Abstract
Previous research has suggested that human herpesvirus-6 (HHV-6) may integrate into host cell chromosomes and be vertically transmitted in the germ line, but the evidence—primarily fluorescence in situ hybridization (FISH)—is indirect. We sought, first, to definitively test these two hypotheses. Peripheral blood mononuclear cells (PBMCs) were isolated from families in which several members, including at least one parent and child, had unusually high copy numbers of HHV-6 DNA per milliliter of blood. FISH confirmed that HHV-6 DNA colocalized with telomeric regions of one allele on chromosomes 17p13.3, 18q23, and 22q13.3, and that the integration site was identical among members of the same family. Integration of the HHV-6 genome into TTAGGG telomere repeats was confirmed by additional methods and sequencing of the integration site. Partial sequencing of the viral genome identified the same integrated HHV-6A strain within members of families, confirming vertical transmission of the viral genome. We next asked whether HHV-6A infection of naïve cell lines could lead to integration. Following infection of naïve Jjhan and HEK-293 cell lines by HHV-6, the virus integrated into telomeres. Reactivation of integrated HHV-6A virus from individuals’ PBMCs as well as cell lines was successfully accomplished by compounds known to induce latent herpesvirus replication. Finally, no circular episomal forms were detected even by PCR. Taken together, the data suggest that HHV-6 is unique among human herpesviruses: it specifically and efficiently integrates into telomeres of chromosomes during latency rather than forming episomes, and the integrated viral genome is capable of producing virions.
Keywords: HHV-6, episome, latency
Human herpesvirus-6 (HHV-6) was first discovered in patients infected with HIV who also suffered from lymphoproliferative disorders (1), although it now is recognized to infect the great majority of humans. HHV-6 is a betaherpesvirus related to the human cytomegalovirus, and there are two distinct subgroups, HHV-6A and HHV-6B (2, 3, 5). More than 90% of children experience HHV-6B infection early in life, and this subtype is the primary cause of exanthem subitum (4, 6).
Following primary infection, both variants of HHV-6 remain in a persistent/latent state for the life of the host (7, 8). However, the virus may reactivate in immunocompetent and, more often, in immunosuppressed hosts. Reactivation of HHV-6 has been associated with seizures, encephalitis, and graft rejection (5, 9–13). HHV-6 can infect CD4+ lymphocytes and may also be a cofactor in AIDS progression, by contributing to the loss of CD4+ cells (14, 15). HHV-6 also has been associated with multiple sclerosis; however, the interpretation of these findings remains uncertain (16–18).
Of the two variants, HHV-6A is more neurovirulent, as is evident by the increased concentration of viral DNA present in the plaques of patients’ brains with MS and its ability to establish latency in oligodendrocytes (5, 16, 17).
Other human herpesviruses achieve latency by persisting as a circular episome in the nucleus. However, some studies using FISH analysis have suggested that both HHV-6A and B can integrate into human chromosomes (19–25) and may be vertically transmitted in the germ line (26). FISH assay, however, cannot distinguish noncovalent linkage from integration, establish the specific integration site, or determine whether the integrated virus can reactivate and produce infectious virions. Our study addressed those questions.
Results
Patients Studied.
Previous research has identified occasional patients with high copy numbers of HHV-6 DNA and FISH results suggesting chromosome integrated HHV-6 (19–25). Often the same findings occur in both parents and children, suggesting vertical transmission in the germline. We studied members of four families in which more than one family member, including a parent and at least one child, had roughly 1 million copies of HHV-6 per milliliter of peripheral blood (Table 1). Several individuals were suffering from neurological symptoms, whereas others were asymptomatic.
Table 1.
Patients from four independent families with chromosome integrated HHV-6
| Family number | Age, sex | Subject | Disease stage | HHV-6 qPCR,* copies per mL | HHV-6 subtype† | Chromosome HHV-6 FISH | Percent sequence identity to HHV-6A (U1102) |
|
| U94 | Direct repeat | |||||||
| 1 | 58, M | Father | Asymptomatic | 629,000 | A | 18q23 | 98% | 98% |
| 1 | 54, F | Mother | PCR negative | Negative | Negative | n/a | n/a | n/a |
| 1 | 24, M | Sibling-1 | Asymptomatic | 1,400,000 | A | 18q23 | 98% | Not done |
| 1 | 22, F | Sibling-2 | CNS dysfunction, hypersomnia | 1,700,000 | A | 18q23 | 98% | Not done |
| 1 | 12, M | Sibling-3 | CNS dysfunction, ataxia | 1,600,000 | A | 18q23 | Not done | Not done |
| 2 | 80, F | Mother | Mild dementia | 625,000 | A | 17p13.3 | 98% | Not done |
| 2 | 45, F | Sibling-1 | CNS dysfunction, fatigue | 4,100,000 | A | 17p13.3 | 98% | 98% |
| 3 | 76, M | Father | Asymptomatic | 2,000,000 | B | 22q13.3 | Not done | Not done |
| 3 | 61, F | Mother | PCR negative | Negative | Negative | n/a | n/a | n/a |
| 3 | 34, M | Sibling-1 | CNS dysfunction, fatigue | 2,000,000 | B | 22q13.3 | Not done | Not done |
| 4 | 62, F | Mother | Asymptomatic | 4,000,000 | B | Not done | Not done | Not done |
| 4 | 36, M | Sibling-1 | Asymptomatic | 4,500,000 | B | Not done | Not done | Not done |
| 4 | 29, F | Sibling-2 | CNS dysfunction, fatigue, ataxia | 4,200,000 | B | Not done | Not done | Not done |
*qPCR on whole blood completed by ViraCor Laboratories (Lee's Summit, MO).
†Subtypes were determined using PCR with subtype-specific primers.
To determine whether HHV-6A or subtype B is present in these patients, sequences of amplified ORF U94 and part of the direct repeat (DR) were sequenced. Viral sequences were identical among members of two families and shared 98% sequence identity with HHV-6A (strain U1102) and only 95% with HHV-6B (Z29) (Table 1 and Figs. S1 and S2).
FISH Analysis of Chromosomal Integration Sites.
To investigate the possibility that HHV-6 had integrated into the family members’ chromosomes, we first performed fluorescence in situ hybridization (FISH) in T cell cultures derived from peripheral blood. The experiments were performed in two independent laboratories (University of Minnesota, Minneapolis, and Children's Cancer Research Institute, Vienna). Each laboratory was blind to knowledge of the individual and family membership from which each specimen had been obtained. In each experiment, HHV-6-specific fluorescence was detected in association with the telomeric regions of chromosomes (Table 1 and Fig. S3). The specific chromosome was identified by cohybridization with viral and chromosome-specific probes. HHV-6-specific FISH signal was detected in chromosomes 17p13.3, 18q23, and 22q13.3, respectively, in the three families studied with FISH. Furthermore, HHV-6 and telomere FISH signals overlapped (Fig. S3).
Specific Integration of HHV-6 into Human Telomeres.
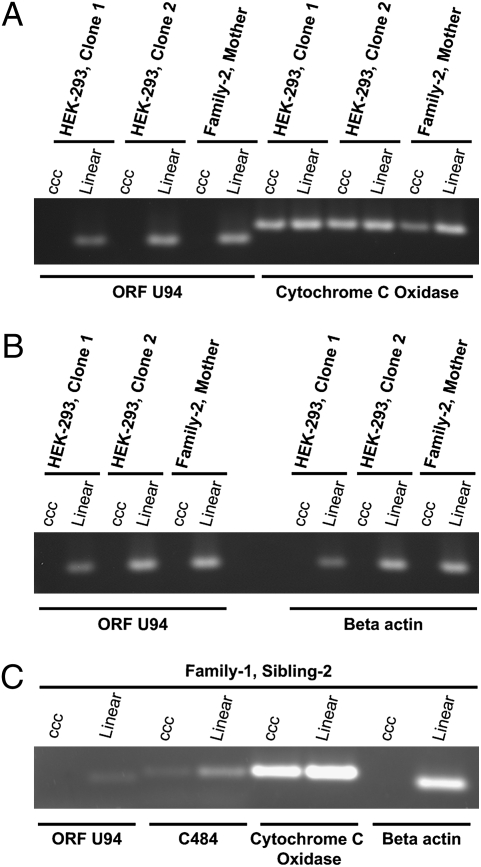
To investigate whether the FISH method truly detects integration of HHV-6 into the human genome, rather than an association of the telomere with episomal viral DNA, we analyzed T cells from Family-1 with the method of Gardella et al. (27). This method uses a vertical agarose gel capable of distinguishing cellular genomic DNA from covalently closed circular DNA (episomes) and from replicating linear viral DNA. Southern hybridization with HHV-6 cosmid probe (Fig. 1A) confirmed the association of the viral genome with cellular DNA located in the loading well of these gels. No circular episomal and unit length linear viral DNA was detected in these experiments.
Fig. 1.
Vertical agarose gel technique identifies HHV-6 present in the host genomic fraction of Family-1 T cells—1 million T cells isolated from Family-1 members, control uninfected PBMCs, and HHV-6 positive Katata cell line (HHV-6B integrated Burkitt's lymphoma cell line) (37). Cells were loaded on a vertical agarose gel and analyzed for episomal, linear, or integrated DNA by the method of Gardella et al. (21). (A Left) Southern hybridization with HHV-6A probe. (A Right) Blot was stripped and hybridized with mitochondrion oligonucleotide probe (41). (B) T cells of family members immortalized with H. saimiri strain C484 were subjected to Gardella gel analysis and hybridized with HHV-6 and H. saimiri probes.
To further validate the Gardella gel results, T cells from three members of Family-1 were immortalized using Herpesvirus saimiri (HVS) strain C484. Southern hybridization with HHV-6 detected signal within the genomic fraction (loading well) of the Gardella gel. Hybridization of the same Southern blot with HVS probe detected both episomal circular and replicating linear HVS DNA, confirming proper release of herpesvirus DNA in the gel (Fig. 1B).
Determination of the Chromosomal Integration Site.
Because the HHV-6A genome encodes a perfect TTAGGG telomere repeat array at the right end direct repeat (DRR) and an imperfect TTAGGG repeat at the end of the left end direct repeat (DRL), we established a working hypothesis that HHV-6A integrates into telomeres via homologous recombination.
To amplify the putative viral–chromosomal DNA junction, we created a primer pair homologous to the DRL and DRR of the viral genome, as well as a primer to the subtelomere of chromosome 17p (Fig. 2A). The subtelomere is the chromosomal region immediately adjacent to the TTAGGG telomere repeat array. DNA from Family-2 was analyzed in these experiments because FISH had identified the integration of HHV-6 into chromosome 17p (Fig. S3) in this family, and the sequence of telomere–subtelomere junction of chromosome 17p is known (28). Amplification with the primer pair designed from DRR and 17p subtelomere successfully amplified the viral–cellular junction as determined by co-Southern hybridization with HHV-6, telomere, and chromosome 17p oligonucleotide probes (Fig. 2B). Furthermore, the absence of amplification with a primer derived from DRL confirmed the integration of HHV-6 into human subtelomere, which is mediated through the perfect TTAGGG found at the end of DRR. Cloning and sequencing of the predominant 1.5-kb amplicon from the DRR-derived primer confirmed the integration of HHV-6 within the telomere of chromosome 17p in the cells of this family (Fig. 2C). The integration site contained five TTAGGG repeats, and integration resulted in the loss of 79 nucleotides from the far right end of the viral genome (GenBank accession no. GU784872).
Fig. 2.
Chromosome 17p subtelomere-specific PCR from DNA of members of Family-2. (A) The putative HHV-6-chromosome 17p-HHV-6A junction. Arrows indicate position of primers (drawing is not to scale) derived from the direct repeat left (DRL) and right (DRR) of the viral genome (Unique long = UL) and to chromosome 17p subtelomere (28). (B) Genomic DNA subjected to amplification from Family-2 Mother and Sibling, HHV-6A-infected and uninfected Jjhan cells. PCR products were separated by electrophoresis and analyzed by Southern blotting using 32P-labeled oligonucleotide probes as indicated at the bottom of each panel. Cohybridization of a 1.5-kb fragment with all three probes suggests HHV-6A chromosome joining with viral DRR telomere repeat. (C) The predominant 1.5-kb amplicon from Family-2/Mother was cloned (n = 3) and sequenced. A GenBank homology search confirmed the presence of chromosome subtelomere 17p sequence (upper) joined with TTAGGG telomere repeats (bold and underlined), and HHV-6 DRR sequence (lower).
In Vitro Integration of HHV-6A into Human T Cell Line Jjhan and Human Embryonic Kidney-293 (HEK-293) Cells.
Surprisingly, the chromosome 17p subtelomere–DRR primer pair amplified DNA fragments from DNA isolated from HHV-6A lytically infected Jjhan cells (Fig. 2B), suggesting that integration takes place during productive infection. To determine the frequency with which new infection of naïve cell lines with HHV-6A leads to chromosomal integration, and whether this is the sole means by which HHV-6A achieves latency, we conducted three in vitro experiments.
First, we evaluated whether HHV-6A strain U1102 can integrate into telomeres of the T cell line Jjhan. These cells are routinely used to propagate HHV-6A, yet we observed that despite supporting lytic infection, Jjhan cells often were not lysed and that many cells survived after the peak of productive infection. We hypothesized that in some of the infected cells, rather than productive infection leading to lysis, the virus had achieved latency through integration. To examine this hypothesis, we prepared DNA from cells at the peak of CPE, containing 103 infectious units/mL virus. We then amplified the putative viral genome–telomere junction using subtelomere-based primers (11q, 17p, and 18q) (28) and a primer derived from near the right end of the DRR. After cloning and sequencing several PCR products (GenBank accession nos. GU784872 and GU784873), the HHV-6A genome was found to be covalently linked with all chromosomes tested (Fig. S4). Thus, the result of this experiment was consistent with the possibility that shortly after infection in this cell line, HHV-6A can integrate into the subtelomeric region of the chromosome.
Second, to monitor the progression and spread of infection in these Jjhan cells, we constructed an HHV-6A recombinant virus (HHV-6AGFP) carrying the green fluorescent protein (GFP) (construction described in Fig. S5A). To accomplish this, we constructed a plasmid in which the GFP expression cassette flanked two 2-kb fragments of ORF U53 and ORF U54 cloned from HHV-6A strain U1102. Then we infected several human monolayer cell lines and screened for those that supported lytic replication and in which transfection of the plasmid was efficient. The most promising cell line was HEK-293 cells (29). Preliminary experiments indicated that HEK-293 cells produce infectious virus at low cell density/confluency. To generate HHV-6AGFP recombinants, a HEK-293 monolayer was then transfected with the ORF U53-GFP-ORF U54 plasmid. The following day, cells were seeded at ∼10% confluency and infected with 103 infectious units of HHV-6A (U1102). After 6 days, we observed the characteristic cytopathic effect (ballooning, refractile giant cells). The virus titer was determined to be 103 infectious units.
Third, the human T cell line Jjhan was cultured in 96-well plates and each well was infected with ∼1 infectious unit of the potential recombinant virus. In one culture, fluorescence microscopy showed a dramatic increase in the number of green fluorescent cells from 7 to 37 days postinfection (Fig. S5B). We observed only a few very bright fluorescent large multinucleated cells, which were presumably producing infectious virus. However, the majority of cells in the well displayed dimmer GFP expression. The distribution of GFP expressing cells is indicated by the FACS analysis (Fig. S5C), showing bright fluorescence in very few cells and overall GFP expression in 60% of the cells. To evaluate whether fluorescence corresponded to production of HHV-6AGFP virions, we examined 200 cells of the culture by transmission electron microscopy (TEM) in the Fred Hutchinson Cancer Center EM core laboratory for virus production: no virions were observed. The viral DNA was reproducibly detected by PCR, indicating the presence of the viral genome (Fig. S5D). However, despite several attempts, we detected no free circular or linear viral genomes by the method of Gardella et al. (27). Therefore, the data are consistent with the hypothesis that HHV-6AGFP established primarily latent infection in Jjhan cells cultured for >1 month, and that latency did not involve the formation of viral episomes.
We infected HEK-293 cells at 50% confluency with HHV-6A (U1102) at 0.1 multiplicity of infection (MOI). The cultures were incubated for 5 days. Then the cells were washed to remove extracellular virus, and single cells were introduced in each well of a 96-well plate. The single cells in each well were then expanded: 10 out of 22 clones were positive for the presence of the viral genome using ORF U94-specific PCR (Fig. S6A). Three PCR-positive clones and one PCR-negative clone were studied for integrated HHV-6 by FISH. In two PCR-positive clones, FISH analysis identified integrated HHV-6A in one chromosome. In the third PCR-positive clone, the virus had integrated into two chromosomes (Fig. S6B).
We could not identify the specific chromosomes into which HHV-6 had integrated in the HEK-293 cells, using standard cytogenetic methods, because of the aneuploidy and chromosomal rearrangements present in these cells. Therefore, we studied the integration event using inverse PCR (IPCR) (30). We digested DNA with the frequent cutter MboI, which cleaves methylated DNA. After digestion and heat inactivation, we diluted the DNA to 2 μg/mL and added T4 ligase to enable self-circularization of the HHV-6 sequence and adjacent chromosomal fragment. We amplified the ligated DNA using primers designed from the extreme right end of the genome (Fig. S7A). Southern blotting identified IPCR products that hybridized with a human telomere probe as well as an HHV-6 probe from integrated HEK-293 and patient T cells (Fig. S7B). No hybridization was observed without ligation of the MboI fragments. In summary, these experiments suggest that HHV-6A can efficiently integrate into the telomeres of in vitro infected HEK-293 cells.
Covalently Closed Circular Viral Episome of HHV-6A Is Not Detectable by PCR.
The majority of herpesviruses establish latency as circular nuclear episome, yet the Gardella assay had not detected HHV-6 episomes. Therefore, we looked for small numbers of episomes that might not have been detectable by the Gardella assay. We isolated DNA from HHV-6A integrated HEK-293, T cells from a member of Family-2, and T cells from a member of Family-1 immortalized using HVS strain C484. We subjected these cells to CsCl/ethidium bromide gradient ultracentrifugation (Fig. 3). HHV-6 DNA was detected only in the linear fraction, and was completely absent from the episomal (ccc) DNA fraction. Because our PCR assay can detect as few as 1–5 molecules, the experiment confirmed that viral episomes cannot be detected even by a highly sensitive method. The detection of mitochondrial sequence in the linear fraction is expected because replicating and relaxed mitochondrial episomes band together with chromosomal linear DNA.
Fig. 3.
PCR amplification fails to detect HHV-6 DNA in episomal fractions of CsCl/ethidium bromide (EtBr) gradients. To search for covalently linked circular viral episomes by a method more sensitive than the method of Gardella et al., 50 μg of DNA from two latently infected HEK-293 clones, T cells from Family-2/Mother, and T cells from Family-1/Sibling-2 immortalized with HVS strain C484 were subjected to CsCl/EtBr gradient ultracentrifugation for 2 days (CsCl density 1.55 g/mL, 10 μg/mL EtBr; VTi 65 Rotor at 40,000 rpm). After centrifugation, fractions were collected, and linear and episomal (ccc) DNA was identified by agarose electrophoresis. Salt and EtBr were removed from combined linear and episomal ccc fractions and subjected to PCR based amplification using primers to HHV-6 ORF-U94 and mitochondrial cytochrome c oxidase (positive episomal control) (A), HHV-6 ORF-U94 and β-actin genomic positive control (B), and HHV-6 ORF-U94, C484 Stp, and cytochrome c oxidase (positive episomal control) (C). ccc, covalently closed circular episomal fraction.
Reactivation of Integrated HHV-6A.
We next asked whether chromosomally integrated HHV-6 can reactivate, making integration a molecular strategy for viral latency. We cultured HEK-293 cells carrying integrated HHV-6A, and T cells from five patients with integrated virus, for 3 days. The cultures were done either in the presence of trichostatin A (TSA)—a compound known to reactivate latent herpesviruses—or in the absence of that compound. Quantitative real-time PCR demonstrated a significant increase in the number of copies of viral DNA, relative to untreated cells. Another compound that causes reactivation of latent herpesviruses, 12-O-tetradecanoyl-13 acetate (TPA), produced similar though milder effects (Fig. 4 A and B and Fig. S8).
Fig. 4.
HHV-6 DNA qPCR analysis of patient T cells and in vitro latently infected HEK-293 cell lines induced by TPA and TSA. T cell cultures from five family members and three latently infected HEK-293 cell lines were cultured in AIM-V or DMEM medium supplemented with 10% FCS and treated with known inducers of herpesvirus lytic replication protein kinase-C inducer TPA (20 ng/mL) and histone deacetylase inhibitor trichostatin-A (TSA) (80 ng/mL) for 3 days (42, 43). DNA isolated from cells (in triplicate) was subjected to quantitative real-time PCR (qPCR) for ORF U94 and fold-change ratios of Ct values normalized to β-actin were relative to untreated control. (A) HEK-293 cells (n = 3) (Fig. S8). (B) T cells (n = 5) (Fig. S8). TSA promoted a significant increase in viral DNA replication, whereas the stimulation with TPA and hydrocortisone had a milder effect. Statistical analysis was based on the Student t test, and P < 0.05 was considered significant. (C) Gardella gel analysis of infected Molt-3 cells 2 weeks after coculturing with PBMCs treated with TPA from Family-1/Sibling-2—Hector 2 T cell line carrying one copy of HHV-6A per a cell.
To determine whether the increase of viral DNA copy number indicated the production of infectious virus, we isolated PBMCs from six members of Family-1 and Family-2 whose cells had been cultured in the presence of TPA and hydrocortisone, leading to a marked increase in copy number. We cocultured these cells with Molt-3 cells in the presence of TPA and hydrocortisone. Syncytia formed in the Molt-3 cells infected with the virus from induced T cells, and replicating linear viral DNA and RNA were detected in these cells by Gardella gel (Fig. 4C and Fig. S9). Sequencing of the virus in the Molt-3 cells confirmed that it was identical to the integrated sequence in the cells of Family-1 and Family-2 (Fig. S2).
Discussion
HHV-6 Achieves Latency Through Chromosomal Integration.
Overall, this study provides substantial evidence that HHV-6A achieves latency in a different way from the other known human herpesviruses. HHV6-A integrates into the telomeres of human peripheral mononuclear cells in vivo, and following infection in Jjhan T cells, and HEK-293 cells (an epithelial cell line) in vitro. It appears that in cell lines capable of supporting productive, lytic infection, some of the cells quickly become latently infected through chromosomal integration, and remain viable.
Previous studies have used FISH to suggest that HHV-6 is capable of chromosomal integration. However, FISH cannot distinguish noncovalent linkage from integration. In our study, evidence that HHV-6 can integrate comes from multiple complementary methods besides FISH: chromosome-specific PCR, sequencing, IPCR, and Gardella gels.
The results suggest that HHV-6 integrates into the host genome via homologous recombination with human telomeres: the perfect TTAGGG repeats encoded in the right and left direct repeats of the viral genome mirror the telomeric repeats. We assume that the left end of the viral genome is joined with a long array of TTAGGG repeats, but confirming this will require additional studies.
We also found that the latent integrated genome of HHV-6A was inducible after being stimulated by TPA and TSA, and cocultivation studies indicated that the latent, integrated genome was capable of producing fully competent virus. Thus, for HHV-6 integration is not an unusual dead-end phenomenon, but a means of achieving latency. Moreover, in neither the in vivo or in vitro studies did we find HHV-6 DNA in episomal form—the strategy by which other human herpesviruses achieve latency.
Integrated HHV-6 Can Be Passed in the Germ Line.
Previous studies, using FISH, have suggested that the viral genome is passed through the germ line in occasional families. Inherited-integrated HHV-6 may have remarkably high prevalence. High HHV-6 viral load (106-107 copies per ml of blood) has been described in several British and US cohorts (33, 34) and attributed to inherited/integrated HHV-6. The prevalence of high viral load in normal blood donors has been reported to range from 0.8% to 1.5% (34). Interestingly, an even higher prevalence—2.9–3.3%—has been reported in hospitalized patients (33).
We studied four families in which multiple members, including parents and children, with remarkably high viral loads of HHV-6. Studies employing not only FISH (Fig. S3) but also sequence data from the viral-chromosomal junction confirmed that integrated HHV-6A can be transmitted vertically via the germ line (Figs. S1 and S2).
Questions Raised by These Studies.
The recognition that HHV-6A achieves latency through chromosomal integration raises important questions. Can HHV-6B and Marek's disease virus, a herpesvirus of fowl do the same, as suggested by FISH data.
What are the molecular switches that dictate whether, following new infection in a cell line capable of sustaining productive lytic infection, the virus instead integrates and establishes latency? What are the switches that activate transcription? Does transcription always lead to production of full virions? If not, might the production of selected viral proteins have pathogenic potential, as appears to be the case with endogenous retroviruses?
Theoretically, the ability of HHV-6 to integrate into the genome could alter the stability of individual chromosomes, and the expression of adjacent subtelomeric genes. Interestingly, subtelomeric rearrangement of chromosomes has been associated with mental retardation (35). These findings might also have relevance for telomerase activity. Mammalian and yeast chromosomes express a telomeric repeat-containing noncoding RNA (TERRA) (36). TERRA is proposed to regulate telomerase and many important telomere functions (36), and TERRA transcription is initiated in the subtelomere. Therefore, it is possible that integration of HHV-6 could alter the expression of TERRA leading to dysfunction of telomerase.
The documentation of inherited HHV-6 also raises important questions. Is the immune system of patients born with HHV-6 tolerant of any viral antigens? Given that the viral genome is present in every cell and capable of producing competent virions, are these patients at particular risk for any diseases? Are the patients susceptible to superinfection with another strain of HHV-6A, or to infection with HHV-6B? Answers to these questions await further study.
Materials and Methods
Primary T Cells, Cell Lines, and Viruses.
Peripheral blood from four families was obtained by the HHV-6 Foundation, after the subjects had given informed consent. PBMCs were isolated through Lymphoprep according to the manufacturer's protocol and incubated in RPMI medium 1640 containing 10% FBS and 10 μg/mL PHA (Sigma-Aldrich) for 72 h followed by culturing in 100 units/μL IL-2 medium. T cell lines Jjhan (HHV-6 Foundation), Molt-3 (ATCC), and Burkitt's lymphoma cell line Raji (ATCC) were maintained in RPMI medium 1640 containing 10% FBS. Human embryonic kidney-293 (HEK-293) cells were maintained in DMEM supplemented with 10% FBS. HHV-6A (U1102 strain) and HHV-6B (Z29 strain) viruses were from P. Pellett (Wayne State University). HHV-6 integrated Burkitt's lymphoma cell line Katata was from M. Daibata (Kochi Medical School, Kochi, Japan) (37). Immortalization of patient T cells with Herpesvirus saimiri strain 484–77 was completed as described in refs. 38 and 39.
Reactivation of HHV-6A.
Freshly isolated PBMCs at 1 × 106 cells per ml cell concentration were cultured in RPMI medium 1640 supplemented with 10% FCS, 20 ng/mL TPA, and 1 × 10−6 M hydrocortisone or 80 ng/mL TSA for 3–5 days. To isolate reactivated virus, 1 × 104 Molt-3 cells were added to 106 PBMCs in 1 mL and cultured for 10–14 days. Reactivation was monitored for cell CPE, Gardella gel, qPCR, and sequencing of ORF U94.
Chromosome-specific PCR.
Chromosome-specific PCR for integration of HHV-6 in chromosome 17p was performed using 400 ng of genomic DNA and primers DRR and 17p or DRL and 17p (28). Cohybridization of PCR products with HHV-6 and telomere sequences was by Southern hybridization.
CsCl/ethidium bromide gradient.
DNA (50 μg) isolated from HEK-293 and family member T cells with integrated HHV-6 were subjected to centrifugation at 40,000 rpm (Beckman L7-65) for 72 h in a solution of CsCl/ethidium bromide (density = 1.55 g/mL). Covalently closed circular DNA and linear DNA fractions were visualized by agarose gel electrophoresis and then subjected to amplification with primers to HHV-6 ORF-U94, β-actin, C484 Stp, and cytochrome c oxidase.
Southern hybridization.
Vacuum blotting and hybridization with 32P-labeled HHV-6A (U1102) cosmids PMF311-12 and PMF335-6 (40) was as described in ref. 44.
Supplementary Material
Acknowledgments
We thank Kristin Loomis, President of the HHV-6 Foundation, for extensive and enthusiastic support of this work. The Cytogenetics Core Laboratory at the University of Minnesota is supported by Masonic Cancer Center–National Institutes of Health Grant P30 CA077598-09. This work was supported by grants from the HHV-6 Foundation (to P.F.A. and P.G.M.) and the National Institutes of Health (Grant 5R01CA111196 to P.G.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913586107/DCSupplemental.
References
- 1.Salahuddin SZ, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez G, et al. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gompels UA, et al. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 4.Yamanishi K, et al. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;1:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 5.Ablashi D, et al. Human herpesvirus-6 strain groups: a nomenclature. Arch Virol. 1993;129:363–366. doi: 10.1007/BF01316913. [DOI] [PubMed] [Google Scholar]
- 6.Asano Y, et al. Clinical features of infants with primary human herpesvirus 6 infection (exanthem subitum, roseola infantum) Pediatrics. 1994;93:104–108. [PubMed] [Google Scholar]
- 7.Lusso P, et al. Productive infection of CD4+ and CD8+ mature human T cell populations and clones by human herpesvirus 6. Transcriptional down-regulation of CD3. J Immunol. 1991;147:685–691. [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989;63:3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Bolle L, Naesens L, De Clercq E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin Microbiol Rev. 2005;18:217–245. doi: 10.1128/CMR.18.1.217-245.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dockrell DH, Paya CV. Human herpesvirus-6 and -7 in transplantation. Rev Med Virol. 2001;11:23–36. doi: 10.1002/rmv.299. [DOI] [PubMed] [Google Scholar]
- 11.Jones CM, Dunn HG, Thomas EE, Cone RW, Weber JM. Acute encephalopathy and status epilepticus associated with human herpes virus 6 infection. Dev Med Child Neurol. 1994;36:646–650. doi: 10.1111/j.1469-8749.1994.tb11903.x. [DOI] [PubMed] [Google Scholar]
- 12.Donati D, et al. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology. 2003;61:1405–1411. doi: 10.1212/01.wnl.0000094357.10782.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamashita N, Morishima T. HHV-6 and seizures. Herpes. 2005;12:46–49. [PubMed] [Google Scholar]
- 14.Lusso P, et al. Human herpesvirus 6A accelerates AIDS progression in macaques. Proc Natl Acad Sci USA. 2007;104:5067–5072. doi: 10.1073/pnas.0700929104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lusso P, et al. Productive dual infection of human CD4+ T lymphocytes by HIV-1 and HHV-6. Nature. 1989;337:370–373. doi: 10.1038/337370a0. [DOI] [PubMed] [Google Scholar]
- 16.Cermelli C, et al. High frequency of human herpesvirus 6 DNA in multiple sclerosis plaques isolated by laser microdissection. J Infect Dis. 2003;187:1377–1387. doi: 10.1086/368166. [DOI] [PubMed] [Google Scholar]
- 17.Goodman AD, Mock DJ, Powers JM, Baker JV, Blumberg BM. Human herpesvirus 6 genome and antigen in acute multiple sclerosis lesions. J Infect Dis. 2003;187:1365–1376. doi: 10.1086/368172. [DOI] [PubMed] [Google Scholar]
- 18.Berti R, Soldan SS, Akhyani N, McFarland HF, Jacobson S. Extended observations on the association of HHV-6 and multiple sclerosis. J Neurovirol. 2000;6(Suppl 2):S85–S87. [PubMed] [Google Scholar]
- 19.Daibata M, Taguchi T, Taguchi H, Miyoshi I. Integration of human herpesvirus 6 in a Burkitt's lymphoma cell line. Br J Haematol. 1998;102:1307–1313. doi: 10.1046/j.1365-2141.1998.00903.x. [DOI] [PubMed] [Google Scholar]
- 20.Hall CB, et al. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics. 2008;122:513–520. doi: 10.1542/peds.2007-2838. [DOI] [PubMed] [Google Scholar]
- 21.Luppi M, et al. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 22.Nacheva EP, et al. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J Med Virol. 2008;80:1952–1958. doi: 10.1002/jmv.21299. [DOI] [PubMed] [Google Scholar]
- 23.Torelli G, et al. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol. 1995;46:178–188. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- 24.Ward KN, et al. Human herpesvirus 6 chromosomal integration in immuno-competent patients results in high levels of viral DNA in blood, sera, and hair follicles. J Clin Microbiol. 2006;44:1571–1574. doi: 10.1128/JCM.44.4.1571-1574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gompels UA, Macaulay HA. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J Gen Virol. 1995;76:451–458. doi: 10.1099/0022-1317-76-2-451. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka-Taya K, et al. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J Med Virol. 2004;73:465–473. doi: 10.1002/jmv.20113. [DOI] [PubMed] [Google Scholar]
- 27.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Britt-Compton B, et al. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum Mol Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- 29.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 30.Ochman H, Gerber AS, Hartl DL. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delecluse HJ, Hammerschmidt W. Status of Marek's disease virus in established lymphoma cell lines: herpesvirus integration is common. J Virol. 1993;67:82–92. doi: 10.1128/jvi.67.1.82-92.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delecluse HJ, Schüller S, Hammerschmidt W. Latent Marek's disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leong HN, et al. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J Med Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- 34.Ward KN, Thiruchelvam AD, Couto-Parada X. Unexpected occasional persistence of high levels of HHV-6 DNA in sera: detection of variants A and B. J Med Virol. 2005;76:563–570. doi: 10.1002/jmv.20399. [DOI] [PubMed] [Google Scholar]
- 35.Baralle D. Chromosomal aberrations, subtelomeric defects, and mental retardation. Lancet. 2001;358:7–8. doi: 10.1016/S0140-6736(00)05300-9. [DOI] [PubMed] [Google Scholar]
- 36.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bandobashi K, et al. Human herpesvirus 6 (HHV-6)-positive Burkitt's lymphoma: establishment of a novel cell line infected with HHV-6. Blood. 1997;90:1200–1207. [PubMed] [Google Scholar]
- 38.Collins CM, Medveczky MM, Lund T, Medveczky PG. The terminal repeats and latency-associated nuclear antigen of herpesvirus saimiri are essential for episomal persistence of the viral genome. J Gen Virol. 2002;83:2269–2278. doi: 10.1099/0022-1317-83-9-2269. [DOI] [PubMed] [Google Scholar]
- 39.Medveczky MM, et al. Herpesvirus saimiri strains from three DNA subgroups have different oncogenic potentials in New Zealand white rabbits. J Virol. 1989;63:3601–3611. doi: 10.1128/jvi.63.9.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neipel F, Ellinger K, Fleckenstein B. The unique region of the human herpesvirus 6 genome is essentially collinear with the UL segment of human cytomegalovirus. J Gen Virol. 1991;72:2293–2297. doi: 10.1099/0022-1317-72-9-2293. [DOI] [PubMed] [Google Scholar]
- 41.Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang LK, Liu ST. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 2000;28:3918–3925. doi: 10.1093/nar/28.20.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu F, et al. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J Virol. 2003;77:11425–11435. doi: 10.1128/JVI.77.21.11425-11435.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medveczky PG, Chang C-W, Oste C, Mulder C. Rapid vacuum driven transfer of DNA and RNA from gels to solid supports. BioTechniques. 1987;5:242–246. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.