Abstract
Neuregulin-1 (NRG1) and Disrupted-in-Schizophrenia-1 (DISC1) are promising susceptibility factors for schizophrenia. Both are multifunctional proteins with roles in a variety of neurodevelopmental processes, including progenitor cell proliferation, migration, and differentiation. Here, we provide evidence linking these factors together in a single pathway, which is mediated by ErbB receptors and PI3K/Akt. We show that signaling by NRG1 and NRG2, but not NRG3, increase expression of an isoform of DISC1 in vitro. Receptors ErbB2 and ErbB3, but not ErbB4, are responsible for transducing this effect, and PI3K/Akt signaling is also required. In NRG1 knockout mice, this DISC1 isoform is selectively reduced during neurodevelopment. Furthermore, a similar decrease in DISC1 expression is seen in β-site amyloid precursor protein cleaving enzyme–1 (BACE1) knockout mice, in which NRG1/Akt signaling is reportedly impaired. In contrast to neuronal DISC1 that was reported and characterized, expression of DISC1 in other types of cells in the brain has not been addressed. Here we demonstrate that DISC1, like NRG and ErbB proteins, is expressed in neurons, astrocytes, oligodendrocytes, microglia, and radial progenitors. These findings may connect NRG1, ErbBs, Akt, and DISC1 in a common pathway, which may regulate neurodevelopment and contribute to susceptibility to schizophrenia.
Linkage and association studies in diverse populations have suggested that the genes encoding for neuregulin (NRG) and ErbB proteins are associated with schizophrenia (1). The NRG family of proteins activates the ErbB family of receptor tyrosine kinases, which play numerous roles in neurodevelopment, in processes such as progenitor cell proliferation, radial and tangential migration, neurite outgrowth, dendritic arborization, and myelination (2, 3). NRG1 is cleaved by β-site amyloid precursor protein cleaving enzyme–1 (BACE1), which was originally found to cleave amyloid-β precursor protein (APP) and thereby contribute to the pathophysiology of Alzheimer's disease (4). This cleavage of NRG1 results in the release of the extracellular EGF-like domain, which in turn activates ErbB receptors and downstream intracellular signaling pathways, including the PI3K/Akt and JAK/STAT cascades, in a context-dependent manner (5). Consequently, decreased levels of cleaved NRG1 and diminished activation of Akt have been reported in BACE1 knockout (BACE1-KO) mice, indicating that these mice represent a loss-of-function model for NRG1 signaling (6).
Disrupted-in-Schizophrenia–1 (DISC1) is another promising genetic susceptibility factor for a wide range of major mental disorders, including schizophrenia (1, 7). DISC1 is expressed in the neocortex and other brain regions, especially during neurodevelopment (8). Like NRG family proteins, DISC1 is multifunctional, and has been shown to have a strong influence on neurodevelopmental processes including progenitor cell proliferation, radial migration, dendritic arborization, and synapse formation, with its effects often mediated by protein–protein interactions (3, 9–13). There is evidence linking DISC1 to cAMP signaling through its ability to bind PDE4 enzymes (14, 15), which can influence brain functioning (16). In addition, PDE4 (17) and cAMP (18) can influence Akt signaling.
Various genetically engineered mice have been generated for NRG proteins, ErbB proteins, DISC1, and BACE1 (19–21). These mice display overlapping abnormal phenotypes. For example, whereas homozygous NRG1 knockout (NRG1-KO) mice lacking the EGF-like domain do not survive past the embryonic stage of development (22), heterozygous NRG1-KO mice show impairments in prepulse inhibition and working memory (23–25). Several types of DISC1 genetically engineered mice have been generated (20, 21); in addition to common deficits of prepulse inhibition, some of these mice display deficits in working memory. Very interestingly, BACE1 knockout mice, originally generated for studies on Alzheimer's disease, also show deficits in prepulse inhibition and working memory, possibly associated with disturbed NRG1 signaling (26). In addition to these results from murine models, a recent report that examined a zebrafish model indicated that knockdown of NRG1 and DISC1 could result in similar phenotypes in development of oligodendrocytes and neurons from olig2-expressing precursor cells (27). Although these data are highly suggestive, a direct link between NRG1 signaling and DISC1 has not yet been elucidated at the molecular level.
Here we present evidence that NRG1 regulates expression of DISC1, mediated via ErbB2/3 and PI3K/Akt signaling. The influence of NRG1 on expression on an isoform of DISC1 is observed in both in vitro neuron cultures and in vivo. Furthermore, we also provide evidence that DISC1 is expressed in glial cells, including astrocytes, oligodendrocytes, and microglia.
Results
NRG Signaling Increases Expression of an Isoform of DISC1 in Primary Neuron Cultures.
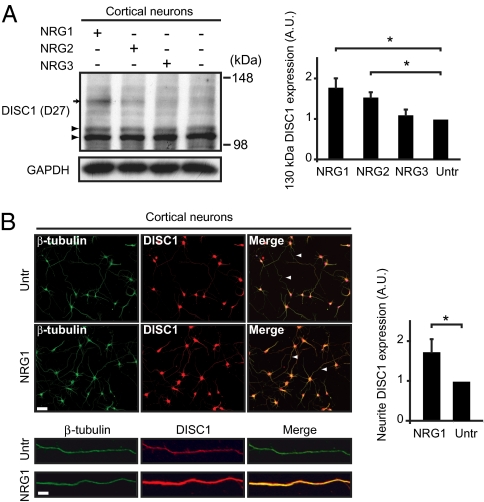
To address a possible link between NRG signaling and DISC1 at the molecular level, we investigated whether NRG ligands might influence the expression of DISC1. We used recombinant NRG1-β-GST (GST), NRG2-β-GST, or NRG3-GST proteins (NRG1, NRG2, and NRG3, respectively, in this study), consisting of the GST-tagged external EGF-like domain of the NRG1 β isoform, NRG2 β isoform, or NRG3, which in all cases is reportedly necessary and sufficient for activation of the ErbB family of receptors and downstream intracellular signaling (28). Equal levels of activation of the downstream signaling were confirmed by assessing ErbB4 phosphorylation in HEK293 cells following exposure to each recombinant NRG protein (Fig. S1A). Western blotting with DISC1 antibody D27 revealed that treatment with NRG1 and NRG2, but not NRG3, increased the levels of DISC1 immunoreactivity at 130 kDa in immature primary neuron cultures compared with treatment with GST alone (Untr) (Fig. 1A). Immature neurons were chosen as the main subject of this study because the NRG and DISC1 cascades play important roles during neurodevelopment (3); however, a similar induction of DISC1 by NRG1 was also observed in mature neuron cultures (Fig. S1B). Strong induction of 130 kDa DISC1 was observed with NRG1 at 3 nM concentration (Fig. S1C), 2 h after treatment, persisting up to at least 24 h (Fig. S1D). DISC1 has multiple isoforms, including an isoform at ∼100 kDa that is thought to represent the full-length protein; therefore, to confirm that the 130 kDa signal reflects an isoform of DISC1, we have used two independent antibodies (D27 and mExon3) generated and published by different research groups (8, 29). Induction of this 130 kDa isoform of DISC1 by NRGs was consistently observed with these two antibodies (Fig. S2A).
Fig. 1.
NRG treatment affects DISC1 expression in an isoform-specific manner. (A) Western blotting for DISC1 in NRG-treated neurons. Western blotting with D27 shows increase in isoform of DISC1 at 130 kDa (arrow) following treatment with NRG1 (mean ± SD, 1.77 ± 0.23-fold increase, P = 0.007, n = 5) and NRG2 (1.53 ± 0.13-fold increase, P = 0.008, n = 4), whereas bands at 100 and 105 kDa were unaffected (arrowheads). GAPDH was used as a loading control. (B) Immunofluorescent cell staining and quantification of DISC1 in NRG1-treated neurons. Cell staining for DISC1 with antibody mExon3 was quantified and normalized to neurite length indicated by β-tubulin staining in primary neurons treated with NRG1. DISC1 in neurites increased after NRG1 treatment (1.72 ± 0.33-fold increase, P = 0.01, n = 3). Magnification, 20×. (Scale bar, 50 μm.)
Immunoprecipitation of neuronal lysates using mExon3, followed by Western blotting with D27, confirmed this 130 kDa band to be DISC1 (Fig. S2B). This 130-kDa band was knocked down by a previously characterized RNAi to DISC1 (9); and this RNAi also prevented induction of the 130-kDa band by treatment with NRG1, further indicating that this band represents DISC1 (Fig. S2 C and D). In the present study, we mainly used D27 for Western blotting and mExon3 for immunofluorescent cell staining, because of minimized background signal with each antibody when optimized for the respective experimental paradigm. Of interest, the influence of NRG1 and NRG2 was observed only on 130 kDa DISC1, but not on previously reported isoforms at 100 and 105 kDa (Fig. S2E). These results suggest that NRG1 and NRG2 may influence DISC1 expression via a signaling pathway not activated by NRG3. We next treated primary cortical neurons with NRG1 as before and performed immunofluorescent cell staining. Although DISC1 expression in the nucleus and cell body was relatively unchanged, the intensity of DISC1 signal in neurites was markedly increased in treated cultures (Fig. 1B).
ErbB2/3 and PI3K/Akt Signaling Mediate the Effect of NRG1 on DISC1 Expression.
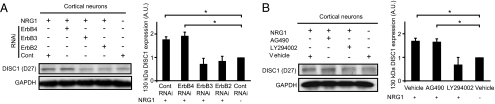
To determine which receptor and secondary signaling pathway activated by NRG1 is responsible for transducing its effect on expression of DISC1 130 kDa isoform, we first used lentivirus-mediated RNAi to knock down expression of each receptor in primary cortical neurons, and treated cultures with recombinant NRG1 as before. RNAi sequences directed against ErbB2, ErbB3, and ErbB4 were confirmed to be specific for each receptor (Fig. S3A). Western blotting with D27 and mExon3 revealed an increase in 130 kDa DISC1 expression in control RNAi- and ErbB4 RNAi-infected neurons following treatment with NRG1 (compared with untreated control RNAi-infected neurons), suggesting that ErbB4 is not necessary for transducing this effect, whereas NRG1-treated ErbB3 RNAi– and ErbB2 RNAi–infected neurons did not show this increase, indicating that these receptors are required (Fig. 2A). To determine the secondary signaling pathway involved, NRG1 treatment was performed in conjunction with blockade of PI3K/Akt or JAK2/STAT3 signaling by using pharmacological agents (10 μM LY294002 and 5 μM AG490, respectively). The efficacy of these agents was verified by Western blotting for phospho-Akt (for LY294002) and phospho-STAT3 (for AG490) (Fig. S3 B and C). Increases in 130 kDa DISC1 were observed after NRG1 treatment in neurons cotreated with vehicle (DMSO) or AG490 compared with untreated cells with vehicle, but this effect was abolished by cotreatment with LY294002, indicating that PI3K/Akt signaling mediates this effect (Fig. 2B). Cotreatment of NRG1 with the PDE4 inhibitor rolipram did not affect induction of 130 kDa DISC1 (Fig. S3D). However, NRG1-induced increase in 130 kDa DISC1 expression was blocked by cotreatment with transcription inhibitor Actinomycin D (Fig. S3E), suggesting that this isoform may arise from a transcriptionally regulated mechanism. These results indicate that NRG1 signaling via ErbB2, ErbB3, and PI3K/Akt signaling is required for the induction of 130 kDa DISC1.
Fig. 2.
NRG1 signaling affects DISC1 expression via ErbB3/ErbB2 and Akt activation. (A) Western blotting for DISC1 after NRG1 treatment with receptor knockdown by RNAi. Primary cortical neurons infected with lentiviral shRNA against ErbB4, ErbB3, and ErbB2 (Fig. S3A) and treated with NRG1. Western blotting was performed for 130 kDa DISC1 with DISC1 antibodies D27 and mExon3 and quantified (*P < 0.01, n = 3). (B) Western blotting for DISC1 after NRG1 treatment with pharmacological blockade of secondary signaling pathways. Primary cortical neurons were treated with LY294002 and AG490, then treated with NRG1. Western blotting was done for 130 kDa DISC1 with DISC1 antibodies D27 and mExon3 and quantified (*P < 0.01, n = 3). Magnification 200× and 400×. (Scale bar, 50 μm and 5 μm.)
DISC1 Expression Is Decreased in NRG1 Knockout Mice.
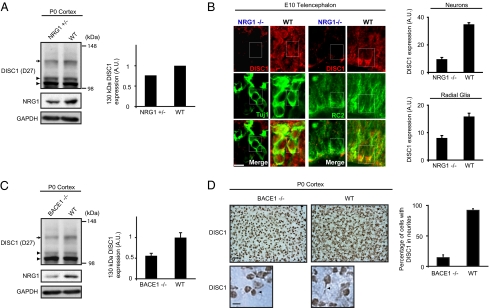
To determine whether NRG1 signaling influences DISC1 expression during neurodevelopment in vivo, we studied DISC1 expression in NRG1-KO mice. In these mice, the EGF-like domain common to all isoforms of NRG1 is replaced with the neomycin resistance gene, thereby eliminating NRG1 signaling (22). Homozygous NRG1-KO mice do not survive beyond embryonic day 11 (E11), whereas heterozygous knockout is not embryonically lethal. We performed Western blotting for DISC1 and NRG1 in cortices dissected from these mice at postnatal day 0 (P0), and confirmed a reduction in cleaved NRG1 (Fig. 3A). In these animals, 130 kDa DISC1 expression was decreased while DISC1 100 and 105 kDa isoforms were unaffected (Fig. 3A and Fig. S4A). This selective influence of NRG1 on the 130 kDa isoform of DISC1 is consistent with observations in primary neuron cultures (Fig. 1A). Immunohistochemistry for DISC1 showed widespread reduction in DISC1 expression in the telencephalon of homozygous NRG1 knockout mice at E10 (Fig. 3B). Costaining with cell type–specific markers for neurons (Tuj1) and radial progenitors (RC2) revealed expression of DISC1 in these cell types in wild-type embryos, and reduction in DISC1 expression in knockout mice, verified by quantification of fluorescent intensity in each cell type (Fig. 3B). These results indicate that NRG1 signaling is important in maintaining expression of 130 kDa DISC1 in the developing mouse brain in vivo.
Fig. 3.
NRG1 signaling knockdown affects DISC1 expression in vivo. (A) Western blotting for DISC1 in NRG1-KO mice. Immunoblotting with DISC1 antibody D27 shows reduced DISC1 expression (arrow) in cortex of heterozygous P0 NRG1-KO mice compared with wild-type littermates (23.6% decrease). A C-terminal–directed NRG1 antibody was used to verify a reduction in cleaved NRG1 in these mice. GAPDH was used as loading control. (B) Immunohistochemistry for DISC1 in NRG1 knockout mice. Neurons (Tuj1) and radial glia (RC2) in E10.5 telencephalon of WT and homozygous NRG1-KO mice were colabeled with DISC1. Both neurons and radial glia express DISC1, as seen in outlined cells; however, DISC1 is down-regulated in NRG1-deficient cells. Quantification of fluorescent intensity (arbitrary units) verifies decreased DISC1 in neurons (NRG1−/−: mean ± SD, 9.62 ± 1.24, WT: 34.91 ± 1.17), and radial glia (NRG1−/−: 8.04 ± 0.9, WT: 15.87 ± 1.2). Magnification, 630×. (Scale bar, 10 μm.) (C) Western blotting for DISC1 in BACE1 knockout mice. Immunoblotting with DISC1 antibody D27 shows reduced DISC1 expression (arrow) in cortex of homozygous P0 BACE1-KO mice compared with wild-type littermates (44.1% decrease, n = 3). A C-terminal–directed NRG1 antibody was used to verify a reduction in cleaved NRG1 in these mice. GAPDH used as loading control. (D) Immunohistochemistry for DISC1 in BACE1 knockout mice. Staining with DISC1 antibody mExon3 shows reduced DISC1 in the cortex of homozygous P0 BACE1-KO mice compared with wild-type littermates, with a reduction in the percent of neurons with visibly stained neurites (76.4% reduction, n = 3). Magnification 630×. (Scale bar, 50 μm and 10 μm.)
DISC1 Expression Is Decreased in BACE1 Knockout Mice.
As NRG1 signaling via Akt activation appears to modulate DISC1 expression (Fig. 2B), we hypothesized that DISC1 expression may also be altered in BACE1-KO mice during neurodevelopment. Consistent with previous publications (6, 26), reduction in cleaved NRG1 and accumulation of uncleaved NRG1 was verified in homozygous BACE1-KO brains at P0 (Fig. S4B). In these brains, a reduction in 130 kDa DISC1 was seen (Fig. 3C and Fig. S4C). This reduction appeared to be specific to the 130 kDa isoform, as expression of the 100- and 105-kDa DISC1 isoforms remained relatively unchanged. Immunohistochemistry using DISC1 antibody mExon3 showed similar DISC1 expression in nuclei and cell bodies but decreased expression in cellular processes in the cortex of homozygous BACE1-KO mice compared with wild-type mice (Fig. 3D and Fig. S4D). Marked reduction in the percentage of DISC1-stained neurons with visible neurites was observed in BACE1-KO mice. This could reflect a loss of DISC1 expression in neurites, consistent with in vitro results showing an increase of DISC1 in neurites following NRG1 treatment (Fig. 1B). Finally, subcellular fractionation revealed that 130 kDa DISC1 was enriched in the P2 (crude mitochondrial/synaptosomal) fraction in the mouse cortex at P0 (Fig. S4E). The expression level of 130 kDa DISC1 was reduced in this fraction in BACE1-KO mice compared with their wild-type littermates (Fig. S4F).
DISC1 Is Expressed in Neurons, Astrocytes, Oligodendrocytes, and Microglia.
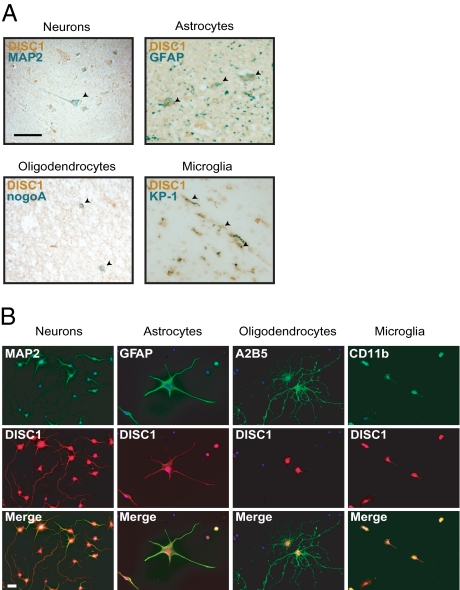
NRG1 signaling plays important roles in both neuronal and glial cells (3, 5). Here we have demonstrated a direct molecular link between NRG1 signaling and DISC1; however, despite some indications of DISC1 expression in glial cells (27), most studies thus far have focused on neuronal DISC1 (3). Therefore, to address whether DISC1 is truly expressed in glial cells, we first used a well-characterized and published affinity-purified monoclonal antibody 3D4 against human DISC1 (30) to investigate DISC1 expression with autopsied human brains. We confirmed high levels of DISC1 expression in pyramidal neurons costained with a neuronal marker MAP2 (Fig. 4A). We also observed equivalent levels of DISC1 expression in astrocytes and microglia, costained with markers glial fibrillary acidic protein (GFAP) and KP-1, respectively (Fig. 4A). Weak staining of DISC1 colocalized with oligodendrocyte marker nogoA was also observed (Fig. 4A). Next, we investigated expression of DISC1 in rat primary cortical neuron, astrocyte, oligodendrocyte, or microglial cultures, with established DISC1 antibody mExon3 (29). In neurons costained with neuronal marker MAP2, DISC1 was seen in the nucleus, cell body, and neurites (Fig. 4B). In astrocytes and microglia costained with markers GFAP and Cd11b, respectively, DISC1 was strongly expressed in the nucleus and cell body, and was seen to a lesser extent in cellular processes, whereas in oligodendrocytes costained with marker A2B5, DISC1 expression was restricted to the nucleus and cell body (Fig. 4B). Colocalization of DISC1 and GFAP were also observed in adult mouse brain, confirming the expression of DISC1 in astrocytes (Fig. S5).
Fig. 4.
DISC1 expression in different cell types. (A) Immunohistochemistry for DISC1 and cell type–specific markers in human cortical sections. Costaining for human DISC1 (brown) with purified monoclonal antibody 3D4 and the following markers (green) was performed: MAP2 (neurons), GFAP (astrocytes), nogoA (oligodendrocytes), and KP-1 (microglia). Magnification 100×. (Scale bar, 50 μm.) (B) Immunofluorescent cell staining for DISC1 and cell type–specific markers in rat primary cortical cultures. Costaining for DISC1 with antibody mExon3 (red) and the following markers (green) was performed: MAP2 (neurons), GFAP (astrocytes), A2B5 (oligodendrocytes), and CD11b (microglia). Magnification 200×. (Scale bar, 10 μm.)
Discussion
This study reports two major findings. First, NRG1 signaling increases the expression of an isoform of DISC1 at 130 kDa and maintains expression of this isoform in the developing mouse cortex. The reduction in DISC1 expression observed in two NRG1 signaling-impaired murine models, NRG1-KO and BACE1-KO mice, is consistent with in vitro results and suggests the possibility that DISC1 deficiency may contribute to their phenotypic abnormalities. This is supported by similarities in behavioral phenotypes among these knockout mice and DISC1 genetically engineered models (3, 20, 21). For example, as DISC1 appears to play an important role in dendritic spine morphogenesis (31), reduction of DISC1 may underlie the alterations in spine morphology and density observed in BACE1-KO mice (26). Clarification of the role of DISC1 in these mice could help to determine the functional significance of the NRG1-PI3K/Akt-DISC1 pathway. Second, DISC1 is expressed in both neuronal and glial lineages, including astrocytes, oligodendrocytes, and microglia, in human brains and rat cortical cultures. We believe that our systematic approach, including costaining with cell type–specific markers to verify neuronal and glial lineage, establishes that DISC1, like NRG and ErbB family proteins, is expressed in several distinct cell types in the human and rodent cortex. DISC1 has been shown to have several functions in neurons, involving multiple subcellular pools capable of interaction with synaptic proteins, centrosomal proteins, and transcription factors (3); however, the role of DISC1 in glia remains to be elucidated. Roles for NRG1 and DISC1 signaling cross-talk across neuronal–glial interactions may well become an important question.
Although we have verified the 130-kDa isoform of DISC1 by Western blotting and immunoprecipitation using well-established antibodies generated from different groups (Fig. S2B) and by verifying its knockdown by DISC1-specific shRNA (Fig. S2 C and D), the true nature of this isoform remains uncertain. These two DISC1 antibodies used have been described previously (29) and were raised against amino acids 360–374 (mExon3) and 734–753 (D27) of mouse DISC1, corresponding to portions of exons 3 and 10, respectively. Full-length DISC1 is 854 amino acids in length and is thought to be detected at ∼100 kDa; however, alternative splicing or posttranslational modification could give rise to the larger DISC1 isoform. The similarity in the magnitude of increase of 130 kDa DISC1 and DISC1 in neurites after NRG1 treatment (Fig. 1 A and B), suggests that this isoform may be neurite-specific or -enriched; this possibility is supported by evidence of selectively decreased 130-kDa DISC1 and corresponding loss of DISC1 in neurites in BACE1-KO mice (Fig. 3 C and D and Fig. S4 C and D). In analysis by mass spectrometry of this 130 kDa signal, following a published protocol (32), we did not observe any DISC1 sequences. Thus, to validate 130 kDa DISC1 signal, we used two antibodies against DISC1 in Western blotting after immunoprecipitation. At least at present, the observation that cotreatment with transcriptional inhibitor actinomycin D prevents induction of 130 kDa DISC1 by NRG1 suggests involvement of a transcriptional mechanism in this induction.
We demonstrate here that NRG1 and NRG2, but not NRG3, can increase expression of 130 kDa DISC1 (Fig. 1A). Interestingly, whereas NRGs can activate receptor ErbB4, only NRG1 and NRG2 can bind ErbB3 (27); furthermore, ErbB3 lacks the receptor tyrosine kinase domain necessary for activation, and ErbB2 lacks an extracellular ligand binding domain, implying that these receptors must heterodimerize to transduce NRG signaling (5). Our results are fully consistent with these observations and directly implicate receptors ErbB2 and ErbB3 as mediators of the effect of NRG1 on DISC1 expression (Fig. 2A). Both receptors are expressed in neurons and radial glia of the developing cortex (33): we also show DISC1 expression in these cell types in vivo and diminished expression of DISC1 in homozygous NRG1-KO mice before embryonic lethality (Fig. 3B). These results suggest that NRG1 maintains DISC1 expression in vivo by signaling via ErbB2/3 heterodimers, in a mechanism that also appears to involve PI3K/Akt signaling (Fig. 2B). As there is evidence associating impaired Akt signaling with schizophrenia (29), this pathway may tie together three prominent schizophrenia susceptibility genes (genes for NRG1, Akt, and DISC1) previously thought to be independent. Although two loss-of-function murine models of NRG1 signaling have been used in this study, previous studies have shown increases in NRG1 mRNA expression and increased NRG1/ErbB4 signaling, but unchanged NRG1 protein levels, in postmortem brains from patients with schizophrenia (34–36); this implies that study of gain-of-function model for NRG1 signaling as well as further examination of NRG1/DISC1 cross-talk at different developmental (including adult) stages could better characterize roles of these genetic susceptibility factors for schizophrenia.
Materials and Methods
Reagents.
Rabbit polyclonal DISC1 antibody D27 (used at 1:50 dilution) was a gift from Dr. Nicholas Brandon (8). Rabbit polyclonal antibody mExon3 antibody (1:200) was raised against AA 360–374 of mouse DISC1 and exhibits similar immunoreactivity to D27 (29). Affinity-purified monoclonal primary antibody 3D4 against human DISC1 was reported previously (30). Antibodies against NRG1 C-term (sc-348, 1:300), ErbB4 (sc-283, 1:200), ErbB3 (sc-285, 1:100), ErbB2 (sc-284, 1:100), and phospho-tyrosine (sc-508, 1:250) were purchased from Santa Cruz Biotechnology. Other antibodies used were mouse anti-A2B5 (R&D Systems. 1:400), mouse anti-GFAP (Millipore, 1:400), rat ant-GFAP (Ambion, 1:400), mouse anti-MAP2 (1:500), mouse anti-Cd11b (1:200), mouse anti–β-tubulin (1:1,000), and mouse anti-GAPDH (1:10,000) (all AbD Serotec), GFAP (DAKO Z0334; 1:2,000), nogoA (Santa Cruz sc-25660; 1:500), anti-CD68, clone KP1 (DAKO M0814; 1:500). Secondary antibodies used were horse anti-mouse biotinylated IgG (Vector BA-2000, 1:1,000), goat anti-rabbit IgG-POD conjugated (Pierce #31460; 1:2,000), goat anti-mouse IgG-POD conjugate (Pierce 31444; 1:2,000), and Alexa Fluor 488 and 568 (Invitrogen, 1:400). Other reagents used were DAB solution (Vector SK-410), streptavidin-peroxidase conjugate (Roche 1089153, 1:2,000), Histogreen (Linaris E109, Wertheim, Germany), and target retrieval solution pH 6.1 (DAKO S1699).
Cell Culture and Transfection.
HEK-293FT cells were maintained in High-Glucose Dulbecco's Modified Eagle's Medium (DMEM) with 10% FBS at 37°C with a 5% CO2 atmosphere in a humidified incubator. 293FT cells were transfected using PolyFect Transfection Reagent (Qiagen) following the manufacturer's protocol, with 5 μg DNA. Cortical primary neuron cultures were prepared from E17-18 Sprague-Dawley rats, and maintained in neuronal medium (Invitrogen neurobasal medium, 2% B27 supplement, 0.5 mM glutamine). Neurons at 5 days in vitro (DIV) were considered immature, and at 21 DIV were considered mature. Glial cell cultures were prepared as follows [adapted from (37) and (38)]: cortical cells from postnatal day 0 (P0) Sprague-Dawley rats were seeded in Poly-D-Lysine (PDL)–coated T-75 flasks in NM-15 medium (MEM with L-glutamine, 15% FBS, 6 mg/mL D-glucose, 100 U/mL penicillin, and 100 μg/mL streptomycin). Homogeneous glial cultures were isolated as follows: after 9 days, medium was replaced and cultures were shaken at 190 rpm for 18 h to separate oligodendrocyte precursor cells (OPCs) and microglia, leaving astrocytes. The medium was removed and added to uncoated 10-cm dishes for 1 h to separate adherent microglia. The remaining supernatant fraction containing OPCs was seeded on PDL-coated dishes, and medium was replaced with differentiation medium the following morning to obtain oligodendrocytes.
Treatment of Primary Neurons.
Immature primary cortical neurons at 3–7 DIV were treated with various compounds (including recombinant proteins, lentivirus, and pharmacological agents) by replacing half of the culture medium with new medium containing the compound. Cells were treated with recombinant NRG proteins at 3 nM concentration for 2 h. Lentiviral infection was carried out by replacing half of the cell culture medium with medium containing lentivirus, incubating cells at 37 °C for 8 h, followed by addition of complete medium and incubation for 5 days. LY294002 in DMSO (Invitrogen) was added to medium at 10 μM concentration concurrently with NRG1. AG490 was reconstituted in DMSO and added to medium at 5 μM concentration 30 min before NRG1 treatment. Rolipram was reconstituted in DMSO and added to medium at 5 μM concentration 1 h before NRG1 treatment. Actinomycin D was reconstituted in DMSO and added to medium 1 h before NRG1 treatment. All pharmacological treatment was conducted based on previously reported protocols (39–42).
Western Blotting and Quantification.
Cells were harvested by scraping into PBS and lysed in RIPA buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 1% deoxycholate, and 5 mM EDTA). Protein concentrations were determined by BSA Protein Assay (Thermo Scientific), and Western blotting was carried out using 8% Tris-Glycine gels, as previously described (29). Exposed films were scanned and densitometric analysis was performed using ImageJ software. Raw intensity was normalized to background and loading control levels.
Immunofluorescent Cell Staining and Quantification.
All cell staining (including neurons and glia) was performed as previously described (13). Briefly, cells cultured on PDL-coated coverslips were fixed (4% formaldehyde, 2% sucrose in PBS) and blocked (2% normal goat serum, 0.1% Triton X-100 in PBS) at room temperature and incubated in primary antibody diluted in blocking reagent overnight at 4 °C. The following day, cells were washed and incubated in secondary antibody and stained with DAPI by incubation for 10 min in 300 ng/mL DAPI (Invitrogen) in PBS. Coverslips were mounted in DABCO anti-fade reagent and observed after drying overnight. To quantify DISC1 expression in neurites, a confocal microscope was used to obtain z-stack images of neurites. MetaMorph software was used to measure integrated signal intensity in the whole cell and intensity in the cell body was subtracted to obtain neurite intensity. Total neurite length was measured by tracing β-tubulin–stained neurites in Metamorph and used to normalize DISC1 signal intensity.
Immunohistochemistry in Human Brains.
Diagnostic human brain samples and authorization for these experiments was obtained from the Ethics Committee of the Heinrich Heine University of Düsseldorf Medical School. Forty-eight hours after incubation in 5% buffered formalin, brain was embedded in paraffin and cut into 5-mm-thick sections. Sections were deparaffinized and endogenous peroxidase was inactivated by incubation in methanol/0.3% H2O2 for 30 min. Sections were then rehydrated in an ethanol gradient and incubated in anti-DISC1 antibody 3D4 overnight at 4 °C, washed, and incubated in secondary antibody, washed and incubated with streptavidin-peroxidase conjugate, and finally developed with DAB solution. For double immunolabeling, before incubation with second antibody, residual peroxidase activity was blocked with PBS/0.3% H2O2 for 30 min. For primary antibodies, incubation was done overnight at 4 °C, followed by secondary antibody and signal development by using Histogreen (Linaris, Wertheim, Germany). For KP-1, pretreatment with target retrieval solution at pH 6.1 (DAKO S1699) was performed before application of primary antibody.
Supplementary Material
Acknowledgments
We thank Yukiko Lema and Dr. P. Talalay for manuscript preparation. We appreciate Dr. Gabrial Corfas for participating in scientific discussions. We thank Dr. Nicholas Brandon and Dr. Cary Lai for providing us with reagents, and Rocky Cheung for technical assistance. This work was supported by Silvo O. Conte Center MH-084018 (to A.S.), MH-069853 (to A.S.), MH-088753 (to A.S. and E.S.A.), NS-39411 (to E.S.A.), and Silvo O. Conte Center MH-51134 (to E.S.A.), as well as by grants from Stanley (to A.S.), Cure Huntington’s Disease Initiative (to A.S.), HighQ (to A.S.), S-R (to A.S.), National Alliance for Research on Schizophrenia and Depression (to A.S., E.S.A., A.K., and A.T.-H.), and DFG Ko 1679/3-1 (to C.K.). S.S. is the recipient of an National Research Service Award fellowship. F31MH081475 from the National Institute of Mental Health. All work is solely the responsibility of the authors.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909284107/DCSupplemental.
References
- 1.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 2.Corfas G, Roy K, Buxbaum JD. Neuregulin 1-erbB signaling and the molecular/cellular basis of schizophrenia. Nat Neurosci. 2004;7:575–580. doi: 10.1038/nn1258. [DOI] [PubMed] [Google Scholar]
- 3.Jaaro-Peled H, et al. Neurodevelopmental mechanisms of schizophrenia: Understanding disturbed postnatal brain maturation through neuregulin-1-ErbB4 and DISC1. Trends Neurosci. 2009;32:485–495. doi: 10.1016/j.tins.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willem M, et al. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 5.Mei L, Xiong WC. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat Rev Neurosci. 2008;9:437–452. doi: 10.1038/nrn2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X, et al. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- 7.Hennah W, Thomson P, Peltonen L, Porteous D. Genes and schizophrenia: Beyond schizophrenia: The role of DISC1 in major mental illness. Schizophr Bull. 2006;32:409–416. doi: 10.1093/schbul/sbj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9:1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- 9.Kamiya A, et al. A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol. 2005;7:1167–1178. doi: 10.1038/ncb1328. [DOI] [PubMed] [Google Scholar]
- 10.Duan X, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camargo LM, et al. Disrupted in Schizophrenia 1 Interactome: Evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 12.Mao Y, et al. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya A, et al. DISC1-NDEL1/NUDEL protein interaction, an essential component for neurite outgrowth, is modulated by genetic variations of DISC1. Hum Mol Genet. 2006;15:3313–3323. doi: 10.1093/hmg/ddl407. [DOI] [PubMed] [Google Scholar]
- 14.Murdoch H, et al. Isoform-selective susceptibility of DISC1/phosphodiesterase-4 complexes to dissociation by elevated intracellular cAMP levels. J Neurosci. 2007;27:9513–9524. doi: 10.1523/JNEUROSCI.1493-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Millar JK, et al. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- 16.Zhang HT, Crissman AM, Dorairaj NR, Chandler LJ, O'Donnell JM. Inhibition of cyclic AMP phosphodiesterase (PDE4) reverses memory deficits associated with NMDA receptor antagonism. Neuropsychopharmacology. 2000;23:198–204. doi: 10.1016/S0893-133X(00)00108-1. [DOI] [PubMed] [Google Scholar]
- 17.Huston E, et al. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci USA. 2008;105:12791–12796. doi: 10.1073/pnas.0805167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monje PV, Bartlett Bunge M, Wood PM. Cyclic AMP synergistically enhances neuregulin-dependent ERK and Akt activation and cell cycle progression in Schwann cells. Glia. 2006;53:649–659. doi: 10.1002/glia.20330. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez D, et al. Phenotypic and biochemical analyses of BACE1- and BACE2-deficient mice. J Biol Chem. 2005;280:30797–30806. doi: 10.1074/jbc.M505249200. [DOI] [PubMed] [Google Scholar]
- 20.Kellendonk C, Simpson EH, Kandel ER. Modeling cognitive endophenotypes of schizophrenia in mice. Trends Neurosci. 2009;32:347–358. doi: 10.1016/j.tins.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Jaaro-Peled H, Sawa A, Brandon NJ. How has DISC1 enabled drug discovery? Mol Cell Neurosci. 2008;37:187–195. doi: 10.1016/j.mcn.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 23.Gerlai R, Pisacane P, Erickson S. Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav Brain Res. 2000;109:219–227. doi: 10.1016/s0166-4328(99)00175-8. [DOI] [PubMed] [Google Scholar]
- 24.Stefansson H, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet. 2002;71:877–892. doi: 10.1086/342734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moy SM, et al. Deficient NRG1-ERBB signaling alters social approach: Relevance to genetic mouse models of schizophrenia. J Neurodev Disord. 2009;4:302–312. doi: 10.1007/s11689-009-9017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savonenko AV, et al. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci USA. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 2009;18:391–404. doi: 10.1093/hmg/ddn361. [DOI] [PubMed] [Google Scholar]
- 28.Buonanno A, Fischbach GD. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol. 2001;11:287–296. doi: 10.1016/s0959-4388(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 29.Ishizuka K, et al. Evidence that many of the DISC1 isoforms in C57BL/6J mice are also expressed in 129S6/SvEv mice. Mol Psychiatry. 2007;12:897–899. doi: 10.1038/sj.mp.4002024. [DOI] [PubMed] [Google Scholar]
- 30.Leliveld SR, et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008;28:3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kvajo M, et al. A mutation in mouse Disc1 that models a schizophrenia risk allele leads to specific alterations in neuronal architecture and cognition. Proc Natl Acad Sci USA. 2008;105:7076–7081. doi: 10.1073/pnas.0802615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawamura N, Sawamura-Yamamoto T, Ozeki Y, Ross CA, Sawa A. A form of DISC1 enriched in nucleus: Altered subcellular distribution in orbitofrontal cortex in psychosis and substance/alcohol abuse. Proc Natl Acad Sci USA. 2005;102:1187–1192. doi: 10.1073/pnas.0406543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anton ES, Marchionni MA, Lee KF, Rakic P. Role of GGF/neuregulin signaling in interactions between migrating neurons and radial glia in the developing cerebral cortex. Development. 1997;124:3501–3510. doi: 10.1242/dev.124.18.3501. [DOI] [PubMed] [Google Scholar]
- 34.Hashimoto R, et al. Expression analysis of neuregulin-1 in the dorsolateral prefrontal cortex in schizophrenia. Mol Psychiatry. 2004;9:299–307. doi: 10.1038/sj.mp.4001434. [DOI] [PubMed] [Google Scholar]
- 35.Hahn CG, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- 36.Law AJ, et al. Neuregulin 1 transcripts are differentially expressed in schizophrenia and regulated by 5′ SNPs associated with the disease. Proc Natl Acad Sci USA. 2006;103:6747–6752. doi: 10.1073/pnas.0602002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barres BA, et al. Cell death and control of cell survival in the oligodendrocyte lineage. Cell. 1992;70:31–46. doi: 10.1016/0092-8674(92)90531-g. [DOI] [PubMed] [Google Scholar]
- 38.Tang DG, Tokumoto YM, Raff MC. Long-term culture of purified postnatal oligodendrocyte precursor cells. Evidence for an intrinsic maturation program that plays out over months. J Cell Biol. 2000;148:971–984. doi: 10.1083/jcb.148.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshii A, Constantine-Paton M. BDNF induces transport of PSD-95 to dendrites through PI3K-AKT signaling after NMDA receptor activation. Nat Neurosci. 2007;10:702–711. doi: 10.1038/nn1903. [DOI] [PubMed] [Google Scholar]
- 40.Barnabé-Heider F, et al. Evidence that embryonic neurons regulate the onset of cortical gliogenesis via cardiotrophin-1. Neuron. 2005;48:253–265. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita N, Yamauchi M, Baba J, Sawa A. Phosphodiesterase type 4 that regulates cAMP level in cortical neurons shows high sensitivity to rolipram. Eur J Pharmacol. 1997;337:95–102. doi: 10.1016/s0014-2999(97)01285-5. [DOI] [PubMed] [Google Scholar]
- 42.Joo CH, et al. Coxsackievirus B4-induced neuronal apoptosis in rat cortical cultures. Neurosci Lett. 2002;326:175–178. doi: 10.1016/s0304-3940(02)00340-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.