Abstract
Bacterial histidine kinases transduce extracellular signals into the cytoplasm. Most stimuli are chemically undefined; therefore, despite intensive study, signal recognition mechanisms remain mysterious. We exploit the fact that quorum-sensing signals are known molecules to identify mutants in the Vibrio cholerae quorum-sensing receptor CqsS that display altered responses to natural and synthetic ligands. Using this chemical-genetics approach, we assign particular amino acids of the CqsS sensor to particular roles in recognition of the native ligand, CAI-1 (S-3 hydroxytridecan-4-one) as well as ligand analogues. Amino acids W104 and S107 dictate receptor preference for the carbon-3 moiety. Residues F162 and C170 specify ligand head size and tail length, respectively. By combining mutations, we can build CqsS receptors responsive to ligand analogues altered at both the head and tail. We suggest that rationally designed ligands can be employed to study, and ultimately to control, histidine kinase activity.
Keywords: agonist, antagonist, quorum-sensing, two-component protein
Quorum sensing is a cell-cell communication process that allows bacteria to synchronize the gene expression of the population in response to changes in cell population density and species composition. Quorum sensing relies on the production, detection, and response to extracellular signaling molecules called autoinducers. In the human pathogen Vibrio cholerae, quorum sensing is mediated by two parallel phosphorelay systems (1). One system, which is the focus of this report, uses the membrane-bound sensor CqsS to detect the autoinducer called CAI-1 (S-3 hydroxytridecan-4-one) (Fig. 1A), which is produced by the aminotransferase enzyme CqsA (2, 3). CqsS also detects another natural ligand called amino-CAI-1 [(S)-3-aminotridecan-4-one] (Fig. 1A). CqsS is a two-component histidine sensor kinase that possesses both kinase and phosphatase activities. In environments in which CAI-1 concentration is below the threshold for detection, such as at low cell density, CqsS functions as a kinase. CqsS shuttles phosphate to the response regulator, LuxO, and phospho-LuxO promotes transcription of the genes encoding four regulatory small RNAs, Qrr1–4 (1, 4). Qrr1–4 inhibit translation of the master quorum-sensing regulator, HapR. Therefore, at low cell density, HapR is not produced (1, 4). At high cell density, when CAI-1 accumulates to an appreciable level, the autokinase activity of CqsS is inhibited. As a consequence, phosphate flow in the circuit is reversed, which leads to dephosphorylation and deactivation of LuxO. Transcription of qrr1–4 terminates, and HapR is thus produced. Production of HapR leads to initiation of the high cell density quorum-sensing gene expression program (1, 5–7).
Fig. 1.
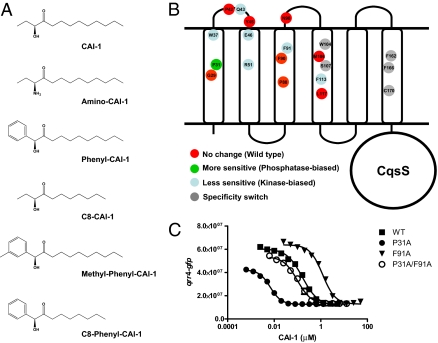
Natural and synthetic CqsS ligands. CqsS mutants and their corresponding phenotypes. (A) CAI-1 and amino-CAI-1 are naturally produced and detected by V. cholerae. P-CAI-1, C8-CAI-1, methyl-P-CAI-1, and C8-P-CAI-1 are synthetic compounds. (B) CqsS is predicted to contain six transmembrane helices (Results). Mutant receptors studied in this work are color coded according to their phenotypes: green, more sensitive to CAI-1 (lower EC50 than WT CqsS); blue, less sensitive to CAI-1 (higher EC50 than WT CqsS); red, no change in sensitivity to CAI-1; gray, specificity switch phenotype. (C) CAI-1 dose–responses of WT and representative kinase-biased and phosphatase-biased CqsS mutants. The qrr4-gfp activity at the specified concentrations of CAI-1 is shown for WT CqsS (▪), CqsS P31A (•), CqsS F91A (▾), and CqsS P31A/F91A double mutant (○). Data were fit with a variable-slope sigmoidal dose–response curve.
Autoinducers, such as CAI-1, represent examples of only a handful of ligands that have been defined for histidine kinases (3, 8–12). Most often, two-component sensory systems respond to environmental information that is poorly understood at the molecular level, such as changes in osmolarity, pH, or redox potential. Furthermore, like CqsS, many two-component sensors have complicated membrane-spanning domains, which has made structural analyses particularly difficult. For these reasons, analyses of ligand-receptor interactions using structural approaches have been limited to a few histidine kinases with well-defined periplasmic sensing domains and known ligands (13–23). Thus, despite intensive effort, it remains unclear how ligand binding affects signaling activity in this important family of receptors. Quorum-sensing systems, because they rely on known molecules as ligands, offer us the opportunity to address questions about sensor kinase-ligand binding. In pairwise fashion, the ligands can be altered by synthetic chemistry and the receptors can be altered by mutagenesis. Here, as a proof of principle, we use this combined chemical-genetic strategy to examine signal recognition in the V. cholerae CqsS quorum-sensing receptor. Particularly interesting in the case of CqsS is that there are two natural ligands, CAI-1 and amino-CAI-1, which differ only by the moiety at the carbon-3 (C3) position (hydroxyl vs. amino) (2, 3). In addition, simple synthetic routes to CAI-1, amino-CAI-1, and related molecules carrying modifications at different positions exist (2, 3) (Fig. 1A). The availability of this set of molecules, the ease of genetic manipulation in V. cholerae, and its robust in vivo readout of quorum-sensing signaling provide us a means to probe how a multipass transmembrane histidine sensor kinase identifies its ligand.
Our analysis shows that conserved amino acids clustered in the first three CqsS transmembrane helices are obligatory for ligand binding and signal transduction. Residues W104 and S107 in the predicted fourth transmembrane helix function to specify the moiety at C3, and thus enable CqsS to discriminate between CAI-1 and amino-CAI-1. Using two CAI-1 analogues, phenyl-CAI-1 (P-CAI-1) containing an enlarged head group and C8-CAI-1 [(S)-3-hydroxyundecan-4-one] containing a shortened tail, we assign residues F162 and C170 the roles of restricting the ligand head group size and tail length, respectively. Based on our ability to parse the CqsS transmembrane domain into distinct regions responsible for sensitivity, specificity for the C3 position, specificity for the head group, and specificity for tail length, we propose a model for the interaction of CAI-1 and CqsS.
Results
CqsS Amino Acid Residues Important for Ligand Detection and Signal Transduction.
By screening random libraries for active CqsS-LacZ and CqsS-PhoA fusion proteins, we found that a six-helix membrane topology prediction (24–28) closely matches the experimentally ascertained functional CqsS fusions (SI Appendix S1). We used this topology model (Fig. 1B) and a mutagenesis approach to investigate the interaction between CqsS and CAI-1 and between CqsS and amino-CAI-1. A plasmid (pDH345) carrying WT cqsS was introduced into a V. cholerae mutant that cannot produce or detect CAI-1. The plasmid-borne cqsS gene allows a response to exogenously provided CAI-1. Plasmid pDH345 also contains a qrr4-gfp transcriptional reporter fusion (29). Thus, CAI-1 response can be monitored by measuring alterations in GFP production. The reporter system works as follows. At low CAI-1 levels, CqsS functions as a kinase, leading to qrr4 transcription and GFP production (Fig. 1C, ▪). At high CAI-1 concentrations, CqsS functions as a phosphatase and qrr4 is not transcribed; thus, GFP production is correspondingly low (Fig. 1C, ▪). Using this strain, we determined the dose–response profiles and the calculated EC50s for CAI-1 and amino-CAI-1 (SI Appendix S2). Consistent with previous findings (2), amino-CAI-1 is slightly more active (i.e., has a lower EC50) than CAI-1.
We determined which regions of CqsS are important for CAI-1 detection and signaling using the above assay as a readout of CqsS function. We aligned multiple CqsS homologues from different bacterial species and identified 17 conserved residues in the transmembrane region of CqsS (SI Appendix S2). We hypothesized that these conserved residues could be important for CAI-1 binding, for signal transduction, or for both activities. Each of these 17 conserved residues was replaced by alanine, and the resulting mutants were assayed for their CAI-1 responses. Many of the mutations, notably those in residues predicted to reside in the periplasm, did not produce significant changes in CAI-1 response (SI Appendix S2). We identified mutants exhibiting decreased sensitivity to CAI-1, mean-ing that more CAI-1 is required to convert the mutant CqsS from a kinase to a phosphatase than is required for WT CqsS (Fig. 1 B and C and SI Appendix S2). These mutations are alanine replacements at W37, Q43, E46, R51, F91, F113, and F162. We also identified one mutant, P31A, with increased sensitivity to CAI-1, meaning that less CAI-1 is required to convert the mutant CqsS from a kinase to a phosphatase than is required for WT CqsS (Fig. 1 B and C and SI Appendix S2). Except for Q43, all the residues that altered CAI-1 sensitivity are located in transmembrane helices of CqsS. All EC50 values are provided in SI Appendix S2. CqsS with alanine substitutions at W37, R51, and F162 do not respond to CAI-1 even at the highest concentration tested (i.e., 20 μM). Thus, we were unable to assign their EC50 values. When selected mutations (e.g., P31A and F91A; Fig. 1C, ○) were combined, the phenotypes of the double mutants were additive, including when the original mutations conferred increased and decreased responses to CAI-1. These data suggest that each residue acts independently to confer its CAI-1 response phenotype.
Recently, it was shown that signaling parameters such as autoinducer dissociation constants (Koffs) can be inferred from dose–response curves using mathematical modeling (30). We automated this earlier approach and performed a series of experiments on WT and mutant CqsS receptors. The automated method, the data, and a full discussion of the parameters we obtained can be found in SI Appendix S3.
Amino Acid Residues That Determine the Moiety at C3 of CAI-1.
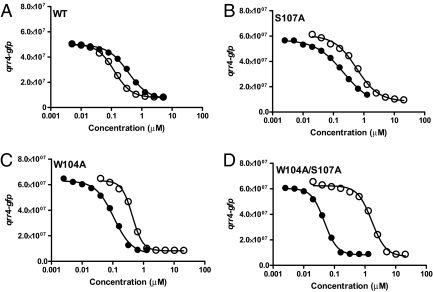
We found that alanine substitution at S107, although decreasing the sensitivity to amino-CAI-1, enables CqsS to detect CAI-1 with greater sensitivity than WT CqsS (Fig. 2 A and B). Thus, unlike WT CqsS, CqsS S107A prefers CAI-1 over amino-CAI-1. To identify additional CqsS mutants possessing altered preference phenotypes, a library of random mutations in the transmembrane domain of CqsS was constructed and screened for those CqsS mutants that detect CAI-1 more avidly than amino-CAI-1. Three CqsS mutants, all with changes at W104 (W104G, W104L, and W104A) were identified (results for W104A are shown in Fig. 2C). Mutations in residues neighboring W104 and S107 did not affect ligand preference. Residues W104 and S107 both reside in the predicted fourth transmembrane helix of CqsS (Fig. 1B). The W104A/S107A double mutant was constructed, and it displays even higher sensitivity to CAI-1 and more drastically reduced sensitivity to amino-CAI-1 than either isogenic single mutant (Fig. 2D). We interpret these results to mean that W104 and S107 contribute additively to the WT CqsS receptor’s ability to differentiate between CAI-1 and amino-CAI-1 and, potentially, other CAI-1 analogues (we return to this point later).
Fig. 2.
Ligand responses of specificity-altered CqsS mutants. GFP production from the qrr4-gfp transcriptional fusion at the specified concentrations of CAI-1 (•) and amino-CAI-1 (○) is shown for WT CqsS (A), CqsS S107A (B), CqsS W104A (C), and CqsS W104A/S107A (D).
To understand the roles played by W104 and S107 in distinguishing between moieties at C3, additional mutations at these two residues were constructed. We engineered CqsS S107C, S107T, W104F, and W104Y. Our rationale for altering Ser to Cys and Thr and for altering Trp to Phe and Tyr was to change the structural features of the two native residues minimally. Analogous to the original specificity-altered CqsS S107A and CqsS W104A mutants, CqsS S107C, CqsS W104F, and CqsS W104Y prefer CAI-1 to amino-CAI-1. By contrast, CqsS S107T retains the WT CqsS preference for the two autoinducers. We do note, however, that CqsS S107T has decreased sensitivity to both molecules (SI Appendix S4). These data suggest that W104 is likely essential for CqsS to prefer amino-CAI-1 over CAI-1, whereas the discriminatory role of S107 relies on the presence of a terminal hydroxyl group on the amino acid side chain that is maintained in the CqsS S107T mutant.
CqsS Mutants That Use P-CAI-1 as an Agonist.
These analyses define amino acids W104 and S107 as essential for proper interaction of CqsS with the moiety at the C3 group of the ligand. Moreover, this initial experiment also suggests that we can use a combined mutagenesis and chemical approach to pair critical amino acid residues in receptors to corresponding chemical features in ligands. Again using CqsS as our example, we explored the amino acid requirements for other structural features (e.g., head group, tail length) of the ligand. We focus on two different synthetic CAI-1 analogues, P-CAI-1 and C8-CAI-1, because they possess modifications at the two extreme ends of CAI-1 (Fig. 1A). P-CAI-1 carries a bulky modification at the CAI-1 head, and C8-CAI-1 lacks the terminal ethyl group in the hydrophobic tail.
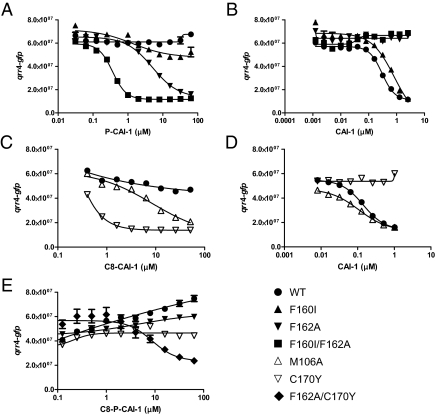
We first describe our analyses using P-CAI-1 to explore the requirements for and limitations on head group size. For this series of experiments, we again use the qrr4-gfp reporter assay. P-CAI-1 is a competitive antagonist of CAI-1; increasing concentrations of P-CAI-1 increase the CAI-1 EC50 for repression of qrr4 transcription, whereas increasing concentrations of CAI-1 alleviate P-CAI-1 antagonism (SI Appendix S5). Because P-CAI-1 is a competitive antagonist, it must bind to the identical site in CqsS as does CAI-1. However, unlike CAI-1, P-CAI-1 must not make the interactions necessary to inhibit CqsS kinase activity. We reasoned that a CqsS mutant using P-CAI-1 as an agonist rather than an antagonist must have acquired unique interactions between P-CAI-1 and CqsS that promote kinase inhibition. To identify such a mutant, we screened our random cqsS mutant library and the set of CqsS mutants with conserved residues altered to alanine for those capable of repressing qrr4-gfp expression in response to P-CAI-1. We identified two classes of mutants. One class of CqsS mutants (M106A and F160I) uses P-CAI-1 as an agonist, and these mutants display no alteration in sensitivity for CAI-1 (F160I is shown as an example in Fig. 3 A and B and SI Appendix S6A). The second class (F162A and F166L) uses P-CAI-1 as an agonist, and these mutants are severely impaired for response to CAI-1 (F162A is shown as an example in Fig. 3 A and B and SI Appendix S6A). M106 is located in the predicted fourth transmembrane helix, and F160, F162, and F166 are located in the predicted sixth transmembrane helix (Fig. 1B).
Fig. 3.
Responses of CqsS mutants specifying CAI-1 head group and tail length. GFP production from the qrr4-gfp transcriptional fusion at the specified concentrations of P-CAI-1 (A), CAI-1 (B and D), C8-CAI-1 (C), and C8-P-CAI-1 (E) is shown for WT CqsS (•), CqsS F160I (▴), CqsS F162A (▾), CqsS F160I/F162A (▪), CqsS M106A (▵), CqsS C170Y (▿), and CqsS F162A/C170Y (◆).
We tested the effects of combining mutations from each class on CAI-1 and P-CAI-1 detection. Combination of F162A with M106A, F160I, or F166L results in double mutants incapable of response to CAI-1 (F160I/F162A is shown as example in Fig. 3B and SI Appendix S6B). Thus, mutation at F162A is epistatic to the other mutations for CAI-1 detection, suggesting that in WT CqsS, F162 plays an absolutely required role in recognition of the native CAI-1 head group. Our findings suggest that M106, F160, F162, and F166 function to prevent the interaction of P-CAI-1 with WT CqsS. F162 is additionally essential for proper interaction with the CAI-1 head group.
CqsS Mutants That Use C8-CAI-1 as an Agonist.
In a series of experiments analogous to those discussed above, we investigated which amino acids in CqsS are important for interaction with the CAI-1 C10 tail. We know that increases or decreases in CAI-1 chain length cause substantial decreases in activity (3). In the present work, we use C8-CAI-1 (Fig. 1A), which has only weak agonist activity (WT) (Fig. 3C). We screened the random CqsS mutant library and the set of mutants with altered conserved residues for those mutants for which C8-CAI-1 acts as a strong agonist. As in the above P-CAI-1 experiment, we identified two classes of CqsS mutants. Mutants in the first class respond to C8-CAI-1 and have WT sensitivity to CAI-1 (W104A, M106A, L117A, and M125A; M106A is shown as an example in Fig. 3 C and D and SI Appendix S7A). By contrast, a single mutant, CqsS C170Y, was identified that, although agonized by C8-CAI-1, has lost the ability to respond to CAI-1 (Fig. 3 C and D). The C170Y mutant also does not respond to CAI-1 carrying a C9 or a C11 tail. The C170Y mutation is in the predicted sixth transmembrane helix. We engineered additional mutations at C170 to probe its function further. Similar to WT CqsS, CqsS C170A prefers CAI-1 over C8-CAI-1. By contrast, similar to the CqsS C170Y mutant, the C170F mutant prefers C8-CAI-1 over CAI-1 (SI Appendix S7B). Thus, a large residue located at position 170 promotes interaction with CAI-1 carrying a shortened tail. We wondered if CAI-1 had been converted into an antagonist in CqsS mutants that are agonized by C8-CAI-1 but not by CAI-1. Our examination of CqsS C170Y shows that, indeed, increasing CAI-1 concentration increases the EC50 of C8-CAI-1. CAI-1 antagonism can be counteracted by increasing C8-CAI-1 levels (SI Appendix S7C). These competition data suggest that CAI-1 and C8-CAI-1 bind to the same site in CqsS; furthermore, residue C170 specifies the ligand tail length to be exactly 10 carbons.
Combining the F162A and C170Y Mutations Allows CqsS to Detect a Unique Molecule.
The F162A mutation that permits a response to P-CAI-1 and the C170Y mutation that allows a response to C8-CAI-1 are both in the predicted sixth transmembrane helix of CqsS. Moreover, F162 and C170 are predicted to reside on the same face of the helix (SI Appendix S1). One plausible model that could account for these data is that CAI-1 interacts, possibly directly, with these residues and the F162A and C170Y mutations alter the configuration of the receptor to accommodate P-CAI-1 and C8-CAI-1, respectively. If so, this would mean that F162 must specify head group size independent of C170 specification of tail length and vice versa. To test this idea, we combined the F162A and C170Y mutations and examined if the resulting double-mutant receptor could respond to a hybrid molecule C8-phenyl-CAI-1 (C8-P-CAI-1) (Fig. 1A). Indeed, although neither the WT, the F162A single mutant, nor the C170Y single mutant responds to C8-P-CAI-1, the CqsS F162A/C170Y double mutant readily detects C8-P-CAI-1 and fully represses qrr4-gfp transcription in response (Fig. 3E).
Discussion
In V. cholerae, the CAI-1 autoinducer is synthesized by CqsA and is detected by the membrane-bound receptor CqsS. Homologues of cqsA and cqsS are almost exclusively found in Vibrio species (1, 31), underpinning the notion that the CAI-1 autoinducer is used for inter-Vibrio communication. Consistent with this idea, many Vibrio species produce an activity that induces the quorum-sensing response in a V. cholerae CAI-1 reporter strain (31). However, the V. cholerae response to cell-free fluids from other Vibrio species is often not as robust as that to cell-free fluids containing endogenously produced CAI-1. This finding suggests that other Vibrio species produce less CAI-1 than V. cholerae or that the molecules produced by other species are not identical to V. cholerae CAI-1 (31). We favor the latter idea, because polymorphisms exist among different CqsA-CqsS pairs (see below). In addition, synthetic CAI-1 analogues carrying slight modifications elicit submaximal quorum-sensing responses in V. cholerae (2, 3), indicating that V. cholerae CqsS has evolved to detect specifically the CAI-1 and amino-CAI-1 molecules. Based on these findings, we wondered whether we could identify specificity determinants in the CqsS receptor that facilitate discrimination among similar molecules. Indeed, we found that the first half of the CqsS transmembrane region is involved in signal detection and sensitivity, amino acids W104 and S107 recognize the moiety at C3, C170 determines the precise C10 tail length, and F162 and F166 limit the ligand head group size.
Among the transmembrane regions of CqsS receptors, the most notable conservation exists in the N-terminal half, indicating that this region is responsible for a function shared by all CqsS receptors. This finding, coupled with our mutagenesis results, implies that the conserved residues in this region interact with chemical moieties that all CAI-1-type molecules have in common. By contrast, the less conserved C-terminal halves of the CqsS transmembrane domains likely carry out functions that vary among Vibrios species, such as interacting with distinct moieties in CAI-1-type molecules. We identified two mutants (W104A and S107A) that convert the CqsS preference from amino-CAI-1 to CAI-1 (Fig. 2), revealing these residues as specificity determinants for the C3 functional group. Moreover, because S107 is completely conserved and W104 is highly conserved among different CqsS proteins, we presume that a blend of hydroxyl- and amino-CAI-1 molecules could exist in nature and that CqsS receptors are capable of responding distinctly to these mixtures. Specifically, CqsS S107/W104 receptors will prefer amino-CAI-1 over CAI-1, whereas CqsS S107/W104X receptors (X = any amino acid other than Trp) will greatly prefer CAI-1 over amino-CAI-1.
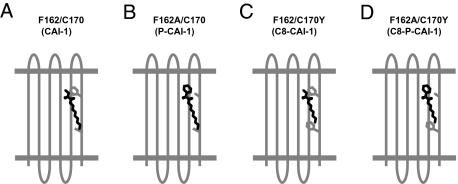
We propose a ligand–receptor interaction model in which the polar head group and the hydrophobic tail of CAI-1 interact, possibly directly, with F162/F166 and C170, respectively, in WT CqsS (Fig. 4A). Mutations in these residues render CqsS responsive only to molecules possessing complementary modifications (Fig. 4 B–D). Thus, we suggest that F162/F166 and C170 act in concert as gatekeepers to impose structural constraints on the alterations in CAI-1 allowable for agonism. This ligand–receptor interaction model also provides us insight into CqsS antagonism. In WT CqsS, P-CAI-1 likely acts as an antagonist because of a nontolerated steric clash between the phenylalanine residues at F162 and F166 in the predicted sixth transmembrane domain and the bulky head group of the synthetic probe molecule (Figs. 3 and 4 and SI Appendix S5 and S6). Similarly, CAI-1 acts as an antagonist for the CqsS C170Y mutant, probably because the bulky Y170 residue compromises a required interaction with the C10 tail (Figs. 3 and 4 and SI Appendix S7). Both F162 and C170 map to the final transmembrane helix, which is directly upstream of the histidine kinase domain of CqsS. Additional mutations in the final transmembrane helix, such as F160I and F166L, also alter the sensitivity of CqsS for P-CAI-1. We speculate that F160, which lies on the opposite face of the helix from F162, F166, and C170, acts as an allosteric determinant to maintain the pocket size of the CAI-1 binding site. Indeed, double mutants carrying F160I display increased sensitivities to P-CAI-1 and its bulkier analogues, such as methyl-P-CAI-1 (Fig. 1A). CqsS lacks a HAMP domain, which is important in signal transduction in many histidine kinases (32–38). Thus, interactions between CAI-1 and the final transmembrane domain suggest that this transmembrane domain serves not only in ligand binding but as a critical regulatory region for CqsS kinase activity.
Fig. 4.
Model for CqsS-CAI-1 ligand-receptor interactions. (A) In WT CqsS, the polar head group and the hydrophobic tail of CAI-1 interact, perhaps directly, with F162 and C170, respectively. These interactions are required to inhibit CqsS kinase activity. (B) Small amino acid residue (e.g., cysteine, alanine) is required at position 162 to permit productive interaction of CqsS with P-CAI-1. (C) Bulky residue (e.g., tyrosine, phenylalanine) is required at position 170 to promote interaction between CqsS and C8-CAI-1. (D) Combination of a small residue at position 162 and a bulky residue at position 170 confers an interaction between CqsS and C8-P-CAI-1.
At a biophysical level, there is no single conformation of the binding pocket of a receptor such as CqsS. Even within the simplest two-state model for signal transduction (30), the receptor must adopt two configurations [kinase active (on) and kinase inactive (off)], and if the addition of CAI-1 is to result in kinase inactivation, CAI-1 must have a higher affinity for the binding pocket in the inactive state of protein. More generally, the effect of a given ligand on a given CqsS variant depends on the ligand-binding pocket interaction in both the kinase-active and kinase-inactive states. Ligands that strongly prefer the inactive state, such as CAI-1 and amino-CAI-1 for WT CqsS, are agonists. Ligands that strongly prefer the active state, such as P-CAI-1, are antagonists. Because only one ligand can occupy the binding pocket at a time, a ligand that is a weak agonist by itself may act functionally as an antagonist by competitively occupying the binding site and displacing a stronger agonist. All these cases can be simply understood by considering the free-energy difference between the active and inactive states of the receptor in the presence of one or more ligands (Materials and Methods and SI Appendix S3). Moreover, the relevant parameters, including the ligand-Koffs in both the kinase-active and -inactive states, can often be extracted from dose–response curves (SI Appendix S3). The gatekeeper model described above specifically concerns the effects of our CqsS mutations on the Koff for various agonists in the kinase-inactive state of CqsS. Studies of ligand-receptor pairs, coupled with quantitative analysis of dose–response curves, can therefore reveal properties of the ligand-binding pocket interaction not only in the receptor state critical for signaling by agonists (the inactive state in the case of CqsS) but in the alternative state. Such studies may prove helpful in developing stronger antagonists to specific WT receptors both in prokaryotes and eukaryotes.
Using synthetic CAI-1 analogues, we identified CqsS mutants with broadened specificity (i.e., they respond to CAI-1 and to other CAI-1 analogues) as well as mutants that display altered specificity (i.e., they respond to a CAI-1 analogue but not to CAI-1). Previous analyses of two-component receptor specificity for ligands have focused on Escherichia coli Tar (a chemoreceptor that transduces signal to the CheA histidine kinase) and Staphylococcus aureus AgrC receptors that, like CqsS, recognize defined molecules. In Tar (responds to aspartate) and AgrC (responds to a cyclic peptide), as in CqsS, point mutants with broadened specificity were easily identified. These and our results suggest that there are multiple routes to make a “sloppier” receptor. For example, in the case of S. aureus AgrC-1, alteration of residue Ile-171 to Lys enables the receptor to respond to its cognate peptide ligand, AIP-1, as well as to additional otherwise inactive AIP peptide variants (39). Similarly, mutation of M106, L117, or M125 allowed CqsS to recognize an increased number of molecules without affecting sensitivity to CAI-1. We propose that M106, L117, and M125 of CqsS play an analogous role to I171 in AgrC in restricting ligand recognition. In terms of altered, rather than broadened, specificity mutants, to achieve a change in Tar specificity, 4 to 12 simultaneous point mutations were required (40). In AgrC, either multiple mutations or exchanges of significant portions of transmembrane domains were required (39, 41, 42). This is not the case in CqsS, because we can change any one of five amino acids (W104, S107, F162, F166, and C170) and alter the receptor specificity. Furthermore, we can map these amino acid changes to specific regions of the ligand, suggesting that each amino acid residue plays a discrete role in defining a particular feature of the ligand. We confirmed that the C170 and F162/F166 gatekeeper residues act independently. Each CqsS mutant harboring a change in only one of the gatekeeper residues responds only to the analogue that carries the complementary modification. Consistent with this, each single gatekeeper mutant does not respond to the analogue possessing the noncomplementary change, nor does each single gatekeeper mutant respond to the hybrid molecule C8-P-CAI-1. Only when we combine the two gatekeeper mutations do we build a receptor that responds to the doubly modified ligand molecule.
We note that residue C170, which is absolutely required for response to CAI-1, is only present in V. cholerae CqsS, whereas CqsS receptors from other Vibrio species carry a phenylalanine in the corresponding position (SI Appendix S2). Based on this observation, we predict that other Vibrio species cannot use CAI-1 as the CqsS ligand. Rather, shorter CAI-1-like molecules, perhaps C8-CAI-1, are more likely their quorum-sensing signals. More importantly, if this is the case, CAI-1 from V. cholerae will likely function as an antagonist for other CqsS receptors; thus, CAI-1 potentially interferes with other Vibrio species’ ability to detect their cognate CAI-1-like molecules (Fig. 3 and SI Appendix S7C). In mixed populations of V. cholerae and other Vibrio species, interspecies communication may depend on the amount of and affinity of CAI-1 and other (shorter) CAI-1-like molecules. Interestingly, in an analogous scenario, some homoserine lactones (HSLs) with long hydrocarbon chains act as antagonists for LuxR-type receptors that naturally detect HSLs possessing short hydrocarbon chains [e.g., 3OC12-HSL antagonizes C4-HSL in RhlR (43); C10-HSL, C12-HSL, and C14-HSL antagonize C6-HSL in CviR (44); several molecules antagonize 3OC8-HSL reception by TraR (45) and 3OC6-HSL detection by LuxR (46)].
Beyond the CqsS receptors, polymorphisms also exist in the sequences encoding the CqsA synthases. Analysis of Vibrio genome sequences shows that cqsS sequences are more related to one another when their corresponding cqsA sequences are similar (SI Appendix S8). Coevolution of cqsS and cqsA likely maintains ligand-receptor fidelity in each species; at the same time, it allows interspecies signal interference through ligand antagonism mechanisms like those described above. This phenomenon appears to be generally applicable to quorum-sensing systems ranging from the canonical HSL system of cytoplasmic LuxR-type receptors, to the peptide quorum-sensing systems in Gram-positive bacteria (e.g., the Agr system in S. aureus and the Com system in Streptococcus pneumoniae) (47–49), to the CAI-1-CqsS system studied here.
One advantage of expanding the repertoire of available ligands by including synthetic molecules in studies such as the one presented here is that they offer exquisite controllability. So-called “orthogonal chemical genetics” in which ligand-receptor pairs are iteratively altered by turns has been successfully used to define ligand-receptor, protein-protein, and enzyme-substrate interactions in a variety of eukaryotic signal-transduction systems, including G protein-coupled receptors and protein kinases (50, 51). Surprisingly, this powerful approach, to our knowledge, has not been routinely adapted for studying prokaryotic two-component signal-transduction systems. The present work, together with the elegant studies on the Agr system by Muir, Novick, and coworkers (39, 41, 52), suggests that rationally designed ligands can be exploited to study and ultimately to control two-component histidine kinase activity.
Materials and Methods
Strains; plasmids; and molecular biological, chemical, and analytical methods used in this study can be found in SI Appendix S9. DNA manipulations were performed using standard methods as described (53).
Supplementary Material
Acknowledgments
We thank Douglas Higgins and Michelle Gonzales for strain construction. This work was supported by the Howard Hughes Medical Institute, National Institutes of Health Grant 5R01GM065859 (to B.L.B.), National Institutes of Health Grant 5R01AI054442 (to B.L.B.), National Science Foundation Grant MCB-0343821 (to B.L.B.), National Institutes of Health Postdoctoral Fellowship GM082061 (to W.L.N.), National Science Foundation Grant PHY-065017 (to N.S.W.), and Defense Advanced Research Projects Agency Grant HR0011-05-1-0057 (to T.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001392107/DCSupplemental.
References
- 1.Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/s0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 2.Kelly RC, et al. The Vibrio cholerae quorum-sensing autoinducer CAI-1: Analysis of the biosynthetic enzyme CqsA. Nat Chem Biol. 2009;5:891–895. doi: 10.1038/nchembio.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 4.Lenz DH, et al. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 6.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–104. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: Sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 9.Milton DL, et al. The LuxM homologue VanM from Vibrio anguillarum directs the synthesis of N-(3-hydroxyhexanoyl)homoserine lactone and N-hexanoylhomoserine lactone. J Bacteriol. 2001;183:3537–3547. doi: 10.1128/JB.183.12.3537-3547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415:545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 11.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Håvarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung J, Hendrickson WA. Structural analysis of ligand stimulation of the histidine kinase NarX. Structure. 2009;17:190–201. doi: 10.1016/j.str.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung J, Hendrickson WA. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J Biol Chem. 2008;283:30256–30265. doi: 10.1074/jbc.M805253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 16.Neiditch MB, et al. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou YF, et al. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J Mol Biol. 2008;383:49–61. doi: 10.1016/j.jmb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Sevvana M, et al. A ligand-induced switch in the periplasmic domain of sensor histidine kinase CitA. J Mol Biol. 2008;377:512–523. doi: 10.1016/j.jmb.2008.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Krämer J, et al. Citrate sensing by the C4-dicarboxylate/citrate sensor kinase DcuS of Escherichia coli: Binding site and conversion of DcuS to a C4-dicarboxylate- or citrate-specific sensor. J Bacteriol. 2007;189:4290–4298. doi: 10.1128/JB.00168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 21.Cho US, et al. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 22.Pappalardo L, et al. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J Biol Chem. 2003;278:39185–39188. doi: 10.1074/jbc.C300344200. [DOI] [PubMed] [Google Scholar]
- 23.Yeh JI, et al. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol. 1996;262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 24.Claros MG, von Heijne G. TopPred II: An improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 25.Tusnády GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 26.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 27.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann K, Stoffel W. TMbase—A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. Available at: www.ch.embnet.org/software/TMPRED_form.html. [Google Scholar]
- 29.Svenningsen SL, Waters CM, Bassler BL. A negative feedback loop involving small RNAs accelerates Vibrio cholerae’s transition out of quorum-sensing mode. Genes Dev. 2008;22:226–238. doi: 10.1101/gad.1629908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swem LR, Swem DL, Wingreen NS, Bassler BL. Deducing receptor signaling parameters from in vivo analysis: LuxN/AI-1 quorum sensing in Vibrio harveyi. Cell. 2008;134:461–473. doi: 10.1016/j.cell.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henke JM, Bassler BL. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appleman JA, Stewart V. Mutational analysis of a conserved signal-transducing element: The HAMP linker of the Escherichia coli nitrate sensor NarX. J Bacteriol. 2003;185:89–97. doi: 10.1128/JB.185.1.89-97.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aravind L, Ponting CP. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 34.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 35.Kishii R, Falzon L, Yoshida T, Kobayashi H, Inouye M. Structural and functional studies of the HAMP domain of EnvZ, an osmosensing transmembrane histidine kinase in Escherichia coli. J Biol Chem. 2007;282:26401–26408. doi: 10.1074/jbc.M701342200. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73:801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, Inouye M. The HAMP linker in histidine kinase dimeric receptors is critical for symmetric transmembrane signal transduction. J Biol Chem. 2004;279:48152–48158. doi: 10.1074/jbc.M401024200. [DOI] [PubMed] [Google Scholar]
- 38.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 39.Geisinger E, Muir TW, Novick RP. agr receptor mutants reveal distinct modes of inhibition by staphylococcal autoinducing peptides. Proc Natl Acad Sci USA. 2009;106:1216–1221. doi: 10.1073/pnas.0807760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derr P, Boder E, Goulian M. Changing the specificity of a bacterial chemoreceptor. J Mol Biol. 2006;355:923–932. doi: 10.1016/j.jmb.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 41.Geisinger E, George EA, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J Mol Biol. 2008;381:300–309. doi: 10.1016/j.jmb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 43.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swem LR, et al. A quorum-sensing antagonist targets both membrane-bound and cytoplasmic receptors and controls bacterial pathogenicity. Mol Cell. 2009;35:143–153. doi: 10.1016/j.molcel.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, et al. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaefer AL, Hanzelka BL, Eberhard A, Greenberg EP. Quorum sensing in Vibrio fischeri: Probing autoinducer-LuxR interactions with autoinducer analogs. J Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 48.Whatmore AM, Barcus VA, Dowson CG. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol. 1999;181:3144–3154. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnsborg O, Kristiansen PE, Blomqvist T, Håvarstein LS. A hydrophobic patch in the competence-stimulating Peptide, a pneumococcal competence pheromone, is essential for specificity and biological activity. J Bacteriol. 2006;188:1744–1749. doi: 10.1128/JB.188.5.1744-1749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bishop A, et al. Unnatural ligands for engineered proteins: New tools for chemical genetics. Annu Rev Biophys Biomol Struct. 2000;29:577–606. doi: 10.1146/annurev.biophys.29.1.577. [DOI] [PubMed] [Google Scholar]
- 51.Shokat K, Velleca M. Novel chemical genetic approaches to the discovery of signal transduction inhibitors. Drug Discov Today. 2002;7:872–879. doi: 10.1016/s1359-6446(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 52.Wright JS, 3rd, Lyon GJ, George EA, Muir TW, Novick RP. Hydrophobic interactions drive ligand-receptor recognition for activation and inhibition of staphy-lococcal quorum sensing. Proc Natl Acad Sci USA. 2004;101:16168–16173. doi: 10.1073/pnas.0404039101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.