Abstract
Residues within processed protein fragments bound to major histocompatibility complex class I (MHC-I) glycoproteins have been considered to function as a series of “independent pegs” that either anchor the peptide (p) to the MHC-I and/or interact with the spectrum of αβ-T-cell receptors (TCRs) specific for the pMHC-I epitope in question. Mining of the extensive pMHC-I structural database established that many self- and viral peptides show extensive and direct interresidue interactions, an unexpected finding that has led us to the idea of “constrained” peptides. Mutational analysis of two constrained peptides (the HLA B44 restricted self-peptide (B44DPα–EEFGRAFSF) and an H2-Db restricted influenza peptide (DbPA, SSLENFRAYV) demonstrated that the conformation of the prominently exposed arginine in both peptides was governed by interactions with MHC-I-orientated flanking residues from the peptide itself. Using reverse genetics in a murine influenza model, we revealed that mutation of an MHC-I-orientated residue (SSLENFRAYV → SSLENARAYV) within the constrained PA peptide resulted in a diminished cytotoxic T lymphocyte (CTL) response and the recruitment of a limited pMHC-I specific TCR repertoire. Interactions between individual peptide positions can thus impose fine control on the conformation of pMHC-I epitopes, whereas the perturbation of such constraints can lead to a previously unappreciated mechanism of viral escape.
Keywords: crystal structure, cytotoxic T cell, MHC class I, viral immunity, epitope
Major histocompatibility complex class I (MHC-I) molecules bind processed peptide (p) fragments (1) within a defined cleft. A conserved hydrogen-bonding network at the N and C termini often, although not exclusively (2), limits the length of bound peptides to 8–10 amino acids, with the specificity of binding being mediated via peptide side chains that interact with six pockets (A to F) within the MHC-I cleft. The extensive polymorphism that characterizes the MHC is largely a function of diversity within the cleft providing the structural basis for a broad spectrum of peptides to bind different MHC-I glycoproteins. The size, hydrophobicity, and electrostatic charge of these pockets impacts the potential peptide repertoire of different MHC-I allotypes by enforcing a marked preference for “anchor residues” at certain positions within a given peptide. For example, P2-Glu and PΩ-Tyr (the C-terminal residue) are the dominant anchor residues for HLA-B44 (3), whereas P5-Asn and PΩ-Met/Leu/Ile represent the anchor motif for H2-Db (4). In addition amino acids at other secondary positions can also influence allele-specific binding (5). Presently, the conceptual perspective for MHC-I-bound peptides is that they align in a linear and extended fashion, with some side chains pointing down to interact with the MHC (anchor residues), while others point up to form potential T-cell receptor (TCR) contact points. Moreover, each residue within the peptide is thought to act as an independent peg and consequently there is considered to be no interplay between the residues within any given peptide. The functional analysis of alanine-scanning mutations of MHC-I restricted peptides relies on this interpretation, which is also the assumption underlying programs that seek to predict immunogenicity.
Immune CD8+ cytotoxic T lymphocytes (CTLs) express αβ-TCRs that recognize pMHC-I complexes. Despite pathogen complexity, CTL responses are often directed against few of the many potential pMHC-I complexes (6). This skewing of effective antiviral immunity to immunodominant peptides puts selective pressure on the virus with the consequence that CTL escape mutants may arise during infection. For example, mutations in dominant viral peptides have been reported to subvert CTL recognition following infection with hepatitis B virus (7), HIV type 1 (HIV-1) (8), and Epstein-Barr virus (EBV) (9). Such mutations may impact on (i) the conformation and flexibility of the bound peptide and alter signaling, as observed for the immunodominant peptide from vesicular stomatitis virus (VSV) (10); (ii) MHC-I binding due to the removal of critical anchor residue interactions; (iii) TCR recognition directly; and (iv) proteosomal cleavage (11). Moreover, mutations within viral proteins can lead to the generation of altered peptide ligands (APLs), including T-cell antagonists (7). What precisely dictates CTL escape is not fully established (10), although maintenance of viral fitness is a key parameter that can limit the capacity of the virus to mutate functionally conserved residues in an immunogenic peptide, thus limiting the molecular potential for immune evasion.
To accommodate extensive pMHC-I diversity, the host possesses a diverse T-cell repertoire generated via somatic rearrangement of gene segments found within the TCRα (TRAV, IMGT nomenclature) and TCRβ (TRBV) gene loci. Although pathogen-specific TCR diversity appears to be of key importance in protective immunity (12, 13), there are examples of TCR bias in antiviral immunity, including Epstein-Barr virus (EBV) (14) and influenza A virus (15). A number of different mechanisms may engender TCR bias (reviewed in refs. 16, 17), including the pMHC-I structural landscape (18–20). Given the importance of understanding immune repertoire selection, mechanisms of viral escape and pMHC-I structure in the design of peptide-based vaccines, we mined the large database of pMHC-I structures to systematically explore the way in which peptides bind to the MHC-I. Surprisingly, we found a large number of MHC-I restricted peptides displaying distinct modes of interaction between residue positions that constrain their conformations. We identified three categories of structural constraints, highlighting the diverse mechanisms by which MHC-oriented peptide residues can simultaneously impact on the conformations of positions important for TCR recognition. Our study provides a conceptual framework for assessing the extent and nature of conformational interdependence that can arise between the residues of MHC-I bound peptides.
Results
pMHC Conformation Is Dependent on Intrapeptide Contacts.
The general view is that MHC-I bound peptide side chains adopt conformations independent of each other to interact with the MHC or serve as potential TCR contact points. However, we noted that the conformation of an endogenous, high-affinity DPα peptide (EEFGRAFSF) bound to HLA-B*4402 (B44) and its closely related allomorphs (3) is characterized by a conserved pattern of intrapeptide side-chain interactions. When in complex with the HLA B44 allomorphs, DPα adopts a constrained conformation within its central region. Namely, the prominently surface-exposed side chain of P5-Arg is supported via interactions with the MHC-orientated, flanking aromatic rings of P3-Phe and P7-Phe (Fig. 1A).
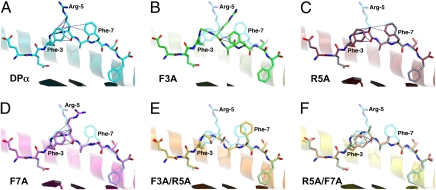
Fig. 1.
Peptide topology for the endogenous DPα and five APLs complexed with HLA-B*4402. In each panel, a cartoon representation is given of the Ag-binding cleft, whereas the bound peptide is presented in stick format. The α2-helix (residues 126–181) has been removed for clarity. In each of the APL panels, the index DPα peptide is superimposed in a semitransparent mode. Vdw interactions between atoms of P5 and other residues of the peptide are shown as blue dashes. (A) HLA-B*4402/DPα (1M6O; cyan carbon atoms). In addition to the interactions displayed, P5-Arg also interacts via its guanidinium head group with the side chain of Gln-155 (removed for clarity). (B) HLA-B*4402/DPα F3A (green carbon atoms). This APL displays an rmsd of 1.80 Å for all peptide atoms with respect to the index peptide and a maximum deviation of 3.27 Å in the side chain of P7-Phe. The side chain of P5-Arg is disordered beyond the Cγ atom. (C) HLA-B*4402/DPα R5A (red carbon atoms). Displayed is an rmsd of 0.24 Å for all peptide atoms with respect to the index epitope and a maximum deviation of 1.15 Å in the side chain of P8-Ser. (D) HLA-B*4402/DPα F7A (magenta). Rmsd for all peptide atoms is 1.06 Å. Maximum deviation is 5.61 Å in the side chain of P5-Arg. Interaction between P5-Arg and Gln-155 is maintained. (E) HLA-B*4402/DPα F3A/R5A (orange). All atom rmsd is 1.09 Å. Maximum deviation is 4.00 Å in the side chain of P5-Ala. (F) HLA-B*4402/DPα R5A/F7A (yellow). All atom rmsd is 0.47 Å. Maximum deviation is 1.59 Å in the side chain of P8-Ser.
We evaluated the degree to which the B44-DPα conformation was dependent on side-chain interactions within DPα itself by solving the pMHC-I structures of five alanine-scan mutants of the peptide (F3A, R5A, F7A, F3AR5A, and R5AF7A) (Fig. 1, Fig. S1, and Table S1). All structures were solved in the same space group, were isomorphous to each other, and the peptides are free from crystal contacts. Thus any conformational changes observed can be attributed to the specific mutation within the peptide. The conformation of the B44 Ag-binding cleft, which constitutes the MHC-I component of the B44-DPα epitope, is conserved between the index B44-DPα structure and each of the five APL complexes (rmsd 0.36–0.61 Å; Fig. S1A). Moreover, alanine substitution of the position P5-Arg (R5A; Fig. 1C) had little effect on the overall peptide conformation when compared to that of the index peptide. In contrast, significant changes in peptide conformation were observed in those variants where the flanking phenylalanines were mutated (Fig. 1 B and D–F). In the B44-DPα-F7A structure the P5-Arg side chain has “flipped” toward the space previously occupied by the P7-Phe aromatic ring (Fig. 1D), whereas in the B44-DPα-F3A structure, the overall peptide conformation had significantly changed with the side chain of P5-Arg becoming disordered (Fig. 1B and Fig. S1B). The loss of intrapeptide interactions between P3-Phe, P5-Arg, and P7-Phe, as observed in the F3A/R5A and R5A/F7A double mutants, also caused the remaining phenylalanine in each case to change conformation. For example, in the B44-DPα-F3A-R5A complex, the aromatic ring of P7-Phe shifted significantly with respect to the index peptide and exhibited greater mobility (Fig. 1E and Fig. S1E). Overall, APL structures displaying the greatest changes in peptide conformation were those with substitutions at P3-Phe enabling the central region of the peptide to shift down toward the base of the antigen binding cleft (maximum deviation in the peptide backbone of up to 2.8 Å; Fig. 1 B and E). Substitution of P7-Phe resulted in less dramatic changes in peptide conformation, whereas the substitution of P5-Arg had little effect. Thus P3-Phe and, to a lesser extent, P7-Phe appear to fix the P5-Arg residue in a constrained position, while also stabilizing their respective conformations. The net consequence of altering these stabilizing interactions is that the structural landscape of the pMHC-I complex is also changed giving a different epitope.
Constrained Peptides and the pMHC-I Database.
To assess the commonality of constrained peptides, we analyzed the pMHC-I entries in the protein structure database (Research Collaboratory for Structural Bioinformatics-Protein Database, RCSB-PDB). As of July 2009, the PDB contained 251 pMHC-I crystal structure entries including 214 binary complexes (Fig. S2A). A total of 103 of the binary pMHC-I entries were excluded from our analyses due to crystal contacts formed by the bound peptides.
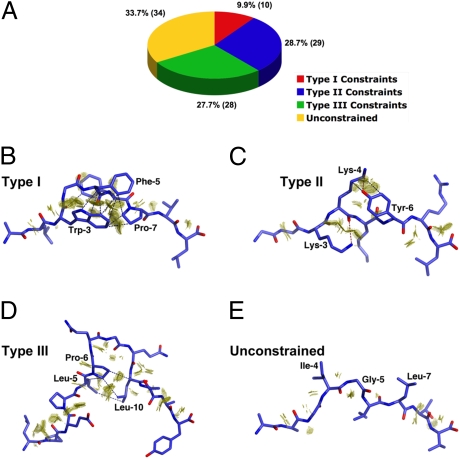
Of the remaining 111 entries all but 3 (1A9B, 3.2 Å; 1E28, 3.0 Å; and 1P1Z, 3.26 Å) have been determined to a resolution higher than 3.0 Å and consist of 87 unique antigenic peptide sequences, bound to 22 different MHC alleles (Table S2). Ten structures were found to have been duplicated at higher resolution and were thus considered redundant. The remaining 101 unique pMHC-I complexes comprised our sample population and were examined for peptide self-constraints. We were able to identify three “constraint” categories (Fig. 2A), each characterized by a distinct set of readily identifiable structural features (Fig. 2 B–D). The three categories (types I, II, and III) are defined below, accompanied by typical examples from the database. A definition of “unconstrained” peptides is also given:
Fig. 2.
MHC class-I restricted epitope self-constraints. (A) Summary of database mining results. Peptide ligands from 101 eligible pMHC-I entries in the PDB were assigned to one of three distinct constraints categories (i) type I, (ii) type II, (iii) type III, or alternatively were designated as being “unconstrained.” (B–E) Examples of “self-constraints” displayed by MHC-I restricted peptides in the PDB database. Peptides are shown in stick format and the interacting vdw surfaces between residues are represented as a discontinuous surface of “contact dots” (dot density, 100 Å−2). Key interactions are further highlighted by blue (vdw) or red (H-bond) dashes. (B) “Type I” constraints observed in the 2.5-Å crystal structure of the p1049 9-mer peptide in complex with HLA-A*0201 (1B0G). (C) “Type II” constraints as observed in the 2.3-Å structure of the K7R mutant of the 8-mer HIV-1 GAG peptide in complex with HLA-B*0801 (1AGE). (D) “Type III” constraints displayed by the 1.6-Å structure of a 13-mer EBV antigen in complex with HLA-B*3501 (1ZHK). (E) The unconstrained 9-mer MART-1 peptide in complex with HLA-A*0201 at 1.9 Å (2GUO).
Unconstrained peptides.
We were surprised during the course of our analysis to find some form of interresidue interaction in nearly all of the peptides examined, including contacts either between main chain atoms of adjacent residues or between the side chain of a given position and the peptide bond atoms immediately either side of it. Although these ubiquitous intrapeptide contacts were discounted during our analysis, we identified a subset of pMHC-I structures (approximately a third of the total analyzed) in which additional interactions were restricted to predominantly van der Waals (vdw) contacts between a side chain at a given position (n) and the main chain atoms of the residues no further than n − 2 and n + 2 (although occasional isolated side chain to side chain interactions were also observed in some of these structures). The positions within these peptides can be considered to act as independent pegs. Peptides displaying these structural characteristics were classed as unconstrained peptides (Fig. 2), an example of which is the structure of a 9-mer peptide derived from MART-1 in complex with HLA-A*0201 [2GUO (21); Fig. 2E]. Within the category of unconstrained peptides are also included pMHC-I structures displaying either (i) more than one distinct backbone conformation, as seen in the B*1501/UbcH6 and B*3508/pp65 complexes [1XR9 (22) and 3BWA (23), respectively], or (ii) significant disorder along their backbones, as in the case of two EBV 11-mer peptides in complex with HLA-B*35 [2FYY (24) and 2NW3 (25)]. The mobility of the peptides observed in these structures is most likely a reflection of an inherent flexibility in their MHC-I-bound states and is thus consistent with the absence of conformational constraints, either self-imposed or MHC-I induced. In summary, 34 “unconstrained” peptides were identified, accounting for 34% of the analyzed pMHC-I entries.
“Type I” constrained peptides.
The HLA-B44-bound DPα peptide represents an archetypal example of a self-constrained peptide (Fig. 1A), characterized by a central solvent exposed residue (n), the side chain of which forms a network of interactions with the side chains of two flanking residues at positions n + 2 and n − 2. Another characteristic example within this category, designated type I, is the HLA-A*0201 restricted p1049 peptide [ALWGFFPVL; 1B0G (26)], where the prominently surface-exposed side chain of P5-Phe is constrained by P3-Trp and P7-Pro (Fig. 2B). This peptide has been shown to elicit a well-defined T-cell response that is dependant on P5-Phe. An analog of the p1049 peptide, designated p1058 (FAPGFFPYL), has also been characterized structurally and biologically [1I7R (27)]: p1058 adopts a conformation equivalent to p1049 within the Ag-binding cleft of HLA-A2 (rmsd <0.55 Å for peptide main chain atoms), the only exception being the side chain of P5-Phe, which has an altered conformation consistent with a loss of type I constraints due to the presence of a P3-proline instead of a tryptophan. Importantly, p1058 has been shown to be antagonistic for p1049/HLA-A2-specific CTL function, with the effect being reversed by a P3W substitution in p1058. In total, 10% of eligible pMHC structures analzyed exhibited type I constraints (Fig. 2A).
“Type II” constrained peptides.
These peptides are also characterized by a solvent accessible position (n), the conformation of which is constrained via interactions with the side chains of two or more side chains. In contrast to type I peptides, however, we noted that these peptides do not display any restrictions on the relative positioning of the residues involved in the constraints. Moreover, in many of these peptides only one of the constraining residues contacts the side chain of n, whereas the other is involved in main chain interactions with n. An example of a peptide classed as type II, is the HLA-B*0801 restricted 8-mer derived from HIV-1 GAG protein p17 [GGKKKYKL; 1AGD (28)], in which the highly solvent exposed side chain of P4-Lys forms vdw and hydrogen bond interactions with P6-Tyr, while its main chain interacts with P3-Lys (Fig. 2C). In summary, 29% of eligible pMHC-I entries exhibited type II constraints.
“Type III” constrained peptides.
Within type III constrained peptides there are at least two flanking residues constraining a central solvent exposed feature (n), consisting of one or more residues itself. However, in contrast to types I and II constraints, these flanking residues interact predominantly with each other, and to a lesser extent, with the backbone of the central constrained feature. No interactions occur with the side chain(s) of n. Although type III constraints are observed in peptides of canonical length, such as the 8-mer H-2Kb restricted HSV glycoprotein B-derived peptide, SSIEFARL [1T0M (29)], they best characterize longer (bulged) pMHC-I peptides. Indeed, 6 of the 8 unique 11–14-mers sampled in our analysis were found to exhibit type III constraints. A 13-mer determinant from EBV (LPEPLPQGQLTAY), for instance, binds to HLA-B*3501 and HLA-B*3508 in a near-identical bulged and rigid conformation (rmsd of 0.625 Å for all peptide atoms), in which the P6-Pro is observed packing against the main chain and side chain of P10-Leu [Fig. 2D; 1ZHK, 1ZHL (30)]. Importantly, although only the backbones of P6-Pro and P10-Leu were observed interacting with the SB27 TCR in the ternary complex structure [2AK4 (18)], substitution of P6-Pro to glycine results in a marked reduction in TCR binding, indicating that maintenance of the conformation of this loop is critical for achieving a sufficient TCR-pMHC-I interaction (18). In summary, a total of 28 pMHC-I entries were assigned to the type III constraints category, representing 28% of the structures analyzed (25% of sampled peptide sequences).
Example Where Loss of Intrapeptide Constraints Resulted in Compromised Antigenicity.
During our analysis we noted several examples where changes in epitope immunogenicity were linked to amino acid substitutions that disrupted intrapeptide contacts. This suggested that targeting residues that stabilize conformations of potential TCR contact residues might represent a unique mechanism for viral escape from CTL-mediated immunity. Variants of the HLA-B*0801 (B8) restricted 8-mer derived from HIV-1 p17 [GGKKKYKL; 1AGD (28)], a type II constrained peptide, represent elegant examples of this mechanism. In addition to the index p17 peptide, the structure of four naturally occurring variants that abrogate recognition by p17-specific CTL have also been solved (28). In the case of the B8p17-K3R variant (1AGB), mutation of the MHC-I-oriented P3-Lys was found to result in subtle but significant changes in peptide conformation. These changes are focused on the predicted TCR contact P4-Lys (all atom rmsd >0.76 Å), which is constrained by interactions with P3-Lys and P6-Tyr in both the index peptide and the two P7-Lys variants (1AGC and 1AGE; Fig. 2C). Importantly, the conformational changes observed in the K3R variant are accompanied by a loss of all stabilizing H-bonds involving P4-Lys, suggesting that the diminished recognition by HLA-B8/p-17-specific CTL for this variant can be attributable to compromised intrapeptide constraints.
Similarly, we previously solved the structure of a H2-Db (Db) restricted influenza peptide derived from the acid polymerase subunit 1 protein (PA224, SSLENFRAYV) that displays type II constraints [1YN6 (19)] similar to those identified for the HIV-1 p17 peptide. The conformation of the PA224 peptide bound to H2-Db is characterized by a pronounced C-terminal arch centered on the critical TCR contact, P7-Arg (Fig. S3 A–C). P7-Arg interacts via its side chain and main chain with the MHC-oriented aromatic rings of P6-Phe and P9-Tyr, respectively. Notably, alanine substitution of either P7-Arg or P6-Phe abrogates DbPA224-specific CTL recognition (19) without influencing pMHC-I stability (Fig. S4). Given the low solvent accessibility of P6-Phe compared to P7-Arg, we proposed that mutations at P6 might alter TCR binding to the pMHC-I indirectly, by influencing the conformation of P7-Arg. To evaluate this, we determined the crystal structure of the H2-Db-PA-F6A complex (Fig. S3 D–F) and compared it to that of the wild-type (WT) epitope (Fig. 3A). We found that whereas the structure of the Ag-binding cleft and backbone configuration of the peptide are very similar in both structures, the DbPA-F6A structure displays a significantly different P7-Arg side chain conformation with respect to the index peptide.
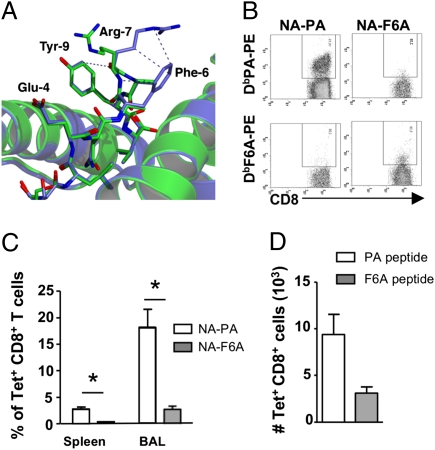
Fig. 3.
Loss of constrained interactions impacts on immunogenicity. (A) Comparison of the H2-Db/PA224 (blue) and H2-Db/PA224-F6A (green) crystal structures reveals that loss of constraints mediated by the aromatic ring of P6-Phe results in a shift of 8.5 Å in the position of the guanidinium head group of P7-Arg. Refined coordinates of the Ag-binding cleft are presented in cartoon format, whereas peptide atoms are shown as sticks. Intrapeptide vdw contacts involving P7-Arg are represented by blue dashes. Coordinates were superimposed using the Cα atoms of residues 1–180 of the heavy chain. (B) Lack of cross-reactivity between DbPA and DbPA-F6A epitopes. Naïve mice were infected with either NA-PA or NA-F6A PR8 influenza A viruses and lymphocytes from spleen and BAL were isolated 9 days later and costained with corresponding DbPA224 or DbPA-F6A –PE tetramer and anti-CD8a-FITC. Representative staining is shown. (C) Proportion of tetramer+CD8+ T cells from the spleen and BAL of mice infected with NA-PA (open bars) or NA-F6A (solid bars) PR8 viruses. Data are mean ± SD for groups of 6 mice. *, P < 0.001. (D) The F6A substitution within PA224 does not affect antigen presentation. Naïve mice were immunized s.c. with either PA224 (open bars) or PA-F6A (solid bars) peptide in CFA. Seven days later splenocytes were stained with either DbPA224 or DbPA-F6A –PE tetramer and anti-CD8a-PercPCy5.5. Shown is the absolute number of tetramer+CD8+ T cells (mean ± SD, n = 5 mice).
To evaluate the functional consequence of the F6A mutation on DbPA224-specific CTL recognition, we used reverse genetics to generate recombinant influenza A viruses where PA224 (31) was disrupted in its normal location and replaced by inserting the wild-type PA224 or PA-F6A peptides into the neuraminidase (NA) stalk of A/Puerto Rico/34/1 (32), referred to as either the NA-PA and NA-F6A viruses, respectively. Both the wild-type NA-PA and the mutant NA-F6A replicated to equivalent titers in vitro (1.58 ± 1.9 × 108 pfu and 1.09 ± 0.8 × 109 pfu/mL, respectively), suggesting these alterations had no impact on viral fitness.
As expected, a robust DbPA224-specific CTL response was observed in mice infected with the NA-PA virus and the responding T cells did not bind the DbPA-F6A tetramer (Fig. 3B). Whereas substantial numbers of DbPA-specific CTLs were found in the bronchoaveolar lavage (BAL) (17.9%) and spleen (2.76%) of mice infected with the NA-PA virus (Fig. 3C), the DbPA-F6A CTL counts (BAL, 2%, P < 0.01; spleen, 0.25%, P < 0.01) from those given NA-F6A (Fig. 3C) were about 10-fold lower. In addition, there was no evidence of DbPA-F6A cross-reactivity for the WT DbPA224 tetramer (Fig. 3B), suggesting that these pMHC-I epitopes induce nonoverlapping TCR repertoires. Analysis of TRBV usage demonstrated that DbPA-F6A-specific CTL preferred usage of the TRBV13-1 gene segment (Table S3) rather than the TRBV29 gene segment used by DbPA224-specific CTL (33). Furthermore, the DbPA-F6A-specific repertoire consisted of a limited number of different TRBV13-1 sequences (Table S4 and Table 1) that was more limited in diversity when compared to the TRBV29+ DbPA224-specific CTL repertoire (Table 1). Intranasal infection with either recombinant virus induced equivalent CTL responses to both the endogenous DbNP366 (Fig. S5A) and DbPB1-F262 (Fig. S5B) epitopes supporting the notion that both viral fitness and antigen load in vivo was equivalent for these two viruses. To exclude the possibility that the diminished F6A-specific CTL response observed in NA-F6A infected mice was due to a defect in processing/presentation, mice were immunized with the PA or F6A peptides emulsified in CFA and the spleen was harvested 7 days later for analysis (Fig. 3D). Similar to our previous findings, DbPA224-specific CTLs were detected in mice immunized with PA224 peptide whereas immunization with the F6A peptide induced a significantly lower percentage and number of DbF6A+CD8+ T cells (Fig. 3D). Overall, this suggests that mutation of the P6-Phe disrupts the constrained nature of the P7-Arg, resulting in a pMHC-I- conformation that is both poorly immunogenic and noncross reactive for the WT DbPA epitope.
Table 1.
Summary of CDR3β TCR repertoire diversity for DbPA224- and DbF6A-specific CTLs
| DbPA224 | DbF6A | |
| Mice analyzed | 12 | 3 |
| TCRs sequenced | 1630 | 73 |
| Predominant Vβ | BV29 | BV13-1 |
| Predominant Jβ region | 2S6, 1S1 | 2S2 |
| Predominant CDR3β length (aa) | 6 | 9 |
| Different clonotypes (aa) | 241 | 11 |
| Repeated clonotypes (aa) | 0 | 1 |
| Clonotypes per mouse (aa) | 20.6 ± 3.8 | 4 ± 1 |
Discussion
The general view has been that immunogenic peptides bind MHC-I in a linear and extended manner, with the peptide residues pointing “up” or “down” and acting independently of each other. Our analysis of peptide conformations within the pMHC-I structural database indicates that this is not necessarily the case. We estimate that >50% of pMHC-I epitopes exhibit self-constraints, in which the conformation and/or positioning of one or more amino acids in the peptide is influenced by the residues flanking it. We defined three types of constrained interactions (types I, II, and III): Types I and II constraints were generally represented by pMHC-I structures that involved presentation of peptides with shorter lengths (8–10 aa), whereas type III constraints were more apparent for longer (≥10 aa) peptides. We formally demonstrated the interdependence of side-chain conformations within constrained peptides by examining the conformations of a self-peptide bound to HLA-B*4402 and an influenza peptide bound to H2-Db. In each case, removal of an MHC-orientated residue specifically impacted on the conformation of a flanking, upward-pointing residue that is available for direct TCR recognition. Moreover, a number of examples of constrained peptides in the pMHC-I database exhibited similar conformational sensitivities to mutations within these constrained regions of the peptide.
There was no consistent correlation between a given MHC allele and the imposed constraints within the bound peptide. Furthermore, we found no particular amino acid biases that were universally characteristic for any given category of constrained peptide, although proline residues (present in 48% of the peptide sequences sampled) were overrepresented within the type I classification (71%). The fact that prolines apply significant torsional limitations on the peptide backbone makes them an obvious candidate for imposing self-constraints. The clearest example to date of proline constraints in an MHC-I restricted peptide whose structure has been solved is the 9-mer IRAAPPPLF derived from the signal sequence of cathepsin A (34). The frequency of prolines in immunogenic peptides is likely to reflect that certain MHC-I alleles, notably the HLA B7 supertype, exhibit specificity for a P2-Pro anchoring motif. Moreover, proline-containing peptides are less readily digestible by the aminopeptidase trimming enzymes in the endoplasmic reticulum (35).
Although the type of constraint is dependent on the peptide sequence, the structural characteristics of the MHC-I molecule will also be a determining factor. For example, the HLA-B35 restricted 11-mer peptide (HPVGEADYFEY) complexed to HLA-B*3501 [2FYY (24)] is partially disordered, but it is well ordered and displays type III constraints when bound to HLA-B*3508 [2FZ3 (24)]. As such, the same, potentially immunogenic peptide can be presented to the available T-cell repertoire as topographically distinct structures. Thus, MHC-I polymorphism ensures that a range of peptides are presented by imposing MHC allele-specific intrapeptide constraints, with the consequence that a diversity of immunogenic structures is presented to the immune system of any one individual (potentially six different MHC-I alleles). The net result is that the “antigenic surface” that can be recognized throughout any given population is greatly enhanced (36).
Our hypothesis that pMHC-I complexes can be modified via differing constraints on peptide binding offers enhanced and unexpected possibilities for viral escape that do not necessarily reflect any direct change in the residues recognized directly by the TCR. An example comes from our analysis of the influenza A virus DbPA224 F6A mutant. Despite being considered an anchor residue, replacing P6-Phe with alanine results in loss of DbPA224-specific CTL reactivity (19), likely due to greater mobility of the P7-Arg. Furthermore, in addition to the loss of DbPA224-specific CTL reactivity, the mutant pMHC-I epitope was less immunogenic following both infection and peptide immunization. The fact that there was very little cross-reactivity between CTLs specific for either the index DbPA224 or the DbPA-F6A variant suggested that different T-cell repertoires were used to recognize each epitope. Analysis of T-cell diversity demonstrated that the DbPA-F6A repertoire draws on a very limiting spectrum of TCRβ chains, in contrast to the very diverse array of T-cell clones that respond to the DbPA224 epitope (33). As such, the limited antigenicity of the variant DbPA-F6A pMHC-I complex appears to be attributable to the increased mobility of the P7-Arg, thereby making this epitope a more difficult target for the available naïve T-cell repertoire.
In conclusion, rather than taking the position that each amino acid within an antigenic peptide functions effectively as an independent entity, our study has defined patterns of intrapeptide interaction that constrain overall structure, and thus antigenicity, for a large number of pMHC-I complexes. This idea of constrained peptides has implications for both peptide prediction programs and for attributing specific roles to particular amino acid residues, as judged by the functional analysis of alanine-scanning mutations. The importance of these constrained interactions is reinforced when considering that “nonobvious” peptide variations can result in the total or partial compromise of immunogenicity. As we move toward the design and improvement of vaccine strategies, an appreciation of such constraints is key for the assessment and selection of peptide targets that function to limit viral escape from TCR/pMHC-I mediated immune control.
Materials and Methods
Database Mining.
A list of deposited pMHC-I structures was obtained by searching the RCSB-PDB and subsequently consolidated by cross-referencing against entries listed in the MGT/3Dstructure-DB. Coordinates and, where available, electron density maps were downloaded from the PDB using COOT (37). Interresidue atomic contacts were identified by applying a cut-off distance of 4.0 Å. Intrapeptide interactions were further evaluated using the COOT implementation of REDUCE and PROBE, thus permitting visualization of the presence and extent of vdw interactions between peptide positions. In the case of structures containing multiple peptide copies within the asymmetric unit, intrapeptide interactions were accepted as legitimate when observed in all copies.
Synthetic Peptides.
Peptides were purchased from Auspep. In general, peptides were supplied as a lyophilized powder at a purity greater than 80%. Peptides were stored at 4 °C and solubilized at 100 mg/mL in DMSO before use.
Preparation of Recombinant MHC-Peptide Complexes.
Recombinant HLA-B*4402 heavy chain (residues 1–274), H-2Db heavy chain (residues 1–274), murine and human β2m were expressed separately as inclusion bodies in Escherichia coli as previously described (3, 19). Correctly folded pMHC complexes were purified by anion exchange and size exclusion chromatography. Protein was buffer exchanged into Tris-HCl (pH 8.0), 150 mM NaCl and concentrated to 3–9 mg/mL for crystallization trials.
Crystallization and Data Collection.
HLA-B*4402/DPα variant complexes: `own at 20 °C using the hanging drop vapor diffusion method. In each case, the protein was mixed with an equal volume of the reservoir buffer (12–30% PEG 4000, 0.1 M trisodium citrate dihydrate (pH 5.2–5.7), 0.2 M ammonium acetate) and incubated over a 1-mL reservoir. Before data collection, crystals were flash-frozen using 10% glycerol as a cryoprotectant. All data sets were collected at 100 K both in-house on an R-Axis IV++ detector and at the BioCars beamline (Advanced Photon Source) using a Quantum 4 CCD detector.
H-2Db/PA224 F6A complex: The structure of H-2Db in complex with the PA224 peptide from influenza A virus acid polymerase has been previously reported (19) (1YN6). Crystals of H-2Db/PA224-F6A were grown by hanging drop vapor diffusion at 20 °C, by mixing an equal volume of protein (9 mg/mL) and reservoir solution, containing 100 mM sodium citrate, pH 5.6, 0.2 M ammonium acetate, and 25% PEG 4000. Before data collection, crystals were cryoprotected against reservoir solution containing 10% glycerol (vol/vol) and flash-frozen at 100 K.
Structure Determination and Refinement.
HLA-B*4402/DPα variant complexes: The five B44DPα variant structures were determined by molecular replacement using previously published coordinates of HLA-B*4402 as a search model [PDB code 1M6O (3)]. All structures were initially subjected to simulated annealing and coordinate/ADP refinement followed by iterative cycles of model building and refinement using COOT and programs within CCP4 (38). In the case of the F3A/R5A and R5A/F7A structures, the final ADP refinement in PHENIX, followed by the REFMAC5 implementation of TLS parameterization. Validation was carried out using programs within CCP4i.
H-2Db/PA224 F6A complex: The structure was solved by molecular replacement using previously deposited H-2Db coordinates as a starting model (3CC5). Structure refinement and validation was carried out in COOT, MOLPROBITY, and programs within CCP4i. A summary of data collection, refinement, and validation statistics for all structures are presented in Table S1.
Mice, Viruses, and Viral Infection.
Female C57BL/6J (B6, H-2b) mice were bred in the animal facility of the Department of Microbiology and Immunology at the University of Melbourne and were held under specific pathogen-free conditions. Naïve mice were infected either intranasally (i.n.) with 102 PFU or intraperitonally (i.p.) with 107 PFU of recombinant A/PR8/34 (H1N1) influenza viruses as previously described (32). The recombinant influenza viruses were generated using an eight-plasmid reverse genetics system (31). A single mutation resulting in alanine substitution in P6-Phe was introduced by PCR into the plasmid encoding acidic polymerase (PA224). Recombinant PR8 viruses expressing either the PA224 (SSLENFRAYV) or PA-F6A (SSLENARAYV) peptide inserted into the NA stalk at amino acid position 42 also contained single amino acid mutations in the native PA224 epitope (N5Q) to disrupt presentation of the endogenous wild-type epitope. Viruses are referred to as NA-PA and NA-F6A. Virus stocks were grown in the allantoic cavity of embryonated hen's eggs at embryonic day 10 and were stored in aliquot at −80 °C. Virus titers were determined as plaque-forming units on monolayers of Madin Darby canine kidney cells.
Tissue Sampling, Cell Preparation, Tetramer Staining, and Intracellular Cytokine Staining.
Lymphocytes were isolated from the spleen, BAL, and mediastinal lymph node (MLN) from mice day 9 after primary PR8 infection and either stained with tetramers or used for intracellular cytokine staining as previously described (32).
Single Cell TCR Sequence Analysis.
The TRBV sequences were obtained from single cell sorted DbPA224+CD8+ and DbPA-F6A+CD8+ CTL using single cell RT-PCR as previously described (19, 33).
Supplementary Material
Acknowledgments
We thank the staff at BioCARS and the Australian Synchrotron Research Program for assistance. A.T. and M.A.D. are National Health and Medical Research Council (NHMRC) Peter Doherty fellows. C.G. is a Marie Curie postdoctoral fellow. S.J.T. is a Pfizer senior research fellow. A.W.P. is a NHMRC senior research fellow. J.R. is an Australian Research Council Federation fellow. A.I.W. and L.K.E are NHMRC C. J. Martin fellows. This work was supported by the NHMRC, ARC, and the Juvenile Diabetes Research Foundation International.
Footnotes
References
- 1.Bjorkman PJ, et al. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 2.Burrows SR, Rossjohn J, McCluskey J. Have we cut ourselves too short in mapping CTL epitopes? Trends Immunol. 2006;27:11–16. doi: 10.1016/j.it.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Macdonald WA, et al. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J Exp Med. 2003;198:679–691. doi: 10.1084/jem.20030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rötzschke O, et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990;348:252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- 5.Burrows JM, et al. The impact of HLA-B micropolymorphism outside primary peptide anchor pockets on the CTL response to CMV. Eur J Immunol. 2007;37:946–953. doi: 10.1002/eji.200636588. [DOI] [PubMed] [Google Scholar]
- 6.Yewdell JW, Haeryfar SM. Understanding presentation of viral antigens to CD8+ T cells in vivo: The key to rational vaccine design. Annu Rev Immunol. 2005;23:651–682. doi: 10.1146/annurev.immunol.23.021704.115702. [DOI] [PubMed] [Google Scholar]
- 7.Bertoletti A, et al. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 1994;369:407–410. doi: 10.1038/369407a0. [DOI] [PubMed] [Google Scholar]
- 8.Phillips RE, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 9.de Campos-Lima PO, et al. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science. 1993;260:98–100. doi: 10.1126/science.7682013. [DOI] [PubMed] [Google Scholar]
- 10.Butler NS, et al. Structural and biological basis of CTL escape in coronavirus-infected mice. J Immunol. 2008;180:3926–3937. doi: 10.4049/jimmunol.180.6.3926. [DOI] [PubMed] [Google Scholar]
- 11.Tenzer S, et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10:636–646. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- 12.Price GE, Ou R, Jiang H, Huang L, Moskophidis D. Viral escape by selection of cytotoxic T cell-resistant variants in influenza A virus pneumonia. J Exp Med. 2000;191:1853–1867. doi: 10.1084/jem.191.11.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 14.Argaet VP, et al. Dominant selection of an invariant T cell antigen receptor in response to persistent infection by Epstein-Barr virus. J Exp Med. 1994;180:2335–2340. doi: 10.1084/jem.180.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss PA, et al. Extensive conservation of alpha and beta chains of the human T-cell antigen receptor recognizing HLA-A2 and influenza A matrix peptide. Proc Natl Acad Sci USA. 1991;88:8987–8990. doi: 10.1073/pnas.88.20.8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Curr Opin Immunol. 2008;20:119–125. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 2006;6:883–894. doi: 10.1038/nri1977. [DOI] [PubMed] [Google Scholar]
- 18.Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol. 2005;6:1114–1122. doi: 10.1038/ni1257. [DOI] [PubMed] [Google Scholar]
- 19.Turner SJ, et al. Lack of prominent peptide-major histocompatibility complex features limits repertoire diversity in virus-specific CD8+ T cell populations. Nat Immunol. 2005;6:382–389. doi: 10.1038/ni1175. [DOI] [PubMed] [Google Scholar]
- 20.Miles JJ, Brennan SR, Burrows SR. T cell receptor bias in humans. Curr Immunol Rev. 2009;5:10–21. [Google Scholar]
- 21.Borbulevych OY, et al. Structures of MART-126/27-35 Peptide/HLA-A2 complexes reveal a remarkable disconnect between antigen structural homology and T cell recognition. J Mol Biol. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Røder G, et al. Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype. Acta Crystallogr D Biol Crystallogr. 2006;62:1300–1310. doi: 10.1107/S0907444906027636. [DOI] [PubMed] [Google Scholar]
- 23.Wynn KK, et al. Impact of clonal competition for peptide-MHC complexes on the CD8+ T-cell repertoire selection in a persistent viral infection. Blood. 2008;111:4283–4292. doi: 10.1182/blood-2007-11-122622. [DOI] [PubMed] [Google Scholar]
- 24.Miles JJ, et al. TCR alpha genes direct MHC restriction in the potent human T cell response to a class I-bound viral epitope. J Immunol. 2006;177:6804–6814. doi: 10.4049/jimmunol.177.10.6804. [DOI] [PubMed] [Google Scholar]
- 25.Tynan FE, et al. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 26.Zhao R, Loftus DJ, Appella E, Collins EJ. Structural evidence of T cell xeno-reactivity in the absence of molecular mimicry. J Exp Med. 1999;189:359–370. doi: 10.1084/jem.189.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buslepp J, et al. T cell activity correlates with oligomeric peptide-major histocompatibility complex binding on T cell surface. J Biol Chem. 2001;276:47320–47328. doi: 10.1074/jbc.M109231200. [DOI] [PubMed] [Google Scholar]
- 28.Reid SW, et al. Antagonist HIV-1 Gag peptides induce structural changes in HLA B8. J Exp Med. 1996;184:2279–2286. doi: 10.1084/jem.184.6.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb AI, et al. The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: Evidence for a conformational switch that governs T cell repertoire selection and viral resistance. J Immunol. 2004;173:402–409. doi: 10.4049/jimmunol.173.1.402. [DOI] [PubMed] [Google Scholar]
- 30.Tynan FE, et al. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J Biol Chem. 2005;280:23900–23909. doi: 10.1074/jbc.M503060200. [DOI] [PubMed] [Google Scholar]
- 31.Webby RJ, et al. Protection and compensation in the influenza virus-specific CD8+ T cell response. Proc Natl Acad Sci USA. 2003;100:7235–7240. doi: 10.1073/pnas.1232449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Gruta NL, et al. A virus-specific CD8+ T cell immunodominance hierarchy determined by antigen dose and precursor frequencies. Proc Natl Acad Sci USA. 2006;103:994–999. doi: 10.1073/pnas.0510429103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner SJ, Diaz G, Cross R, Doherty PC. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity. 2003;18:549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 34.Kumar P, et al. Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens. J Biol Chem. 2009;284:29784–29797. doi: 10.1074/jbc.M109.038497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25:795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tynan FE, et al. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J Exp Med. 2005;202:1249–1260. doi: 10.1084/jem.20050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.