Abstract
Soil salinity limits agricultural production and is a major obstacle for feeding the growing world population. We used natural genetic variation in salt tolerance among different Arabidopsis accessions to map a major quantitative trait locus (QTL) for salt tolerance and abscisic acid (ABA) sensitivity during seed germination and early seedling growth. A recombinant inbred population derived from Landsberg erecta (Ler; salt and ABA sensitive) × Shakdara (Sha; salt and ABA resistant) was used for QTL mapping. High-resolution mapping and cloning of this QTL, Response to ABA and Salt 1 (RAS1), revealed that it is an ABA- and salt stress-inducible gene and encodes a previously undescribed plant-specific protein. A premature stop codon results in a truncated RAS1 protein in Sha. Reducing the expression of RAS1 by transfer-DNA insertion in Col or RNA interference in Ler leads to decreased salt and ABA sensitivity, whereas overexpression of the Ler allele but not the Sha allele causes increased salt and ABA sensitivity. Our results suggest that RAS1 functions as a negative regulator of salt tolerance during seed germination and early seedling growth by enhancing ABA sensitivity and that its loss of function contributes to the increased salt tolerance of Sha.
Keywords: abscisic acid response, germination, natural variation, quantitative trait locus
About 20% of irrigated agricultural land is affected by salinity, and 10 million ha of the land is abandoned each year because of salinity (1). Despite this, crop production must be increased by 50% by 2025 to meet the demand of a burgeoning population (2). Soil salinity causes both ion toxicity and osmotic stress, and plants have evolved complex salt tolerance mechanisms (3). The salt overly sensitive (SOS) pathway of salt tolerance is crucial for maintaining ion homeostasis under salt stress. This pathway is conserved across diverse plant species, including rice (3, 4). The SOS2 ser/thr protein kinase regulates both SOS1-mediated Na+ extrusion from the cytosol and vacuolar Na+/H+ antiporter-mediated Na+ sequestration into the vacuole (5, 6). The high-affinity K+ transporter (HKT) type transporters are also important for Na+ homeostasis under salt stress. A genetic screen for second-site suppressor mutations of sos3 led to the identification of the gene AtHKT1;1/AtHKT1 (7). AtHKT1;1 was also identified from a genetic screen for mutants that overaccumulate Na+ in the shoot (8), and it has been shown to contribute to natural variations for Na+ accumulation in the shoot (9).
Natural variations in plants can help to unravel the molecular basis of agronomically important traits. These natural variations are often complex and require quantitative trait locus (QTL) analysis. Using natural variations among Arabidopsis accessions, QTLs have been cloned for a number of traits such as flowering time, seed dormancy, light response, biomass production, hybrid incompatibility, embryogenesis, root growth, low phosphate response, copper toxicity, sulfate assimilation, and molybdate accumulation (10). Natural variation has also been used to map salt tolerance QTLs in Arabidopsis (11, 12) and in some crop plants (13). However, the underlying genes for only a few salt tolerance QTLs have been identified. Ren et al. (14) cloned and characterized the rice SKC1 QTL for salt tolerance. SKC1 encodes an HKT-type Na+-selective transporter involved in unloading Na+ from the xylem (14). In du-rum wheat, Na+ exclusion was linked to the Nax1 (Na+ exclusion 1) and Nax2 loci, which likely correspond to the Na+ transporters HKT1;4 (HKT7) and HKT1;5 (HKT8), respectively (15, 16).
The plant stress hormone abscisic acid (ABA) regulates many genes involved in plant salt tolerance (3). Although considerable progress has been made in identifying genes encoding components of ABA response pathways by molecular cloning of induced variations, little is known about QTLs controlling ABA sensitivity in plants.
The Arabidopsis accessions Landsberg erecta (Ler, NW20) and Shakdara (Sha, N929) show substantial phenotypic difference in their seed dormancy and tolerance to heat, salt, and osmotic stress. A recombinant inbred line (RIL) population obtained from a cross between the Ler and Sha accessions has been used to map QTLs for seed dormancy and for heat, salt, and osmotic stress tolerance during germination (12). In the present study, we mapped a major QTL conferring salt tolerance and ABA insensitivity in seedlings using an RIL population from Ler × Sha. Positional cloning of this Response to ABA and Salt 1 (RAS1) QTL revealed a previously undescribed gene that is induced by ABA and salt stress. Genetic and molecular data from our study demonstrate that RAS1 is a negative regulator of salt tolerance and an important modulator of ABA sensitivity.
Results
Mapping of Salt Tolerance QTLs Using Ler × Sha RILs.
We evaluated the salt tolerance of the Arabidopsis accessions Ler and Sha in Murashige–Skoog (MS) agar medium supplemented with NaCl and in soil irrigated with NaCl. Sha is more tolerant to NaCl stress than Ler under both salt-stress conditions (Fig. 1A). To dissect this natural variation for salt tolerance by using a QTL approach, we employed Ler × Sha RILs (12). Salt tolerance was measured in terms of two phenotypic traits, namely, the percentage of green seedlings (GSs) and root length (RL); these traits were measured on MS agar medium containing 120 mM NaCl. For the GS trait, one major QTL was detected on chromosome 1 (Table S1). The GS1 QTL on chromosome 1 explained 76.6% of the total phenotypic variance (PVE). The additive effect of the Sha allele at this locus was very large for GSs and contributed to increasing GSs. For the RL trait, four QTLs were found on three chromosomes. Two RL QTLs were located on chromosome 1 and one each on chromosomes 3 and 4, respectively (Table S1). The PVE values of these QTLs were consistent with the coefficient of multiple determination (R2) from ANOVA, suggesting that the putative QTLs were mapped close to specific markers (Table S1). The RL1 QTL on chromosome 1 was localized at the same region as the GS1 QTL. The additive effect of the Sha allele at the RL1 locus was the highest and contributed to increasing RL. The results suggest that GS1 and RL1 may be the same QTLs involved in salt tolerance of Arabidopsis. Later experiments revealed that salt tolerance and ABA insensitivity cosegregate with the GS1 locus; hence, the locus was renamed RAS1. We confirmed the salt tolerance function of RAS1 by QTL mapping of the GS trait under 120 mM NaCl treatment in another RIL population from a Bay-0 × Sha cross (17). In this population, the RAS1 QTL explained 89.4% of the total PVE, with a logarithm of odds (LOD) value of 45.5 (Fig. S1A).
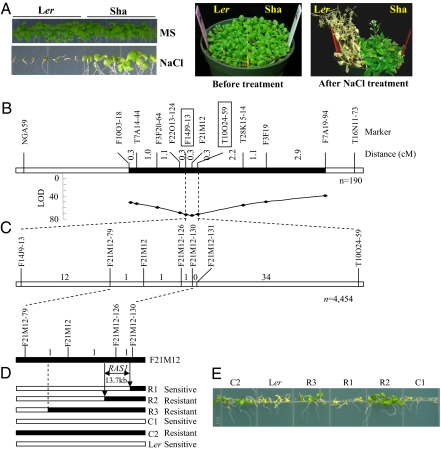
Fig. 1.
Map-based cloning of RAS1. (A) Salt tolerance of the parents, Ler and Sha accessions. Seedlings were germinated and grown on vertical MS agar medium (control) or MS supplemented with 120 mM NaCl for 2 weeks, or 2-week-old seedlings grown in soil (before treatment) were irrigated with 50 mM NaCl for 4 days, 150 mM NaCl for the next 3 days, and then 200 mM NaCl for 19 days. (B) Map location of RAS1 in a small region (filled bar) on Arabidopsis chromosome 1 based on 190 BC2F2 plants. (C) High-resolution linkage map of the RAS1 region produced with 4,454 BC2F2 plants. The number of recombinants between the adjacent markers is indicated above the linkage map. Filled bar shows a part of BAC clone F21M12. (D) Progeny testing of fixed recombinant plants (BC2F3) narrowed the RAS1 locus to the region between markers F21M12 and F21M12-130. Two recombinant lines (R2 and R3) and control 2 (C2) homozygous for Sha in the target region were resistant to 120 mM NaCl. In contrast, one recombinant line (R1) and control 1 (C1) homozygous for Ler in the target region were sensitive to 120 mM NaCl. Filled and open bars represent homozygous chromosomal segments for Sha and Ler, respectively. (E) Salt-stress response phenotype of the recombinant lines on MS medium containing 120 mM NaCl.
Map-Based Cloning of the RAS1 QTL.
Because the RAS1 QTL was identified as a major QTL conferring salt tolerance to Arabidopsis in both the Ler × Sha and Bay-0 × Sha RIL populations, we chose this QTL for map-based cloning. To perform fine mapping, CS24560, an RIL derived from the Ler × Sha cross, was selected. This line possessed the Ler genetic background, except for chromosome 1 and a ≈9-cM region of the lower arm of chromosome 3 from Sha (Fig. S1B). Fine mapping in the F2 population derived from the CS24560 × Ler cross placed the RAS1 locus between the markers F14J9-13 and T10O24-59 on chromosome 1, where the QTL was mapped originally (Fig. 1B). Further high-resolution mapping of RAS1 was carried out using 4,454 BC2F2 plants (Fig. 1C). RAS1 was localized to a high-resolution linkage map by progeny testing of BC2F4 homozygous recombinant plants (Fig. 1 D and E). By sequencing two key recombinant plants, the RAS1 locus was narrowed down to a 13.7-kb region between markers F21M12 and F21M12-130 (Fig. 1D). This region contains four predicted ORFs, namely, At1g09932 (phosphoglycerate/bisphosphoglycerate mutase-related), At1g09935 (phosphoglycerate/bisphosphoglycerate mutase family protein), At1g09940 (glutamyl-tRNA reductase), and At1g09950 (transcription factor-related). We sequenced this region from Ler and Sha and then identified At1g09950 as the candidate gene for RAS1. Comparison of the nucleotide sequences of the Col-0, Ler, and Sha alleles of RAS1 revealed a premature stop codon in the C-terminal region of the Sha RAS1 allele. In addition to the premature stop codon, the Ler and Sha alleles differ at 16 nucleotide positions, including 14 silent nucleotide substitutions and two nucleotide substitutions that result in two amino acid variations (Fig. S2A).
RAS1 Near-Isogenic Line Exhibits Salt Tolerance and ABA Insensitivity.
To investigate the effects of RAS1 on salt tolerance, we bred a near-isogenic line, NIL(RAS1), in the Ler genetic background with a 4.9-cM region between markers F21M12 and F7A19-94 that contains the RAS1 genomic region from Sha. NIL(RAS1) exhibited substantially higher salt tolerance and ABA insensitivity than the Ler isogenic control (Fig. 2A and Fig. S2 B and C).
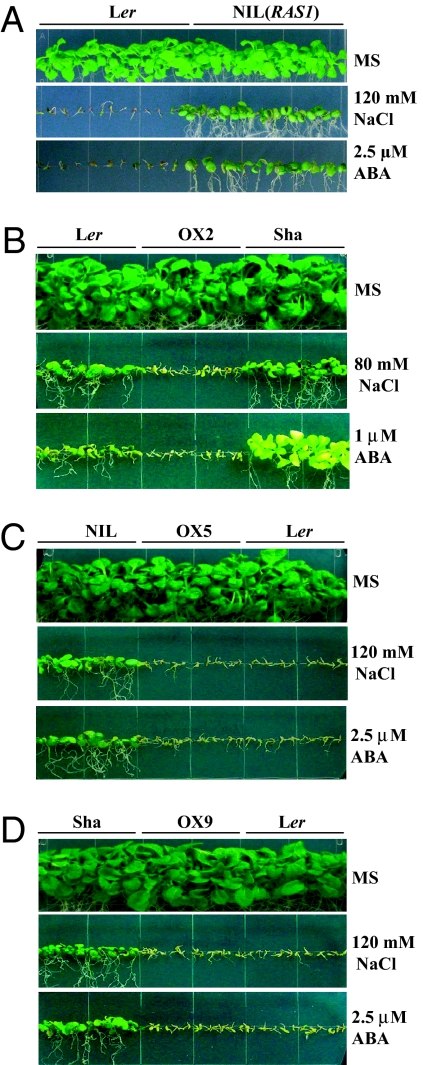
Fig. 2.
Salt and ABA sensitivity of NIL(RAS1) and transgenic complementation lines. (A) Germination and seedling growth phenotype of NIL(RAS1) and Ler on MS medium supplemented with 120 mM NaCl or 2.5 μM ABA after 14 and 45 days, respectively. Effect of ABA and NaCl on germination and early seedling growth of a representative transgenic (OX) Ler (B), NIL(RAS1) (C), or Sha (D) line transformed with the Ler RAS1 genomic DNA (LerRAS1g).
Complementation Analysis.
To confirm that the RAS1 gene is responsible for the GS phenotype under salt and ABA treatment, we performed complementation experiments. The F1 plants from a cross between Ler and NIL(RAS1) exhibited NaCl and ABA sensitivity similar to that of Ler (Fig. S3). This result suggests that the RAS1 allele from Ler is dominant. We cloned a 2.7-kb genomic fragment from Ler, which contains the promoter, ORF, and 3′UTR of the RAS1 gene, and introduced it into NIL(RAS1), Sha, and Ler. Northern blot analysis showed greater RAS1 expression in transgenic plants than in their respective untransformed parental genotypes (Fig. S4). The transgenic plants expressing Ler RAS1 in NIL(RAS1), Sha, and Ler backgrounds showed hypersensitivity to NaCl and ABA relative to the nontransgenic control plants (Fig. 2 B–D).
ABA and NaCl Induce RAS1 Expression.
Northern blot analysis showed that RAS1 expression was virtually undetectable under control conditions, but the expression was induced transiently by NaCl or ABA treatment (Fig. 3A). Although the RAS1 expression level was somewhat lower in Ler, the expression kinetics were similar among Ler, NIL(RAS1), and Sha during the time course of NaCl or ABA treatment (Fig. 3A). The results suggest that the salt tolerance of NIL(RAS1) and Sha is probably not attributable to any difference in the expression level of RAS1 but rather to differences in the RAS1 protein between Ler and Sha (Fig. 2 and Fig. S2).
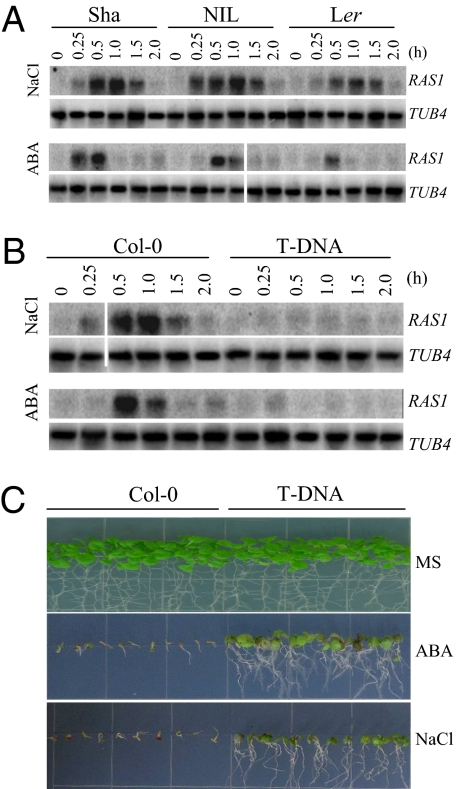
Fig. 3.
Characterization of RAS1 expression and T-DNA insertion mutant. (A) RAS1 expression in Sha, NIL, and Ler. (B) RAS1 expression in the T-DNA line and its WT. Seedlings were treated with 150 mM NaCl or 100 μM ABA for 0–2 h. TUB4 expression serves as the RNA loading control. (C) Germination and seedling growth of the T-DNA insertion mutant of RAS1 on MS medium supplemented with 150 mM NaCl or 2.5 μM ABA.
RAS1 Is a Negative Regulator of Salt Tolerance.
Because the salt-tolerant Sha allele has a premature stop codon, we predicted that a loss of function, reduction, or knockdown of RAS1 may increase the salt tolerance of Arabidopsis. To test this hypothesis, we obtained a transfer-DNA (T-DNA) insertion mutant (Salk_058470) for the RAS1 candidate gene At1g09950 in the Col-0 background. The T-DNA insertion was found in the 5′ region at 113 bp upstream of the start codon of At1g09950. The T-DNA insertion substantially reduced RAS1 expression (Fig. 3B). The T-DNA mutant showed enhanced salt tolerance and ABA-resistant seedling growth compared with Col-0 (Fig. 3C). This phenotype is similar to that of NIL(RAS1).
Results from the analyses of NIL(RAS1), the T-DNA mutant, and RAS1 complementation lines suggested that RAS1 is a negative regulator of salt tolerance and a positive regulator of ABA sensitivity. To confirm this further, we generated RNA interference (RNAi) lines of RAS1 in Ler, Sha, and NIL(RAS1). Northern blot analysis showed that RAS1 expression was reduced in the RNAi lines (Fig. S5 A, C, and E). Ler RAS1 RNAi lines exhibited enhanced salt tolerance and reduced ABA sensitivity compared with WT Ler plants (Fig. S5B). This phenotype of the Ler RNAi line is consistent with that of the T-DNA knockdown mutant in the Col-0 background (Fig. 3). RAS1 RNAi lines of NIL(RAS1) and Sha did not differ from their respective WTs in salt or ABA sensitivity (Fig. S5 D and F). In addition, Ler, NIL(RAS1), Sha, the T-DNA mutant, and Col-0 plants were transformed with a construct in which the Ler RAS1 cDNA was driven by the strong CaMV 35S promoter. Overexpression of full-length Ler RAS1 conferred salt and ABA sensitivity to NIL(RAS1), Sha, and the T-DNA mutant plants. The salt and ABA sensitivity of Col-0 and Ler was further increased by the RAS1 overexpression (Fig. S6). In contrast, CaMV 35S promoter-driven overexpression of the C-terminally truncated Sha allele of RAS1 or a mutated C-terminally truncated version of Ler RAS1 (209 amino acids) in Ler, Col, Salk_058470, and NIL(RAS1) did not affect salt and ABA responses (Fig. S7).
RAS1 Effects Require a Functional ABA Signaling Pathway.
RAS1 loss of function reduced ABA sensitivity, whereas its overexpression increased ABA sensitivity (Figs. 2 and 3 and Figs. S5, S6, and S7), suggesting that RAS1 is a positive regulator of ABA responses. The dominant abi1-1 mutation blocks ABA signaling by disrupting the interaction between ABSCISIC ACID INSENSITIVE 1 (ABI1) and ABA receptors PYRABACTIN RESISTANCE 1 (PYR1)/PYR1-like (PYLs) (18). To probe the genetic interaction between abi1-1 and RAS1, we made reciprocal crosses between abi1-1 and the ABA hypersensitive RAS1-overexpression line of Ler. F1 plants of the abi1-1 × Ler reciprocal crosses exhibited NaCl and ABA responses similar to those of abi1-1 (Fig. S8A). F1 plants obtained from reciprocal crosses between abi1-1 and the RAS1-overexpression line also showed ABA insensitivity (Fig. S8B), suggesting that a functional ABA signaling pathway is required for the effects of the overexpressed RAS1.
Natural Variation in Salt Tolerance and RAS1 Sequence Polymorphism.
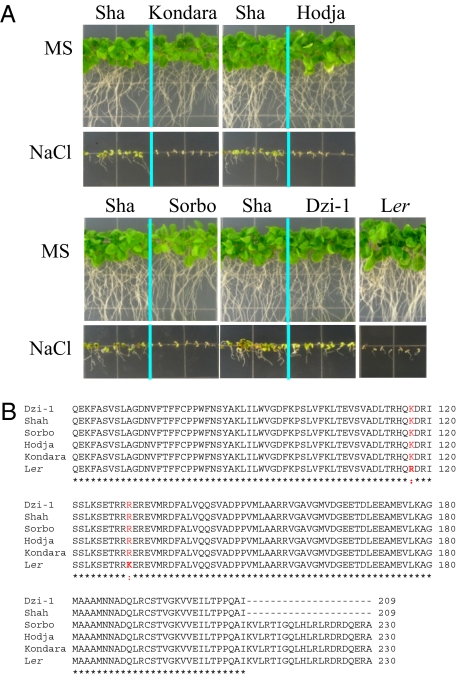
To investigate the association between RAS1 sequence polymorphism and natural variation in salt tolerance among Arabidopsis accessions, we evaluated the salt tolerance of 38 accessions and compared their RAS1 sequences. Phenotypic analysis showed that 5 of the accessions (i.e., Sha, Chi-0, CO-1, Litva, Wil-1) were tolerant to NaCl (Fig. S9). However, only the RAS1 of Sha encodes a truncated protein, whereas the RAS1 in the remaining 37 Arabidopsis accessions encodes a full protein with 230 amino acid residues. Because the Sha accession was from Tadjikistan, we evaluated the salt tolerance and RAS1 sequence polymorphism from 4 additional accessions from Tadjikistan [i.e., Kondara, Hodja-Obi-Garm (Hodja), Sorbo, Dzhiland (Dzi-1)]. Of these, only Dzi-1 exhibited a salt-tolerant phenotype similar to that of Sha (Fig. 4A). Sequence comparisons of RAS1 from the Tadjikistan accessions revealed that like Sha, Dzi-1 RAS1 had a premature stop codon that causes a truncated RAS1 protein with 209 amino acid residues (Fig. 4B). Interestingly, all 5 Tadjikistan accessions differed from Ler at two amino acid residues (amino acid 117 and amino acid 130) (Fig. 4B). These results further support that the premature stop codon rather than the two amino acid substitutions is responsible for the salt tolerance of Sha.
Fig. 4.
Salt tolerance and RAS1 sequence polymorphism of Arabidopsis accessions from Tadjikistan. (A) Germination and early seedling growth of the Tadjikistan accessions as compared with Ler. Seeds were sown on MS medium or MS medium supplemented with 120 mM NaCl and allowed to grow for 2 weeks. The Arabidopsis Biological Resource Center seed stock numbers for Kondara, Hodja, Sorbo, and Dzi-1 are CS916, CS922, CS931, and CS28980, respectively. (B) Alignment of RAS1 amino acid sequences from the Tadjikistan accessions and Ler. The two residues with polymorphisms are colored in red.
Discussion
In this study, we have identified the RAS1 locus involved in salt tolerance and ABA responses in Arabidopsis by QTL mapping of progenies from a cross between Ler × Sha. By analyzing the QTLs in the Bay-0 × Sha RIL population and by studying NIL(RAS1) in the Ler background, we confirmed the involvement of the RAS1 QTL in salt tolerance and ABA insensitivity at germination and early seedling stages. We mapped and cloned RAS1 and found that it encodes a previously undescribed plant-specific protein with a high similarity to At1g58330, a predicted transcription factor-related protein (www.arabidopsis.org). In the salt-sensitive Ler, RAS1 encodes a polypeptide with 230 amino acid residues. In Sha, a premature stop codon led to a truncated RAS1 with 209 amino acid residues. Northern blot analysis showed a fast and transient induction of RAS1 expression in response to NaCl and ABA treatments. The inducible expression is consistent with a role for RAS1 in salt stress and ABA responses. Ler, Sha, and NIL(RAS1) plants genetically transformed with the Ler RAS1 genomic sequence showed NaCl and ABA hypersensitivity. Genetic crosses and transgenic complementation analysis showed that the full-length RAS1 is dominant over the C-terminally truncated RAS1 in the response of Arabidopsis to salt and ABA. Consistent with these findings, our results with the T-DNA knockdown mutant of RAS1 in the Col-0 genetic background and with RAS1 RNAi lines in Ler, Sha, and NIL(RAS1) backgrounds confirmed that the knockdown or loss of RAS1 leads to enhanced salt tolerance and ABA insensitivity. Furthermore, CaMV 35S promoter-driven overexpression of full-length but not truncated RAS1 conferred NaCl and ABA hypersensitivity to transgenic plants. Importantly, our analysis of the salt tolerance and RAS1 sequences of Arabidopsis accessions from Tadjikistan revealed that the premature stop codon but not the two amino acid substitutions at residues 117 and 130 is associated with salt tolerance and ABA insensitivity. Together, our results suggest that a loss of function of RAS1 attributable to the C-terminal truncation is responsible for the decrease in ABA sensitivity and increase in salt tolerance of Sha. On the other hand, it is possible that the function of the Sha allele may depend on interactions with the Sha alleles of other genes involved in salt and ABA responses. The truncated protein may have evolved a function that is beneficial for salt tolerance specifically in Sha.
Salt stress induces ABA accumulation, which, in turn, inhibits germination and seedling growth (19). Previous genetic studies have shown that recessive mutations in positive regulators of ABA signaling, such as abi3, abi4, and abi5, confer ABA insensitivity and NaCl-resistant germination capacity to Arabidopsis (20). The results from our QTL analysis are consistent with the findings from mutational studies. Genetic analysis showed that the abi1-1 mutation, which disrupts ABA signaling, can suppress the salt and ABA sensitivity conferred by RAS1 overexpression. This suggests that the function of RAS1 requires the core ABA signaling pathway. ABI1 and its related type 2C protein phosphatase family members interact with the ABA receptors, and negatively regulate ABA signaling during germination (18, 21). Our results suggest that although RAS1 is not essential for ABA signaling, it plays an important role in enhancing ABA sensitivity. The induction of RAS1 by ABA suggests that RAS1 is part of a feed-forward loop for modulating ABA sensitivity.
Our finding of RAS1 as a previously undescribed regulator of NaCl and ABA responses is an initial step toward determining the molecular basis of this natural variation in stress responses. Future studies will clarify the molecular mechanisms of RAS1 in modulating ABA sensitivity and salt-stress tolerance. Abiotic stresses such as salinity reduce germination and seedling establishment, resulting in poor crop establishment and yield. Further studies on RAS1 will increase our understanding of the cellular basis for regulation of seed germination and seedling establishment under salt stress.
Materials and Methods
Plant Materials.
RILs for QTL mapping were from a cross between the Sha (salt-tolerant) and Ler (salt-sensitive) accessions. One of the RILs, CS24560, was crossed with Ler. The resultant F1 seeds were selfed to produce F2 seeds for fine mapping and were back-crossed with Ler to produce BC1F1 seeds. By repetitive back-crossing and marker-assisted foreground and background selection, we selected several plants in which the region around RAS1 was heterozygous, although the rest of the genomic regions were homozygous for Ler, so as to develop the segregating populations for high-resolution mapping of RAS1. From the BC2F2 generation, we developed NIL(RAS1), which contains a very small Sha chromosomal region containing the RAS1 locus in the Ler genetic background. The Bay-0 × Sha RIL population (17) was obtained from the Arabidopsis Biological Resource Center (stock no. CS57921).
Evaluation of Physiological Traits for Salt Tolerance.
The parents, Sha and Ler, and 114 F9 RILs (16–20 plants in each line) were used to evaluate salt tolerance. The seeds were sterilized for 5 min with 4% (wt/vol) sodium hypochlorite in 0.01% (wt/vol) Triton × 100 and were sown on MS nutrient medium supplemented with 120 mM NaCl. After stratification at 4 °C for 4 days, the seeds on the agar plates were placed in a growth chamber with 16-h light/8-h dark conditions at 23 °C for 16 days. Resistance to salt stress was measured as the percentage of GS and RL for QTL mapping.
Construction of a Linkage Map and QTL Analysis.
The genotype of each plant in the Ler × Sha RIL population was determined by a set of 66 markers covering the whole Arabidopsis genome. These data were kindly provided by Maarten Koornneef (Wageningen, The Netherlands). A linkage map was established using the MAPMAKER/EXP 3.0 program (Whitehead Institute, Cambridge, MA) based on the genotype data of the F2 population. Map distances between marker loci were presented in centimorgans, which were derived using the Kosambi function of the program. A LOD score of 2.0 was used to determine both the linkage groups and the order of markers. The MAPMAKER/QTL program (22) was used to identify QTLs affecting salt tolerance on the basis of interval analysis. The proportion of phenotypic variation explained by individual marker loci associated with specific QTLs was also determined by ANOVA (R2) (23). A LOD score of 2.0 was used to determine the presence of putative QTLs in a given genomic region. The percentages of total phenotypic variation explained by each QTL, and the additive effect, were estimated by MAPMAKER/QTL (22).
Fine Mapping and High-Resolution Mapping.
To perform fine mapping of the RAS1 QTL, we used the GS percentage calculated for 190 F2 plants growing on MS agar medium with 120 mM NaCl, 12 markers in a target region containing RAS1, and 13 markers in other regions. The molecular markers F14G9-13 and T10O24-59, which flank RAS1, were used to detect recombinants in 4,454 BC2F2 plants. To determine the location of the recombination nearest to RAS1, we developed markers on the basis of the sequence of BAC clone F21M12 and determined genotypes of the recombinants with these markers. The BC2F3 progeny derived from recombinant plants were used to screen for homozygous recombination products. We used fixed recombinant plants (BC2F4) to test the salt tolerance and determine the RAS1 genotypes. The chromosomal region between markers F21M12 and F21M12-130 from the recombinants R1 and R2 were sequenced to identify the fine position of recombination.
Complementation Test.
We amplified the genome region of RAS1, including the 2.72-kb promoter region upstream of the ATG start codon and the 0.32-kb region downstream of the stop codon, from Ler genomic DNA by PCR with PfuUltra High-Fidelity DNA Polymerase (Stratagene) and the following primers: RAS1gF, GAGCTCTTTCTCGCTTCACAGTCGTC, and RAS1gR, TCTAGAGCGACCACATCATAAGCTAC. The PCR product was digested with SacI and XbaI (restriction sites underlined in the primer sequences) and cloned into the plant binary vector pCAMBIA1305.1. This construct was introduced into Agrobacterium tumefaciens stain GV3101 and used for transformation of Ler, NIL(RAS1), and Sha. NIL(RAS1) was also crossed with Ler, and the salt tolerance of F1 plants was evaluated.
We amplified the 630-bp truncated coding sequence (CDS) from Sha or Ler with the following primers: RAS1cdsF, CACCATGCCAAACACTAGCAGCTC, and RAS1truncatedR, TTAAATCGCTTGCGGCGGAG. The CDS was cloned into pMDC32 by Gateway Technology (Invitrogen), under the transcriptional control of the CaMV 35S promoter. The constructs were used for Agrobacterium-mediated transformation of Ler, NIL(RAS1), Col, and Salk_058470. The 693-bp full-length CDS from Ler was amplified with the primers RAS1cdsF and RAS1cdsR (TTAAGCTCTTTCTTGGTCTCTG) and cloned into pMDC32. The construct was used for Agrobacterium-mediated transformation of Ler, NIL(RAS1), Col, Salk_058470, and Sha.
RNAi of RAS1.
A 374-bp RAS1 fragment was amplified by PCR with the following primers: RAS1cdsF and RAS1RNAiR, GACTTAAGACTCGAGATCCG. This fragment was cloned into pANDA35HK using Gateway Technology.
RNA Analysis.
Seedlings (2-week-old) were treated with 150 mM NaCl or 100 μM ABA for the indicated durations. Total RNA was extracted using TRIZOL Reagent (Invitrogen) according to the manufacturer's protocol. Ten micrograms of total RNA was fractionated on 1.2% (wt/vol) formaldehyde agarose gel and transferred onto a Hybond N+-Nylon membrane (GE Healthcare). After transfer, RNA was fixed by UV cross-linking and was hybridized with [α-32P]dCTP-labeled RAS1 probe. Blots were rehybridized using [α-32P]dCTP-labeled TUB4 as a control for RNA loading.
Supplementary Material
Acknowledgments
We thank Maarten Koornneef for kindly providing the Ler × Sha RIL population and Hong-Xuan Lin for valuable advice on QTL analysis. This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 and by National Science Foundation Grant IBN0420152 (to J.-K.Z). Z.Z. was supported by the China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910798107/DCSupplemental.
References
- 1.Szabolcs I. Salt-Affected Soils. Boca Raton, FL: CRC Press; 1989. [Google Scholar]
- 2.Khush GS. Challenges for meeting the global food and nutrient needs in the new millennium. Proc Nutr Soc. 2001;60:15–26. doi: 10.1079/pns200075. [DOI] [PubMed] [Google Scholar]
- 3.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez-Atienza J, et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007;143:1001–1012. doi: 10.1104/pp.106.092635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu QS, et al. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the SOS pathway. J Biol Chem. 2003;279:207–215. doi: 10.1074/jbc.M307982200. [DOI] [PubMed] [Google Scholar]
- 7.Rus A, et al. AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA. 2001;98:14150–14155. doi: 10.1073/pnas.241501798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berthomieu P, et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rus A, et al. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2006;2:e210. doi: 10.1371/journal.pgen.0020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alonso-Blanco C, et al. What has natural variation taught us about plant devel-opment, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quesada V, García-Martínez S, Piqueras P, Ponce MR, Micol JL. Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol. 2002;130:951–963. doi: 10.1104/pp.006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clerkx EJ, et al. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 2004;135:432–443. doi: 10.1104/pp.103.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins NC, Tardieu F, Tuberosa R. Quantitative trait loci and crop performance under abiotic stress: Where do we stand? Plant Physiol. 2008;147:469–486. doi: 10.1104/pp.108.118117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren ZH, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, et al. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006;142:1718–1727. doi: 10.1104/pp.106.088864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrt CS, et al. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007;143:1918–1928. doi: 10.1104/pp.106.093476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0 × Shahdara recombinant inbred lines population: A powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 18.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of ABA binding START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiong L, Zhu JK. Salt tolerance. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. Available at http://www.aspb.org/publications/arabidopsis/ [Google Scholar]
- 20.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seed-lings. Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 22.Lincoln S, Daly M, Lander ES. Mapmaker/QTL 1.1 Manual. Cambridge, MA: Whitehead Institute; 1993. [Google Scholar]
- 23.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.