Abstract
Hose in Hose mutants of primrose and cowslip have been cultivated since the early 17th century and show dominant homeotic conversion of sepals to petals. The phenotype shows variable penetrance and expressivity and is linked to the S locus, which controls floral heteromorphy in Primula species. Here we demonstrate that the homeotic conversion of sepals to petals in Hose in Hose is associated with up-regulation of both Primula B-function MADS box genes PvDef and PvGlo in the first floral whorl. We have defined a restriction fragment length polymorphism associated with PvGlo that cosegregates with the Hose in Hose phenotype and have also identified and characterized a retrotransposon insertion in the PvGlo promoter which is associated with the up-regulated expression of PvGlo. Excision of this retrotransposon, associated with epigenetic changes at the locus, causes reversion toward normal calyces and restores wild-type flower development. These data define the molecular basis of the Hose in Hose mutation and provide an explanation for its long-documented phenotypic instability.
Keywords: flower development, floral heteromorphy, transcription-associated recombination
Floral homeotic mutants have captured imaginations for more than 2,000 years and have been well documented in numerous species (1), with those of primrose and cowslip among the first to be described in detail. Hose in Hose is one such homeotic mutation which shows conversion of sepals to petals that features in various early herbals and florilegia (2–5) and influenced the thinking of both Lyell (6) and Darwin (7). The ABC model of flower development (8) predicts that ectopic expression of the B-function MADS box genes PvDef and PvGlo (9) in the first whorl would transform sepals to petals, and phenocopy Hose in Hose and previous transgenic studies have demonstrated this (10–12). Ectopic expression of Antirrhinum Glo in tobacco leads to petaloid sepals, and ectopic expression of both Def and Glo leads to the almost complete conversion of sepals to petals (10). Similar studies in Petunia (11) and Arabidopsis (12) provide further examples of dominant engineered homeotic conversions. The demonstration that in some cases up-regulation of a single B-function MADS box gene can lead to the development of petaloid sepals is consistent with inheritance of Hose in Hose as a single dominant locus. In contrast, the choripetala and despenteado mutants of Antirrhinum are inherited as recessive mutations which also result in the conversion of sepals to petals (13).
Primula species also exhibit floral heteromorphy, with development of distinct pin and thrum flowers on separate plants (14, 15). Pin flowers produce a long style which presents the stigma at the mouth of the flower and anthers halfway down the floral tube. The reciprocal elevation of male and female reproductive structures in thrum flowers creates a breeding system which facilitates cross-pollination between the different forms of flower. An incompatibility system reinforces this complex breeding system. Developmental control is mediated by the S locus (16, 17), which comprises a coadapted linkage group of as yet unknown genes. Pin plants are homozygous for the recessive s allele (s/s); thrum plants are heterozygous and carry a dominant S allele (S/s). The Hose in Hose mutation is linked to the S locus (18, 19), and following rare recombination events can be either in coupling or repulsion to the dominant S allele. These observations demonstrate that a gene responsible for floral organ identity is linked to the S locus, although this gene does not influence any aspects of heteromorphic development. The Hose in Hose phenotype is dominant (18, 19) and shows variable penetrance and expressivity (18); the unexplained nature of this phenotypic instability, together with the genetic linkage to the S locus, prompted us to investigate the molecular basis of this early homeotic mutant.
Results and Discussion
To investigate the causal mechanism underpinning the dominant nature and phenotypic instability of Hose in Hose, we used a mutant line that normally shows complete penetrance, with the dominant Hose in Hose allele in coupling with the recessive s allele of the S locus. A cross between a pin Hose in Hose heterozygote and a wild-type thrum plant yielded four progeny classes (Fig. 1A); Southern analysis with PvGlo as a probe revealed restriction fragment length polymorphisms (RFLPs) for PvGlo which cosegregate with different phenotypes (Fig. 1B). One such PvGlo RFLP is a 2.4-kbp band that is Hose in Hose-specific, as determined by Southern analysis of 28 segregating plants. We have previously shown by RFLP analysis of 157 pin and thrum plants that PvGlo is linked to the S locus (9). These data, together with the fact that Hose in Hose is linked to the S locus (18, 19), suggest that PvGlo and Hose in Hose may be allelic; PvDef is not linked to the S locus (9).
Fig. 1.
Genetics, organization, and expression of PvGlo in Hose in Hose and wild-type plants. (A) Phenotypes and genotypes of parents and progeny classes segregating for dominant Hose in Hose and thrum phenotypes. (B) Southern analysis of XbaI-digested genomic DNA from sibling Hose in Hose (Hose) or wild-type (WT) pin (P) and thrum (T) plants using PvGlo cDNA (GenBank accession no. DQ381428) as probe. The Hose in Hose allele is indicated (*). (C) RNA gel blot analysis of PvGlo and PvDef in leaves (L) and the four floral whorls (W1–W4) from wild-type and Hose in Hose plants alongside ethidium-bromide-stained RNA gel, probed by PvDef cDNA (GenBank accession no. DQ381427) and PvGlo cDNA (GenBank accession no. DQ381428), respectively.
Expression analysis of PvDef and PvGlo in leaves and individual floral whorls of mature wild-type and Hose in Hose flowers revealed expression of both PvDef and PvGlo primarily in the petals (whorl 2) of mature wild-type flowers (Fig. 1C). In contrast, both PvDef and PvGlo are up-regulated in whorl-1 petals in Hose in Hose flowers and retain normal expression in whorl-2 petals. Neither gene is up-regulated in the third and fourth floral whorls or in the leaves of Hose in Hose plants, indicating that up-regulation of both B-function genes is limited to the first whorl of mutant flowers.
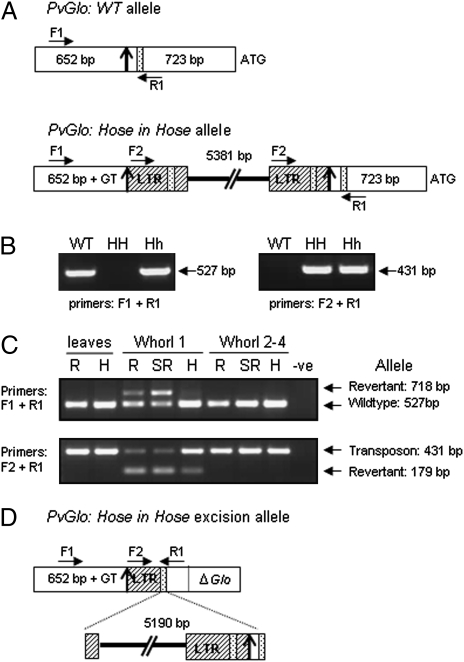
Comparison of the PvGlo genomic sequence from Hose in Hose with the wild-type locus (9) revealed a 5381-bp gypsy/Ty-3-like retrotransposon (20) 723 bp upstream of the translation initiation codon with a 2-bp GT sequence insertion site duplication (Fig. 2A). A 10-bp promoter motif downstream of the retrotransposon would have been unremarkable had it not also been present in each of the long terminal repeats (LTRs). Modulation of tissue-specific expression of genes next to retrotransposons has been reported previously (21–23); we therefore predicted that the retrotransposon was up-regulating PvGlo in the first whorl, which in turn caused up-regulation of PvDef, as found in Antirrhinum and Arabidopsis (24, 25).
Fig. 2.
Hose in Hose is caused by a retrotransposon insertion in the promoter of PvGlo. (A) Structure of wild-type (GenBank accession no. DQ381430) and Hose in Hose alleles (GenBank accession no. DQ38144) of PvGlo. The 1373-bp sequence upstream of the translation initiation codon (ATG) is bisected into regions upstream (650 bp) and downstream (723 bp) of the transposon insertion site (vertical arrow). The 2-bp target site duplication (GT) and the 10-bp motif AGCAATTTTA (stippled) in the native promoter and present in both long terminal repeats (LTR, hatched) of the 5381-bp retrotransposon (GenBank accession no. DQ381432) (thick line) are shown together with PCR primers. (B) PCR analysis of DNA from wild-type (WT), homozygous Hose in Hose (HH), and heterozygous Hose in Hose (Hh) plants. (C) Agarose gel electrophoresis of PCR products from Whorl 1 only and combined whorl 2, 3, and 4 tissue of revertant (R), semirevertant (SR), and Hose in Hose flowers (H) alongside a no-DNA control (-ve). DNA from leaves of the crowns producing semirevertant and revertant (R) and Hose in Hose (H) flowers was also amplified with primers as shown. (D) Revertant allele structure derived from sequence analysis (GenBank accession no. FJ897502) of PCR products from C, showing deletion (ΔGlo) after retrotransposon excision. Labels are as in A.
The identification of a spontaneous Hose in Hose revertant in our population, as first illustrated by van de Passe in 1614 (4) (Fig. 3A), provided the opportunity to define the relationship between PvGlo and Hose in Hose. This plant produced two crowns, one with stable Hose in Hose calyces; the other produced two scapes, one with semirevertant petaloid sepals and one with revertant almost wild-type calyces (Fig. 3 B and C). PCR primers that distinguish between wild-type and retrotransposon insertion alleles of PvGlo (Fig. 2 A and B) were used to amplify genomic DNA from leaves of Hose in Hose and wild-type sectors of the revertant plant, as well as DNA from dissected floral whorls of Hose in Hose, semirevertant, and revertant flowers (Fig. 2C).
Fig. 3.
Hose in Hose mutant and revertant flowers. (A) Woodblock print (4) of Hose in Hose showing sepal-to-petal conversion and revertant wild-type flower (arrow) exhibiting a normal calyx. (B) Two crowns from a single Hose in Hose plant with mutant, semirevertant, and revertant flowers on sectors of the same plant. (C) Individual Hose in Hose, semirevertant, and revertant flowers from B.
The revertant plant is heterozygous for Hose in Hose, so all samples contain both wild-type and transposon alleles of PvGlo. The primers also produced amplification products restricted to whorl 1 of revertant, semirevertant, and to a lesser extent Hose in Hose flowers (Fig. 2C). Sequence analysis of these products revealed an allele in which 5190 bp of retrotransposon, together with 101 bp of promoter sequence, had been deleted, leaving a 293-bp footprint comprising part of the upstream LTR; we refer to this as the excision allele. The sequence of this excision allele is consistent with a homologous recombination event between the 10-bp motif in the upstream LTR and the identical motif downstream of the transposon insertion site (Fig. 2 A and D), reminiscent of some gypsy excisions in Drosophila (26).
Further analysis of the excision allele revealed the associated loss of PvGlo sequences downstream of the 10-bp recombination site (Fig. 2D). Reversion of Hose in Hose therefore results from loss of the transposon responsible for up-regulation of PvGlo as well as deletion of the PvGlo coding sequence from this dominant allele, leaving the wild-type recessive allele as an appropriately regulated functional gene. Because PvGlo is not disrupted in the previously described S locus-linked sepaloid mutant which lacks petals (9), these data demonstrate that Hose in Hose and sepaloid represent distinct S locus-linked genes that affect petal identity.
Unlike two-component DNA transposons where excision is associated with their nomadic behavior (27), retrotransposons do not routinely, but do occasionally, excise (26, 28). The whorl-1-specific presence of the excision allele reveals that reversion has occurred at or after commitment of the first floral whorl (29). The presence of identical excision events in semirevertant and revertant flowers shows that the same sequence-specific recombination event has occurred in different flowers.
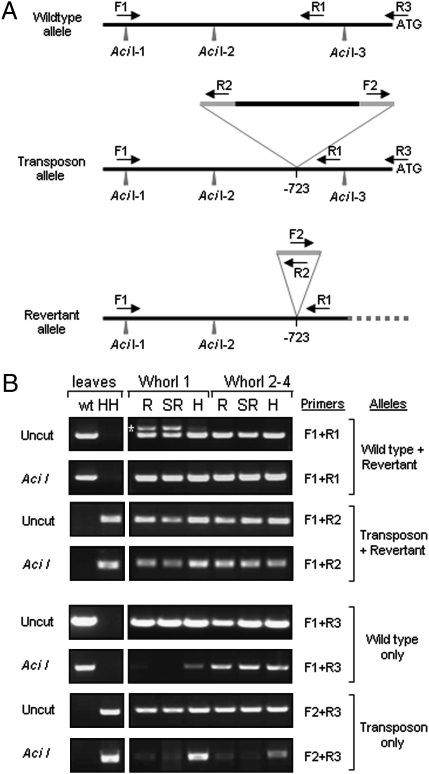
The variable expressivity and penetrance of Hose in Hose (18, 19) suggest an epigenetic influence on the mutant phenotype. To explore a possible mechanism for the whorl-specific excision of the retrotransposon that was consistent with the potential epigenetic basis of the event, we used a PCR-based assay to investigate the methylation status of the wild-type, transposon, and revertant alleles. In this assay, parallel genomic DNA samples, one digested with the methylation-sensitive restriction enzyme AciI, the other undigested, were used as templates for PCR amplification with primers that flank the AciI cleavage sites (Fig. 4A). The presence of an amplification product reveals CpG methylation of the CCGC cleavage site; absence of a PCR product shows that the restriction site is demethylated and therefore cleavable by AciI, which cuts the PCR template between primer sites.
Fig. 4.
Retrotransposon excision is associated with demethylation of the PvGlo locus. (A) Organization of wild-type, transposon, and revertant alleles of PvGlo showing location of upstream methylation-sensitive AciI restriction enzyme sites. The PvGlo-specific primers (F1, R1, R3) and transposon-specific PCR primers (R2, F2) are indicated. The retrotransposon insertion site is shown (-723); LTRs are shown in gray. (B) Methylation-sensitive PCR analysis of the wild-type, transposon, and revertant alleles of PvGlo using allele-specific primer combinations as indicated. Primer specificity is demonstrated on DNA from leaves of wild-type (wt) and homozygous Hose in Hose (HH) plants. DNA samples from whorl-1 tissue or combined whorl-2, -3, and -4 tissue from revertant (R), semirevertant (SR), and Hose in Hose (H) flowers from the same plant were either digested with Aci1 or uncut as indicated before PCR amplification using allele-specific primer combinations as shown before fractionation by agarose gel electrophoresis. The different alleles amplified by each primer combination are indicated and the excision allele-specific PCR product is highlighted (*). PCR products generated from uncut genomic DNA that are absent after predigestion with AciI reveal unmethylated sites.
PCR amplification of genomic DNA from leaves of wild-type and homozygous Hose in Hose plants revealed the allelic specificities of relevant primer combinations (Fig. 4B). In addition, the presence of PCR amplification products using both cut and uncut DNA template revealed that in leaves, all three AciI sites proximal to the retrotransposon insertion site are methylated (Fig. 4B). We then assayed the methylation status of upstream AciI sites (AciI-1 and AciI-2) (Fig. 4B Upper) and the downstream AciI-3 site (Fig. 4B Lower) separately using genomic DNA from the first whorls and the combined second, third, and fourth whorls of Hose in Hose, semirevertant, and revertant flowers. These assays revealed both whorl-specific and reversion-specific differences in the methylation status of the different alleles, as shown by the absence of amplification products from some AciI-digested templates (Fig. 4B).
The wild-type and revertant Hose in Hose alleles in whorl 1 are revealed by two PCR products using uncut genomic DNA as template with primers F1 and R1; the revertant allele-specific product (indicated by an asterisk) is absent from whorl 1 of Hose in Hose flowers and whorls 2–4 of revertant, semirevertant, and Hose in Hose flowers (Fig. 4B; see also Fig. 2C). The loss of the excision allele-specific PCR product following AciI digestion of template DNA from whorl 1 of revertant and semirevertant flowers revealed demethylation of either AciI-1 or AciI-2, or both, in this allele (Fig. 4B). The consistent amplification of the wild-type allele following AciI digestion of all templates reveals that both AciI-1 and AciI-2 are methylated in the wild-type allele in all four whorls. PCR amplification of the same samples using primers F1 and R2, which produce the same product from both the transposon insertion and revertant excision allele, reveal that the upstream AciI-1 and AciI-2 are methylated in all whorls of revertant, semirevertant, and Hose in Hose flowers, as digestion of template DNA does not affect PCR amplification. However, as demonstrated by using primers F1 and R1 above, the excision allele (indicated by an asterisk) is in fact demethylated, as the product is lost following AciI digestion. The observed F1 and R2 primer-derived PCR products following template digestion with AciI must therefore come from the transposon and not the excision allele. In combination, these data show that upstream AciI sites are methylated in both the wild-type and transposon alleles, and that the first whorl-specific excision allele has undergone demethylation (Fig. 4B).
Analysis of the methylation status of AciI-3 revealed a different story (Fig. 4B). Using undigested template DNA and primers F1 and R3, which amplify the wild type but not the transposon or excision alleles, across all three AciI sites, uniform amplification was observed in all samples. However, AciI digestion of template DNA from whorl 1 of revertant and semirevertant flowers disrupted amplification and also reduced amplification from whorl 1 of Hose in Hose flowers. As the upstream AciI-1 and AciI-2 sites are methylated and cannot be cut by AciI (see above), these data demonstrate whorl-1-specific demethylation of the wild-type allele at the downstream AciI-3 site. Similar analyses with primers F2 and R3, which only amplify across AciI-3 in the transposon allele, reveal that demethylation of this site is not whorl-specific but is associated with all whorls of flowers showing reversion (Fig. 4B). The primer R3 in combination with F1 or F2 does not amplify the excision allele, as sequences downstream of the transposon insertion and R1 primer site are deleted in this allele (Fig. 4A).
We conclude that during reversion, demethylation of the AciI-3 site downstream of the transposon occurs in all whorls of flowers predisposed to reversion (Fig. 4B; primers F2 and R3). Whorl-specific transposon excision then occurs by homologous recombination between the 10-bp motif present in the upstream LTR and the promoter, with the concomitant loss of downstream sequences in this allele (Fig. 4A). It is not possible to determine whether whorl-specific demethylation of AciI-1 and AciI-2 in the excision allele (Fig. 4B; primers F1 and R1) occurs as a consequence of whorl-specific excision or whether whorl-specific demethylation facilitates excision. The first whorl-specific demethylation of AciI-3 in the wild-type allele (Fig. 4B; primers F1 and R3) suggests a paramutagenic postexcision event, as this allele is not fully demethylated in whorl 1 of Hose in Hose flowers (Fig. 4B; primers F1 and R3). These epigenetic changes lead to loss of the dominant Hose in Hose transposon insertion allele and restoration of the wild-type phenotype mediated by the remaining wild-type allele. Previous examples of naturally occurring epi-mutations affecting plant development have been documented (30–32). Our findings reveal that epigenetic events in wild populations can work in both directions to drive plant development, either by causing or by restoring mutant phenotypes.
Although we would not anticipate the germline transmission of the revertant phenotype, as the transposon excision allele cannot be detected in either the third or fourth whorls which give rise to the gametes, previous studies on Hose in Hose reversion (33) demonstrated that homozygous Hose in Hose flowers showing partial reversion could yield both Hose in Hose and wild-type progeny when crossed to wild-type flowers. The molecular basis of reversion in these plants is not known, leading us to speculate that either the reversion events were not limited to the first whorl in the line used for genetic analysis, or transmission of the genetic instability does not rely on transmission of an excision allele but on the epigenetic predisposition for the whorl-specific loss of the transposon in subsequent generations.
Perhaps one of the most surprising findings arising from these studies is the whorl-specific homologous recombination that leads to excision of the retrotransposon. There are, however, several examples in prokaryotes, yeast, and mammalian systems of transcription-associated recombination in which mitotic recombination rates are increased in transcriptionally active DNA (34). The observation that the retrotransposon causes whorl-1-specific up-regulation of PvGlo, rather than ubiquitous overexpression, coupled with whorl-specific homologous recombination, leads us to speculate that the instability of Hose in Hose represents an example of transcription-associated recombination arising from the aberrant expression of the PvGlo locus.
These observations not only identify PvGlo as the S locus-linked gene responsible for Hose in Hose, they demonstrate that Hose in Hose and sepaloid (9) are nonallelic linked floral organ-identity loci. Our findings may also reveal one of the first examples of transcription-associated recombination in plants as an explanation for the phenotypic instability of Hose in Hose (18) which was so presciently illustrated by van de Passe in 1614 (4).
Experimental Procedures
Plant Materials.
Hose in Hose plants were provided by M. Webster, who holds the National Collection of Primula (British floral variants). The unstable Hose in Hose plant was a spontaneous revertant that produced two crowns; one produced stable Hose in Hose flowers, and the second produced semirevertant and almost wild-type revertant flowers.
Genomic DNA Extraction and Genome Walker Analysis.
Genomic DNA was extracted from frozen material using a Nucleon Plant DNA Extraction Kit (Amersham Pharmacia). PvGlo promoter sequences were obtained from wild-type and Hose in Hose homozygote and heterozygote plants using a Genome Walker Kit (Clontech).
DNA Methylation Analysis.
For PCR and methylation analysis, genomic DNA (1 μg) was digested overnight with 20 U AciI (New England Biolabs), and digested DNA was used as templates for PCR. Nondigested DNA extracted from the same tissue was used as uncut controls.
Southern and Northern Blot Analysis.
Genomic DNA was digested with XbaI, fractionated by gel electrophoresis, and analyzed as described previously using radiolabeled PvGlo cDNA as probe (9). RNA was extracted as described previously (35). Both DNA and RNA gel blots were washed at a stringency (0.2× SSC, 0.1% SDS at 60 °C) at which PvGlo and PvDef do not cross-hybridize (9).
PCR Amplification, Cloning, and Sequencing.
DyNAzyme II Hot Start DNA Polymerase (Finnzymes) was used with primers F1, 5′-CGGTATATATG-CCCGCTTCCGTCTAA-3′; F2, 5′-CGATGTAGTATCAGTAGCCATAGGATTAGT-3′; R1, 5′-TTGCATGGTGAGTTGGTGACAC-3′; R2, 5′-CCCACGTTGTCGTAACGTTGATGAT-3′; R3, 5′-CCTCTACCCATCTCTTTCTTTTTCTCTTCCTCTCTA-3′. PCR products were cloned into pGEM-T Easy Vector (Promega) using standard protocols and sequenced on Applied Biosystems 373 and Li-Cor sequencers.
Acknowledgments
We thank Martin Lappage for growing plants, Peter Meyer for methylation analysis advice, various colleagues for comments on the manuscript, the BBSRC and Gatsby Foundation for funding, and Mike Ambrose for the image from Hortus Floridus (4).
Footnotes
The authors declare no conflict of interest.
References
- 1.Meyerowitz EM, Smyth DR, Bowman JL. Abnormal flowers and pattern formation in floral development. Development. 1989;106:209–217. [Google Scholar]
- 2.Gerard J. In: The Herball or Generall Historie of Plantes. Johnson T, editor. London: John Norton; 1633. [Google Scholar]
- 3.de Reneaulm P. Specimen Historiae Plantarum: Plantae Typis Aneis Expressae. Paris: Beys; 1611. [Google Scholar]
- 4.van de Passe C. Hortus Floridus. Utrecht: J. Jansson; 1614. [Google Scholar]
- 5.Parkinson J. Paradisus in Sole Paradisus Terrestris. London: Humfrey Lownes and Robert Young; 1629. [Google Scholar]
- 6.Lyell C. Principles of Geology. London: John Murray; 1830. [Google Scholar]
- 7.Darwin C. The Variation of Animals and Plants Under Domestication. London: John Murray; 1868. [Google Scholar]
- 8.Coen E. Goethe and the ABC model of flower development. C R Acad Sci III. 2001;324:523–530. doi: 10.1016/s0764-4469(01)01321-x. [DOI] [PubMed] [Google Scholar]
- 9.Li J, et al. The S locus-linked Primula homeotic mutant sepaloid shows characteristics of a B-function mutant but does not result from mutation in a B-function gene. Plant J. 2008;56:1–12. doi: 10.1111/j.1365-313X.2008.03584.x. [DOI] [PubMed] [Google Scholar]
- 10.Davies B, DiRosa A, Eneva T, Saedler H, Sommer H. Alteration of tobacco floral organ identity by expression of combinations of Antirrhinum MADS-box genes. Plant J. 1996;10:663–677. doi: 10.1046/j.1365-313x.1996.10040663.x. [DOI] [PubMed] [Google Scholar]
- 11.Halfter U, Ali N, Stockhaus J, Ren L, Chua NH. Ectopic expression of a single homeotic gene, the Petunia gene GREEN PETAL, is sufficient to convert sepals to petaloid organs. EMBO J. 1994;13:1443–1449. doi: 10.1002/j.1460-2075.1994.tb06398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krizek BA, Meyerowitz EM. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development. 1996;122:11–22. doi: 10.1242/dev.122.1.11. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson M, Silva ED, Zachgo S, Saedler H, Schwarz-Sommer Z. CHORIPETALA and DESPENTEADO: General regulators during plant development and potential floral targets of FIMBRIATA-mediated degradation. Development. 2000;127:3725–3734. doi: 10.1242/dev.127.17.3725. [DOI] [PubMed] [Google Scholar]
- 14.Darwin C. On the two forms, or dimorphic condition, in the species of Primula, and on their remarkable sexual relations. J Proc Linn Soc Bot. 1862;6:77–96. [Google Scholar]
- 15.Ernst A. Further investigations towards the analysis of fertilization and genetics of heterostyly in Primula. I. Primula viscose (Translated from German) Arch Julius Klaus Stift Vererbungsforsch Sozialanthropol Rassenhyg. 1933;8:1–215. [Google Scholar]
- 16.Bateson W, Gregory RP. On the inheritance of heterostylism in Primula. Proc R Soc Lond B Ser. 1905;76:581–586. [Google Scholar]
- 17.Lewis D, Jones DA. The genetics of heterostyly. In: Barrett S.C.H., editor. Evolution and Function of Heterostyly. Berlin: Springer; 1993. pp. 129–150. [Google Scholar]
- 18.Ernst A. Transmission by unstable genes (Translated from German) Arch Julius Klaus Stift Vererbungsforsch Sozialanthropol Rassenhyg. 1942;17:1–567. [Google Scholar]
- 19.Webster MA, Grant CJ. The inheritance of calyx morph variants in Primula vulgaris (Huds) Heredity. 1990;64:121–124. [Google Scholar]
- 20.Havecker ER, Gao X, Voytas DF. The diversity of LTR retrotransposons. Genome Biol. 2004;5:225. doi: 10.1186/gb-2004-5-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung H, et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175:1071–1077. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conte C, Dastugue B, Vaury C. Promoter competition as a mechanism of transcriptional interference mediated by retrotransposons. EMBO J. 2002;21:3908–3916. doi: 10.1093/emboj/cdf367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerman DN, Feder ME. Naturally occurring transposable elements disrupt hsp70 promoter function in Drosophila melanogaster. Mol Biol Evol. 2005;22:776–783. doi: 10.1093/molbev/msi063. , and erratum (2005) 22:1160. [DOI] [PubMed] [Google Scholar]
- 24.Jack T, Fox GL, Meyerowitz EM. Arabidopsis homeotic gene APETALA3 ectopic expression: Transcriptional and post-transcriptional regulation determine floral organ identity. Cell. 1994;76:703–716. doi: 10.1016/0092-8674(94)90509-6. [DOI] [PubMed] [Google Scholar]
- 25.Trobner W, et al. GLOBOSA—A homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nefedova LN, Ljubomirskaya NV, Ilyin YV, Kim AI. Precise excision of long terminal repeats of the gypsy (mdg4) retrotransposon of Drosophila melanogaster detected in Escherichia coli cells is explained by its integrase function. Russ J Genet. 2006;42:1398–1404. [PubMed] [Google Scholar]
- 27.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: Where genetics meets genomics. Nat Rev Genet. 2002;3:329–341. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 28.Carbonare BD, Gehring WJ. Excision of Copia element in a revertant of the white-apricot mutation of Drosophila melanogaster leaves behind one long terminal repeat. Mol Gen Genet. 1985;199:1–6. doi: 10.1007/BF00327501. [DOI] [PubMed] [Google Scholar]
- 29.Webster MA, Gilmartin PM. A comparison of early floral ontogeny in wild-type and floral homeotic mutant phenotypes of Primula. Planta. 2003;216:903–917. doi: 10.1007/s00425-002-0942-y. [DOI] [PubMed] [Google Scholar]
- 30.Bastow R, et al. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- 31.Cubas P, Vincent C, Coen E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature. 1999;401:157–161. doi: 10.1038/43657. [DOI] [PubMed] [Google Scholar]
- 32.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 33.Webster MA. Floral morphogenesis in Primula: Inheritance of mutant phenotypes, heteromorphy, and linkage analysis. UK: PhD thesis (Univ of Leeds, Leeds; 2005. [Google Scholar]
- 34.Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: Link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Webster MA, Furuya M, Gilmartin PM. Identification and characterization of pin and thrum alleles of two genes that co-segregate with the Primula S locus. Plant J. 2007;51:18–31. doi: 10.1111/j.1365-313X.2007.03125.x. [DOI] [PubMed] [Google Scholar]