measuring ≥2.5 μm were counted in ΔstpX ΔphoB cells, whereas stalks of ΔphoB cells often exceeded 3 μm in length (Fig. 2B).
Fig. 2.
Stalk elongation control by StpX. (A) Stalk length distribution in NA1000 WT cells containing either the control vector (pMT375) or pPxyl-StpX, a low-copy plasmid encoding StpX under Pxyl control, after overnight growth in PYEX (PYE supplemented with 0.3% xylose). (B) Stalk length distribution in ΔphoB single mutants and ΔphoB ΔstpX double mutants grown in PYE. (C) Stalk length distribution of NA1000 WT and ΔstpX cells grown in phosphate-limiting conditions (HIGG medium + 30 μM phosphate).
The stalk is thought to fulfill a role in nutrient acquisition (8, 18), but conditions in which the stalk and/or its contents are critical for growth and/or viability are not known. Because growth of ΔstpX cells in PYE or in HIGG is not notably different from that of WT cells, we conclude that the principal discernible role of StpX is to promote stalk elongation in cells that perceive phosphate limitation.
Determinants of StpX Localization.
How might StpX be directed to the stalked organelle? To dissect the underlying mechanism, we determined the localization of truncated StpX-GFP derivatives [StpX(Δper)-, StpX(ΔperΔtm)-, and StpX(Δcd)-GFP] expressed in place of WT StpX at the stpX locus. These mutant derivatives lack the predicted periplasmic domain (residues 16–363; Δper), the periplasmic domain and the TM domain (residues 16–387; ΔperΔtm), or the predicted cytoplasmic C-terminal domain (residues 387–643, Δcd), respectively (Figs. 3 A and B and 4A, Fig. S4). Immunoblotting revealed StpX-GFP, StpX(Δper)-GFP, and StpX(Δcd)-GFP (but not free GFP) are in the membrane fraction (Fig. S5). Whereas StpX(ΔperΔtm)-GFP proved unstable during the fractionation procedure, the experiments described below (Fig. 3) support the idea that the TM domain is required for membrane anchoring of StpX.
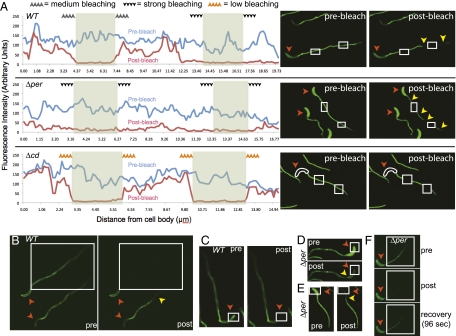
Fig. 4.
Mobility of StpX-GFP and mutant derivatives in vivo as determined by FLIP and/or FRAP analysis in HIGG (30 μM phosphate). (A–E) FLIP analysis of cells expressing StpX-GFP (A–C; “WT”), StpX(Δper)-GFP (A and D–E; “Δper”) and StpX(Δcd)-GFP (A; “Δcd”). Fluorescence images were acquired before (“pre”) and after (“post”) bleaching (52 sec) the area defined by the white boxes. Yellow arrowheads denote the loss in fluorescence (bleaching) in areas of the stalks that were not directly illuminated by the laser. Red arrowheads denote cell bodies. (F) FRAP analysis of cells expressing StpX(Δper)-GFP (“Δper”). Images were captured before (“pre”) and immediately after (“post”) bleaching (13 sec) the white boxed area and finally again after 96 sec of recovery (“recovery”). A Left shows the quantification of fluorescence signals of the images in the Right before (“pre-bleach”, blue line) and after (“post-bleach”, red line) illumination with the laser. The gray sections denote the bleached regions. The extent of fluorescence loss in regions adjacent to bleached regions is indicated above the relevant areas by arrays of triangles.
StpX(Δper)-GFP is delocalized and dispersed throughout the entire cell periphery, including the stalk (Figs. 3B and 4A). By contrast, StpX(ΔperΔtm)-GFP is restricted to the cell body (i.e., absent from the stalk, Fig. 3B) akin to cytoplasmic GFP. To further explore the role of the TM domain in StpX localization, a 54-residue polypeptide harboring the StpX SS and TM domains but lacking periplasmic and cytoplasmic domains was fused to sEGFP (TM-GFP). TM-GFP expressed from a low-copy plasmid under Pvan control was observed throughout the cell periphery including the stalk (Fig. 3F). By contrast, the fluorescence conferred by a derivative lacking 14 residues of the TM domain (ΔTM-GFP) was restricted to the cell body (Fig. 3F). Remarkably, a chimera of GFP to the C terminus of the TM domain (TM-GFP) from the flagellar positioning factor PflI (27) was also observed in the stalk (Fig. 3F). As PflI is predicted to adopt a comparable bitopic orientation to StpX in the membrane (i.e., a cytoplasmic C terminus), we conclude that TM domains like that of StpX or PflI can provide access to GFP and other moieties (Fig. 3 B and F) to the stalked compartment/organelle.
Cells expressing StpX(Δcd)-GFP in lieu of WT StpX-GFP exhibit an apparent identical stalk localization. On these grounds we propose that (i) the StpX TM domain facilitates entry into the stalked compartment, (ii) the periplasmic domain is required for retention in the stalk, and (iii) the cytoplasmic domain is dispensable for sequestration of StpX to the stalk.
Distinct StpX Mobilities in the Stalk.
If the periplasmic domain is needed for retention in the stalk, the StpX(Δper)-GFP version might exhibit an increased degree of mobility between the stalk and the cell body compared to that of WT StpX-GFP. To test this idea, we measured the mobility of WT StpX-GFP and StpX(Δper)-GFP, using fluorescence-loss-in-photobleaching (FLIP) and fluorescence-recovery-after-photobleaching (FRAP) experiments (Fig. 4 A–C, Figs. S6 and S7). First, quantitative FLIP (qFLIP) analysis was used to determine the mobility of WT StpX-GFP by bleaching different fluorescent areas of the stalk and the cell body using a focused laser beam (Fig. 4A). Unexpectedly, these experiments disclosed distinct mobilities of WT StpX-GFP as a function of its position within the stalk: Whereas StpX-GFP is essentially immobile near the base of the stalk, it is comparatively mobile near the tip. When the laser was aimed at the stalk tip, loss of StpX-GFP fluorescence extended beyond the site of bleaching (arrowheads in Fig. 4 A and B), indicating that StpX-GFP is mobile at this location, moving between the region illuminated by the laser and adjacent regions. By contrast, when the laser was positioned proximal to the stalk base (Fig. 4 A and B), bleaching into the neighboring region was not evident, indicating that StpX-GFP is immobile near the base and does not exchange with the cell body. In support of this result, when the laser was used to bleach the cell body, fluorescence in the stalk did not diminish (Fig. 4C). In summary, these experiments provide evidence that two distinct forms of StpX exist: an immobile one proximal to the stalk base and another one with increased mobility near the stalk tip.
To identify determinants of StpX mobility, we conducted qFLIP analyses on strains expressing StpX(Δper)-GFP and StpX(Δcd)-GFP (Fig. 4A). These experiments revealed that StpX(Δper)-GFP is mobile at both stalk proximal and distal positions, as inferred from the observation that fluorescence was lost in the vicinity of the bleached areas in both areas after laser illumination (Fig. 4A). Unlike WT StpX-GFP, StpX(Δper)-GFP is also mobile in the cell body, exchanging between the stalk and the cell body (Fig. 4 C–E). FRAP experiments also indicate that StpX(Δper)-GFP is mobile (Fig. 4F, Fig. S6 A and C) and that it has markedly slower recovery kinetics than cytoplasmic GFP (as expected for an integral membrane protein, Fig. S7 A and B). After bleaching all StpX(Δper)-GFP fluorescence in the stalk, significant recovery was measured in the stalk after 96 sec (Fig. 4F and Fig. S7C). This recovery was due to diffusion of StpX(Δper)-GFP from the cell body into the stalk and not from new biosynthesis, as indicated by the fact that bleaching of an entire cell (i.e., stalk and cell body) did not yield fluorescence recovery. Thus, StpX(Δper)-GFP has more mobility than WT StpX-GFP and diffuses throughout the entire stalk and the body.
By contrast, qFLIP analyses (Fig. 3B) revealed that deletion of the C-terminal domain rendered StpX less mobile than WT StpX-GFP, even near the tip of the stalk (Fig. 4A). Moreover, bleaching of StpX(Δcd)-GFP at cell body distal or proximal regions did not extend into the neighboring areas. Thus, whereas the periplasmic domain inhibits mobility, the cytoplasmic C-terminal domain promotes StpX mobility at the stalk tip.
Cell-Cycle Dependent Accumulation of StpX Isoforms.
Immunoblotting experiments with a polyclonal antibody to GFP revealed that StpX(Δcd)-GFP-expressing cells accumulate only a single GFP-tagged translation product. By contrast, the other StpX-GFP derivatives yield shorter products (Fig. 3D). In cells with StpX-GFP, derivatives with apparent molecular masses of 85 and 120 kDa (Fig. 3 C and D) are present (as well as a fragment slightly larger than GFP). Because GFP is appended to the C terminus of StpX, we hypothesize that the 120-kDa version corresponds to full-length StpX-GFP (predicted molecular mass 94 kDa), whereas the 85-kDa StpX-GFP derivative reflects an N-terminal truncation into the putative periplasmic domain. The accumulation of these forms is strongly and dissimilarly cell-cycle regulated (Fig. 3C): Whereas the appearance of the 120-kDa form coincides with the elaboration of the stalk, the 85-kDa form is present during the SW cell phase, the time during the cell cycle when both stalks and StpX-GFP foci are absent (Fig. 1O). However, the 85-kDa StpX-GFP is also discernible throughout the predivisional (PD) cell stage. Immunoblotting with a polyclonal antibody raised against the C-terminal domain of StpX (α-StpX) also detected two derivatives in extracts of WT (but not of ΔstpX) cells with molecular masses of 80 and 50 kDa (Fig. 3E), consistent with the idea that the 50-kDa protein is an N-terminally truncated derivative of StpX that accumulates in WT cells and lacks a significant region of the periplasmic domain. The result that the periplasmic domain is required for localization to the stalk, along with the finding that the N-terminally truncated 85-kDa StpX-GFP derivative is present in PD cells in which dim fluorescence can be detected in the cell body, suggests that it is present throughout the cell envelope [akin to StpX(Δper)-GFP], whereas the 120-kDa form of StpX-GFP is confined to the stalk. Because no detectable truncated derivatives were detected in StpX(Δcd)-GFP cells (Fig. 3D and Fig. S5), the C-terminal domain influences not only the mobility of StpX (Fig. 4A), but also its stability and/or expression.
Conclusion.
Using a high-throughput localization screen, we identified StpX as a protein that is sequestered to the stalk organelle of C. crescentus. Photobleaching and deletion analyses revealed that the mobility of StpX is spatially regulated within the cell. The stalk-specific localization and the mobility of StpX are both influenced by the N-terminal (periplasmic) domain (NTD) and the C-terminal (cytoplasmic) domain (CTD). The NTD is rich in prolines that have the potential to organize into polyproline helices that could mediate interactions with putative anchoring proteins in the stalk and/or with motor proteins in the periplasm (28, 29). It has been suggested that the composition of peptidoglycan (PG) in the stalk is distinct from that surrounding the cell body. Perhaps retention of StpX by the NTD relies, directly or indirectly, on the recognition of such hypothetical chemically distinct PG. StpX mobility could also be modulated by the formation of a higher-order StpX protein complex that has a low diffusion coefficient and whose formation is specifically nucleated in the stalk. The CTD promotes StpX mobility at the tip of the stalk and it facilitates the formation of N-terminally truncated StpX derivatives, raising the possibility of a causal relationship between the prevalence of these StpX isoforms and elevated mobility. The CTD might direct the formation of such isoform(s) through proteolysis or via another mechanism specifically at or near the tip of the stalk to yield mobile StpX at this site.
Although stalks lack ribosomes, StpX might be translated and inserted into the stalk envelope at the base (the stalked pole) as the stalk elaborates. Continued insertion during the stalk elongation phase and subsequent immobilization (retention) of StpX might then result in stalk-specific localization. Alternatively, it is possible that after translation and membrane insertion at random positions along the cell body, StpX diffuses into the stalk or is actively transported to that site by a motor tracking along a cytoskeletal structure. The outgrowth of StpX-containing stalks is dependent on the MreB actin homolog and an MreB homolog (MreBH) mediates the correct localization of the LytE cell wall hydrolase in Bacillus subtilis (30). Thus, potentially superimposed on the mechanisms regulating the mobility of proteins in subcellular space, specialized insertion mechanisms aided by cytoskeletal components might contribute to the disposition of selected proteins within minute subcellular compartments or highly specialized organelles.
Experimental Procedures
Growth Conditions.
C. crescentus was grown in PYE, PYEX (PYE supplemented with 0.3% xylose), M2G, or HIGG medium (1) at 30 °C. Unless otherwise stated, cells were grown in PYE. Escherichia coli was grown at 37 °C in LB with the appropriate antibiotics (2, 3).
Microscopic Techniques.
An upright Nikon 90i fitted with a 100× TIRF objective (na 1.45) and a Photometrics QuantEM 512SC CCD camera operated through Metamorph 7.2 were used. FLIP/FRAP was done on a Leica TCS SP5 scanning confocal microscope equipped with an HCX PL APO Lambda Blue 63× 1.4 oil objective (Leica Microsystems). Excitation was at 4% of the maximum laser power for nonbleaching frames. For bleaching frames, 100% laser power was used in user-specified regions, while fluorescence in the rest of the field was simultaneously monitored using 4% power. Bleaching was for 52 sec (FLIP) and 13 sec (FRAP) and recovery was monitored for 100–200 sec in 10-sec intervals.
FLIP/FRAP, immunoblotting, and strain and plasmid constructions are described in SI Text.
Supplementary Material
Acknowledgments
We thank Piet de Boer for discussions. Funding was from the US Department of Energy (DE-FG02-05ER64136 to P.H.V.), the Swiss National Science Foundation (31003A_127287 to P.H.V.) and the National Institutes of Health (GM51986 to Y.V.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909119107/DCSupplemental.
References
- 1.Thanbichler M, Shapiro L. Getting organized—how bacterial cells move proteins and DNA. Nat Rev Microbiol. 2008;6:28–40. doi: 10.1038/nrmicro1795. [DOI] [PubMed] [Google Scholar]
- 2.Möll A, Thanbichler M. FtsN-like proteins are conserved components of the cell division machinery in proteobacteria. Mol Microbiol. 2009;72:1037–1053. doi: 10.1111/j.1365-2958.2009.06706.x. [DOI] [PubMed] [Google Scholar]
- 3.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Werner JN, et al. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poindexter JS. Biological properties and classification of the Caulobacter group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ireland MM, Karty JA, Quardokus EM, Reilly JP, Brun YV. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol Microbiol. 2002;45:1029–1041. doi: 10.1046/j.1365-2958.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- 9.Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci USA. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poindexter JS. The caulobacters: Ubiquitous unusual bacteria. Microbiol Rev. 1981;45:123–179. doi: 10.1128/mr.45.1.123-179.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skerker JM, Laub MT. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat Rev Microbiol. 2004;2:325–337. doi: 10.1038/nrmicro864. [DOI] [PubMed] [Google Scholar]
- 12.Russell JH, Keiler KC. Screen for localized proteins in Caulobacter crescentus. PLoS One. 2008;3(3):e1756. doi: 10.1371/journal.pone.0001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 14.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 15.Sommer JM, Newton A. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics. 1991;129:623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 17.Nierman WC, et al. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci USA. 2001;98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner JK, Setayeshgar S, Sharon LA, Reilly JP, Brun YV. A nutrient uptake role for bacterial cell envelope extensions. Proc Natl Acad Sci USA. 2006;103:11772–11777. doi: 10.1073/pnas.0602047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JC, et al. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biondi EG, et al. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2006;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- 21.Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radhakrishnan SK, Pritchard S, Viollier PH. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell. 2010;18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Reisinger SJ, Huntwork S, Viollier PH, Ryan KR. DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J Bacteriol. 2007;189:8308–8320. doi: 10.1128/JB.00868-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poindexter JS, Staley JT. Caulobacter and Asticcacaulis stalk bands as indicators of stalk age. J Bacteriol. 1996;178:3939–3948. doi: 10.1128/jb.178.13.3939-3948.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J Bacteriol. 2005;187:544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonin M, Quardokus EM, O'Donnol D, Maddock J, Brun YV. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol. 2000;182:337–347. doi: 10.1128/jb.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obuchowski PL, Jacobs-Wagner C. PflI, a protein involved in flagellar positioning in Caulobacter crescentus. J Bacteriol. 2008;190:1718–1729. doi: 10.1128/JB.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rath A, Davidson AR, Deber CM. The structure of “unstructured” regions in peptides and proteins: Role of the polyproline II helix in protein folding and recognition. Biopolymers. 2005;80:179–185. doi: 10.1002/bip.20227. [DOI] [PubMed] [Google Scholar]
- 29.Creamer TP, Campbell MN. Determinants of the polyproline II helix from modeling studies. Adv Protein Chem. 2002;62:263–282. doi: 10.1016/s0065-3233(02)62010-8. [DOI] [PubMed] [Google Scholar]
- 30.Carballido-López R, et al. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.