Abstract
To identify genetic factors that interact with social environments to impact human health, we used a bioinformatic strategy that couples expression array–based detection of environmentally responsive transcription factors with in silico discovery of regulatory polymorphisms to predict genetic loci that modulate transcriptional responses to stressful environments. Tests of one predicted interaction locus in the human IL6 promoter (SNP rs1800795) verified that it modulates transcriptional response to β-adrenergic activation of the GATA1 transcription factor in vitro. In vivo validation studies confirmed links between adverse social conditions and increased transcription of GATA1 target genes in primary neural, immune, and cancer cells. Epidemiologic analyses verified the health significance of those molecular interactions by documenting increased 10-year mortality risk associated with late-life depressive symptoms that occurred solely for homozygous carriers of the GATA1-sensitive G allele of rs1800795. Gating of depression-related mortality risk by IL6 genotype pertained only to inflammation-related causes of death and was associated with increased chronic inflammation as indexed by plasma C-reactive protein. Computational modeling of molecular interactions, in vitro biochemical analyses, in vivo animal modeling, and human molecular epidemiologic analyses thus converge in identifying β-adrenergic activation of GATA1 as a molecular pathway by which social adversity can alter human health risk selectively depending on individual genetic status at the IL6 locus.
Keywords: gene–environment interaction, inflammation, social epidemiology, stress, transcription
Social factors are believed to interact with genetic polymorphisms to influence human health and longevity (1–4), but the specific genetic loci involved and the molecular basis for their interaction with social conditions remain poorly understood. Social factors can alter gene expression via neural and endocrine activation of cellular transcription factors (TFs) such as NF-κB/Rel, CREB/ATF, AP-1, and STAT family proteins (5–8). Those TFs bind to gene cis-regulatory sequences to activate transcription and thereby modify the cellular protein complement (9). One mechanism by which genetic characteristics could modulate social influences on health involves regulatory polymorphisms in gene promoter sequences (10–14), which can affect the binding of socio-environmentally activated TFs and thereby alter transcriptional responses to changing social conditions (3, 15, 16).
To systematically identify Gene × Social Environment (GxSE) interactions, we developed a novel bioinformatics strategy that models the molecular mechanism by which regulatory polymorphisms alter the capacity of environmentally responsive TFs to activate the expression of health-relevant genes. Regulatory GxSE interactions can be conceptualized as the Boolean product of two distinct information streams involving socio-environmental activation of a specific TF, and DNA sequence-conferred ability of a gene to be transcribed in response to an activated TF. Both biological information streams can be analyzed computationally through (i) reverse inference of TF activity from empirical gene expression profiles (17), and (ii) in silico modeling of differential TF binding to alternative regulatory sequences created by DNA polymorphism (18). We integrated these two approaches to identify candidate regulatory polymorphisms that might modulate socio-environmental activation of the human IL6 gene. IL6 was targeted for analysis because (i) it is known to be relevant to human health (IL-6 has been linked to several prevalent causes of morbidity and mortality including cardiovascular disease, neurodegeneration, and some types of cancer) (4, 19, 20); (ii) it is sensitive to socio-environmental conditions (adverse conditions are associated with increased levels of IL-6 and its biomarker C-reactive protein) (21–27), and (iii) the IL6 promoter is genetically polymorphic, including a G/C transversion (rs1800795) (28) that has been linked to inflammation-related diseases in some direct association analyses but not others. Inconsistent results from simple genetic association studies may stem from an unidentified modifying interaction with environmental factors (3), raising the possibility that IL6 promoter polymorphism might have a functional impact on gene expression only in the presence of environmental conditions that induce TF activity. To determine whether IL6 promoter polymorphism might interact with adverse socio-environmental conditions to affect inflammation-related disease risk, we computationally analyzed all known SNPs in the human IL6 promoter for potential modulation of environmentally responsive TF activity. We then tested those in silico predictions in biochemical analyses of TF/promoter interactions in vitro, in cellular models of IL6 gene regulation in vitro, in in vivo laboratory animal models, and in human molecular epidemiologic analyses of inflammation-related mortality risk.
Results
Computational Prediction of Gene × Environment Interaction.
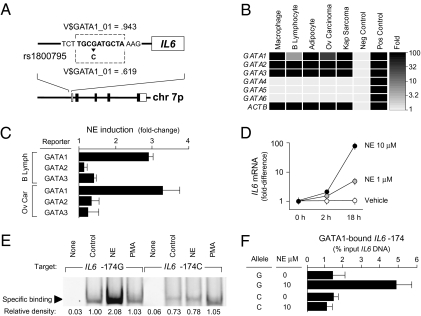
To identify SNPs that might modulate induction of the human IL6 gene by environmentally responsive TFs, we scanned the human genome reference sequence spanning the region −1,000 bp upstream to +200 bp downstream of the RefSeq-designated IL6 transcription start site for predicted TF-binding motifs and compared that distribution with results from parallel analysis of IL6 promoter sequences bearing each known single nucleotide substitution, insertion, or deletion cataloged in the dbSNP database (29). A G/C transversion 174 bp upstream of the transcription start site (rs1800795) emerged as the sole predicted functionally active regulatory SNP (rSNP) (Fig. 1A). Promoters bearing the ancestral G allele of rs1800795 (30) were predicted show high-affinity binding of the GATA1 TF in the region spanning −177/−168 bp. However, that predicted binding site was largely abrogated in sequences bearing the variant C allele at IL6 −174. To the extent that adverse socio-environmental conditions can increase IL6 gene expression by activating GATA1, those results imply that the rs1800795 polymorphism might provide a molecular substrate for gene−environment interaction in inflammation-related biological processes.
Fig. 1.
(A) Computational modeling of the ancestral IL6 promoter sequence (−174G) identified a high-affinity GATA1-binding motif (TRANSFAC V$GATA1_01 mat_sim > 0.90) that was predicted to be abrogated by the rs1800795 G/C transversion (mat_sim < 0.75). (B) RT-PCR detection of GATA factor mRNA in IL6-producing cell types including macrophages, B lymphocytes, adipocytes, ovarian carcinoma cells, and B lymphoid cells latently infected with human herpesvirus 8 (Kaposi's sarcoma–associated herpesvirus). Data represent the mean fold-increase above negative controls in triplicate determinations. (C) Luciferase reporter assays assessed the capacity of the sympathetic neurotransmitter norepinephrine (NE) to activate GATA1, GATA2, or GATA3 transcriptional activity. Data represent the mean ± SE of three independent experiments using reporter constructs specifically responsive to individual GATA factors, with GATA1 showing greatest NE-induced activation in each cell type studied (P < 0.01). (D) NE induction of IL6 gene transcription was confirmed by RT-PCR in primary macrophages homozygous for the G allele of rs1800795. Data are mean ± SE of three replicate determinations in one experiment, with results representative of three independent experiments. (E) NE-induced binding of nuclear transcription factors to the IL6 promoter sequence (−187/−163) bearing either the −174G allele (NE induction, P < 0.001) or the −174C allele (NE induction, P = 0.46). Specificity of effects to the NE-activated PKA signaling pathway was tested by parallel PMA stimulation of PKC. Analyses also verified decreased binding of transcription factors to the −174C sequence under basal conditions (difference from −174G, P = 0.023). Data are representative of results from three independent experiments. (F) Allele-specific chromatin immunoprecipitation assays were preformed in rs1800795 heterozygous cells (primary macrophages shown) to confirm NE induction of GATA1 binding to the −174G allele of the IL6 promoter, but not to the −174C allele. GATA1-bound IL6 promoter DNA was quantified as a fraction of total IL6 promoter DNA. Data represent mean ± SE of three independent experiments.
Molecular Analysis GATA1-Mediated IL6 Transcription.
To determine whether GATA1 might modulate IL6 transcription in response to adverse environmental conditions, we examined the expression and activation of GATA family TFs in multiple IL-6–producing cell types. Experimental studies confirmed that primary macrophages, B lymphocytes, adipocytes, ovarian carcinoma cells, and B lymphocytes latently infected with human herpesvirus 8 all expressed high levels of mRNA for GATA1, GATA2, and GATA3 but not other GATA family members (Fig. 1B). To determine whether GATA TFs might be activated by adverse socio-environmental conditions, we examined the ability of the sympathetic nervous system (SNS) neurotransmitter norepinephrine (NE) to stimulate GATA-mediated gene transcription in luciferase reporter assays. Each cell type examined showed up-regulation of GATA1-mediated gene transcription (but not GATA2- or GATA3-mediated transcription) following exposure to 1–10 μM NE (Fig. 1C). Consistent with potential GATA1 induction of the IL6 promoter, NE also enhanced IL6 mRNA expression in each cell type tested (Fig. 1D). As in previous studies (31–33), β-adrenergic receptors were found to mediate these effects (Fig. S1). Given that many types of socio-environmental stress have been linked to increased SNS activity (7, 8, 34–41), NE activation of GATA1 could constitute one molecular pathway by which adverse environments influence IL6 gene expression.
Molecular Analysis of IL6 Regulatory Polymorphism.
To verify computational indications that the C allele of rs1800795 might abrogate NE-induced TF binding to the IL6 promoter, we assayed binding of nuclear TFs to oligonucleotides bearing C vs. G allele promoter sequences. Results verified that both constitutively active TFs and NE-induced TFs bound less efficiently to the IL6 −174C promoter sequence than to the G allele sequence (Fig. 1E). To confirm that GATA1 bound to the IL6 −174 locus, and to determine whether that interaction occurred preferentially for the G allele sequence, we carried out allele-specific chromatin immunoprecipitation assays in cells exposed to NE vs. vehicle control stimulation. Allele-specific PCR analysis of GATA1-immunoprecipitated chromatin confirmed specific interaction between GATA1 and the IL6 −174 locus (Fig. 1F). That interaction was present at low levels under basal conditions, was significantly up-regulated in response to NE, and was up-regulated significantly more strongly on promoter sequences bearing the −174G allele than on those bearing the −174C allele (Fig. 1F).
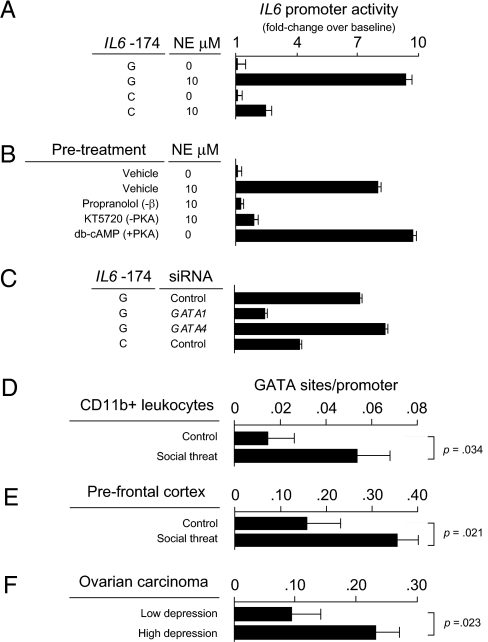
β-Adrenergic Mediation of GATA1 Activation.
To assess the functional significance of altered GATA1 binding for SNS-induced transcriptional response, we quantified the magnitude of NE-induced transcription from IL6 −174G vs. C allele promoters. Across multiple IL6-producing cell types, NE markedly enhanced transcription from the ancestral −174G promoter, whereas the variant −174C promoter showed signficantly less induction (Fig. 2A). Pharmacologic agonist and antagonist studies confirmed that NE induction of the IL6 promoter was mediated by β-adrenergic activation of the protein kinase A (PKA) signaling pathway (Fig. 2B). To determine whether GATA1 specifically mediated PKA induction of the IL6 promoter, we used siRNA to selectively reduce GATA1 protein levels. GATA1 inhibition retarded PKA induction of the G allele promoter by more than 50% (Fig. 2C), reducing PKA sensitivity to levels comparable to those observed for the GATA1-insensitive C allele promoter.
Fig. 2.
(A) Luciferase reporter assays gauged NE-induced activation of human IL6 −174G vs. C allele promoters (data represent mean ± SE from three replicate determinations in ovarian carcinoma cells; difference in NE-mediated induction, P < 0.001). (B) The role of beta-adrenergic / PKA signaling in mediating NE induction of the IL6 −174G promoter was tested by pre-exposing cells to the beta-adrenergic antagonist propranolol or the PKA antagonist KT5720 before NE exposure. Sufficiency of PKA activation alone to induce the IL6 −174G promoter was tested using the pharmacologic PKA agonist db-cAMP. All data represent the mean ± SE from three replicate determinations. (C) The role of GATA1 in PKA-induced IL6 −174G promoter activation was tested by GATA1-targeted siRNA inhibition. Specificity of GATA1’s effect was tested by parallel siRNA inhibition of other GATA family members (e.g., GATA4 shown). Data represent mean ± SE of three replicate determinations. (D) In vivo activation of GATA-mediated gene transcription was assessed by TELiS promoter-based bioinformatic analysis of genome-wide transcriptional profiles in CD11b+ myeloid spleen cells harvested from adult male C57BL/6 mice after six daily cycles of 2-h exposure to social threat (n = 10) vs. control home caged animals (n = 10). Data represent the average (± SE) prevalence of GATA1 transcription factor-binding motifs in promoters of 100 genes showing the greatest magnitude of up-regulation following social threat relative to 100 genes showing the greatest magnitude of up-regulation in controls. (E) Parallel analyses of GATA1 transcription factor activity in brain prefrontal cortex from the same n = 10 control and n = 10 socially threatened animals as in B. (F) Differential prevalence of GATA1 transcription factor-binding motifs in the promoters of 220 human genes up-regulated ≥ 50% in ovarian carcinoma cells from 10 women experiencing adverse social conditions (high depressive symptoms and low social support) vs. 46 genes up-regulated in grade- and stage-matched ovarian carcinomas from 10 women experiencing favorable social conditions (low depressive symptoms and high social support).
Socio-Environmental Activation of GATA1 in Vivo.
To determine whether GATA1 mediates in vivo transcriptional responses to socio-environmental adversity, we analyzed genome-wide transcriptional activity of promoters bearing GATA1-binding motifs (17). In adult male C57BL/6 mice subject to three daily cycles of 2-h exposure to social threat vs. nonthreatening control conditions, microarray transcriptional profiling of brain prefrontal cortex and myeloid-lineage CD11b+ spleen cells indicated transcriptional modulation of more than 200 promoters in each tissue (Tables S1 and S2). Promoter-based bioinformatic analyses indicated activation of GATA1 in both myeloid spleen cells (Fig. 2D) and brain prefrontal cortex (Fig. 2E). To confirm the relevance of those findings for human social adversity, we carried out similar analyses in primary ovarian carcinoma tissues resected from women experiencing high levels of depression and low social support vs. minimal depressive symptoms and high social support (42) (Tables S3 and S4). Results again indicated up-regulated GATA1 activity in association with social adversity (Fig. 2F). Consistent with NE induction of GATA1 activity in the present biochemical studies (Fig. 1 and Fig. 2), intratumor NE levels were also elevated in tissues from patients experiencing high levels of social adversity (mean = 19.5 ± 6.9 pg NE /mg tissue vs. <0.1 ± 0.1 in low-adversity conditions; difference, P = 0.0482) (42). Histological mapping showed that perivascular nerve fibers with occasional parenchymal radiations constituted the primary source of intratumor NE (Fig. S2).
Molecular Epidemiologic Analysis of Gene–Environment Interaction.
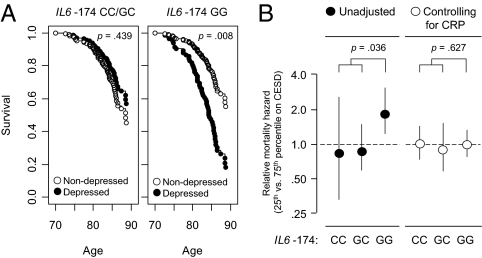
To confirm the health significance of rs1800795 modulation of IL6 transcriptional response to socio-environmental stress, we carried out molecular epidemiologic analyses of late-life mortality risk in the MacArthur Study of Successful Aging (20, 43). IL6 is involved in several major drivers of all-cause mortality, including cardiovascular disease, Alzheimer's disease, and certain types of cancer (4, 19, 20). Progression of each of those diseases has also been linked to adverse social conditions (44, 45). We therefore analyzed the risk of all-cause mortality over 11.7 years following the initial MacArthur Study assessment of socio-environmental conditions in 1988, at which point all study participants were healthy and high functioning. Based on results linking GATA1 activity to depressive symptoms (Fig. 2F), measures of depression served as indicators of subjectively significant socio-environmental adversity across multiple social domains (15, 46). In an ethnically homogenous sample of men and women aged 70–80 years in 1988 (Table S5), high levels of depressive symptoms were associated with significantly increased subsequent mortality risk among those homozygous for the GATA1-sensitive IL6 −174G allele, but no increased mortality risk among those homozygous for the −174C allele or among heterozygotes (Table 1). This GxSE interaction in mortality risk emerged regardless of whether depressive symptoms were measured by the Center for Epidemiologic Studies–Depression scale (CES-D) or the Hopkins Symptom Checklist Depression scale (SCL-D), and regardless of whether analyses focused on crude mortality rates or risks adjusted for age at study entry, gender, body mass index, smoking, alcohol consumption, and socio-economic status. Among rs1800795 G homozygotes (39% of the sample), the presence of depressive symptoms in late life was associated with a 2.8-year shorter average lifespan, whereas the presence of late-life depressive symptoms in those carrying one or more C allele was associated with a nonsignificant 0.2-year greater lifespan (interaction P = 0.036 for CES-D and P = 0.014 for SCL-D) (Fig. 3A). The rs1800795 genotype was not associated with the level of depressive symptoms at study entry in 1988 or at 3-year follow-up in 1991 (CES-D: difference among genotypes, 4.3% of mean score in 1988 and 2.9% in 1991, both P > 0.40; SCL-D: difference among genotypes, 4.1% in 1998 and 3.7% in 1991, both P > 0.30).
Table 1.
All-cause mortality risk
| Model | Depression measure | IL6 −174 genotype | Hazard ratio* (95% CI) | P† |
| Simple‡ | CES-D | CC | 0.87 (0.30–2.47) | 0.7855 |
| GC | 0.89 (0.57–1.40) | 0.6209 | ||
| GG | 1.76 (1.16–2.68) | 0.0079 | ||
| SCL-D | CC | 1.02 (0.44–2.39) | 0.9584 | |
| GC | 0.76 (0.43–1.32) | 0.3198 | ||
| GG | 2.13 (1.14–3.95) | 0.0166 | ||
| Adjusted§ | CES-D | CC | 0.95 (0.25–3.55) | 0.9343 |
| GC | 0.76 (0.43–1.33) | 0.3331 | ||
| GG | 2.06 (1.29–3.27) | 0.0024 | ||
| SCL-D | CC | 0.86 (0.19–3.88) | 0.8442 | |
| GC | 0.52 (0.27–1.01) | 0.0521 | ||
| GG | 7.93 (2.71–23.2) | 0.0002 |
CES-D, Center for Epidemiologic Studies Depression scale; SCL-D, Hopkins’ Symptom Checklist Depression scale.
*Hazard ratio for 75th percentile vs. 25th percentile on depression measure.
†Two-tailed significance level.
‡Simple (unadjusted) association between depression and mortality risk.
§Multivariate analysis controlling for age at study entry, sex, body mass index, smoking, alcohol consumption and socio-economic status.
Fig. 3.
Modification of late life socio-environmental mortality risk by rs1800795 genotype. (A) Relationship between depressive symptoms and subsequent all-cause mortality risk for IL6 −174G homozygotes (Right) vs. carriers of one or more C allele (Left). Data represent estimated survival functions from Cox proportional hazards regression analyses controlling for sex and age at study entry (range 70–80 years) in 184 initially healthy Caucasian participants in the MacArthur Study of Successful Aging. Open circles represent mortality risk given cohort average levels of depressive symptoms at study entry (CES-D = 4); filled circles represent mortality risk given high depressive symptoms at study entry (CES-D = 16). (B) Effect of controlling for chronic inflammation (plasma C-reactive protein ≥3 mg/L) on rs1800795 genotype modification of relative mortality hazard associated with low vs. high depressive symptoms at study entry (same cohort of n = 184 MacArthur Study participants as in A). Values represent point estimate (± 95% confidence interval) of relative hazard associated with 75th vs. 25th percentile values of cohort CES-D distribution.
To determine whether rs1800795 genotype modulated depression-associated mortality risk specifically via effects on inflammatory biology, we conducted multivariate mediation analyses using plasma C-reactive protein (CRP) as a measure of IL-6-related chronic inflammation (21, 22). As would be expected if mortality differences were mediated specifically by variations in chronic inflammation, inclusion of CRP in survival analyses rendered the Genotype × Depression interaction term nonsignificant (Fig. 3B; CES-D, P = 0.627; SCL-D, P = 0.601), but CRP levels continued to significantly predict mortality (P < 0.001 in all analyses). Competing hazards analyses of cause-specific mortality also supported a mediating role of inflammatory biology by showing that the IL6 −174 Genotype × Depression interaction pertained specifically to inflammation-related causes of death (cardiovascular, neurodegenerative, and neoplastic) and did not affect other causes of death (Tables S6 and S7).
Discussion
The present data show that SNS activation can induce IL6 gene expression via β-adrenergic activation of GATA1. These data also identify the rs1800795 G/C transversion as a regulatory SNP (rSNP) that can moderate the impact of SNS/GATA1 signaling in ways that ultimately protect C allele carriers from the heightened inflammation-related disease risk associated with adverse socio-environmental conditions. These findings are consistent with previous indications that rs1800795 might affect IL6 gene expression by modulating binding of unspecified TFs (28, 47, 48), and with data showing that β-adrenergic activation of the cAMP/PKA pathway can phosphorylate other GATA family TFs and modulate their transcriptional activity (49–54). The present results are also consistent with previous molecular epidemiologic analyses linking rs1800795 to differential risk of inflammation-related disease (4, 28, 55). Confirmation that rs1800795 is a functional rSNP with environmentally contingent implications for human mortality risk also shows that the present in silico analytic approach can accurately predict in vivo GxSE interactions. Genome-wide application of this strategy could potentially accelerate the discovery GxSE interactions throughout the human genome.
Because the −174G/C transversion gates GATA1 access to the IL6 promoter, it provides the molecular basis for an intrinsic Gene × Environment interaction that links rs1800795 to inflammatory disease risk only under environmental conditions that activate GATA1. Consistent with that intrinsic interaction, −174G homozygotes showed no increased mortality risk in the absence of late-life depressive symptoms and rs1800795 genotype was unrelated to the prevalence of late-life depressive symptoms. Thus, rs1800795 does not constitute a cause of depressive symptoms but, rather, a genetic vulnerability factor that renders G homozygotes more sensitive to IL6-related health risks when those individuals encounter adverse environmental conditions sufficient to trigger depressive symptoms (3, 15, 46). Given that IL6 −174G appears to represent the ancestral human genome sequence (30), the present mortality results suggest that the variant C allele may have been retained in the human gene pool because it conferred a selective advantage in limiting hyperinflammatory IL6 responses during periods of chronic SNS activation.
Identification of rs1800795 as the genetic component of a GxSE interaction may explain why some previous genetic association studies have failed to confirm relationships between that polymorphism and disease risk. To the extent that those analyses failed to consider the environmentally contingent activation of GATA1 as a necessary precondition for phenotypic manifestation of rs1800795 genetic influences, false negative findings could result from functional mis-specification of data analytic models (resulting in loss of statistical power and biased parameter estimation) (2, 56). The risk of overlooking such conditionally penetrant SNPs might be reduced in future studies by applying the present computational approach at a genome-wide level to predict specific genetic loci likely to interact with environmental conditions. Incorporation of the resulting GxSE hypotheses into genome-wide association studies would (i) enhance statistical power by appropriately including outcome-determining effects in the deterministic portion of the statistical model, and (ii) reduce potential downward bias in the estimation of genetic effect parameters that would otherwise result from averaging differing genetic effects expressed under alternative socio-environmental conditions. The later advantage is particularly significant, given the potential for antagonistic pleiotropy to induce “net zero” estimates of genetic effects that are actually significantly beneficial under some socio-environmental conditions and significantly detrimental under others. Routine incorporation of mechanistically derived GxSE interaction hypotheses into analytic models for genome-wide association studies might considerably increase the number of genetic influences discovered and the magnitude of their identified effects. Such results also have the potential to suggest socio-environmental alterations that could mitigate genetically linked health risks.
Additional studies will be required to clarify the mechanism, scope, and health implications of the present results. The biochemical and animal model data reported here involve causal effects from controlled experiments, but the human health effects of the IL6 GxSE interaction were estimated by association in a longitudinal cohort study of moderate size and limited geographic diversity. The present analyses were also restricted to white adults to avoid confounding by population ethnic stratification. The relevance of these findings to other ethnic or age groups should not be assumed until these results are replicated in larger and more diverse study populations. Additional molecular biological studies are also needed to clarify the mechanistic basis for the dominant protective effect of the variant rs1800795 C allele. It is conceivable that the presence of one C allele promoter is sufficient to block the development of an IL6 positive feedback cycle. Given that siRNA knock-down of GATA1 abrogated only ∼50% of total NE-mediated IL6 promoter induction, transcriptional regulation at the IL6 −174 locus may also involve additional TFs that remain to be identified. The present analysis focuses solely on transcription factor activity within the core promoter and would not detect more distant regulatory polymorphisms greater than 1 kb upstream or downstream in the gene coding sequence or introns. It is also important to note that the present bioinformatic approach is not intended to provide a comprehensive model of IL6 gene regulation, but seeks only to identify functional rSNPs involved in GxSE interactions. However, the Boolean conjunctive model applied here could be expanded to include empirical measures of other known gene regulatory influences such as promoter methylation, epistatic interactions with other genes, chromatin dynamics, and micro-RNA inhibition. Expansion of the present model to encompass a broader array of regulatory dynamics could provide a promising direction for future research.
The basic approach of combining computational models of gene regulation with empirical bioinformatic discovery of environmentally sensitive TFs provides a rational strategy for prioritizing candidate GxSE interactions in a search space involving billions of candidate combinations (∼107 SNPs crossed with tens to hundreds of potentially relevant environmental conditions). Although the present model is relatively simple, its conjunctive Boolean structure could easily be extended to incorporate other molecular regulators of gene expression (e.g., promoter methylation or other epigenetic mechanisms, epistatic interactions with other genes, micro-RNA inhibition, distant promoter/enhancer elements). Genome-wide application of such integrated structural/functional genomic models could provide a powerful strategy for accelerating discovery of other GxSE interactions.
Methods
Computational Modeling of Gene × Environment Interaction.
To identify gene polymorphisms that might modulate the induction of IL6 gene expression by environmentally responsive transcription factors (ERTFs), we carried out bioinformatic analyses to map the conjunctive set of (i) transcription factors (TFs) that are empirically activated by socio-environmental stress, and (ii) TFs with predicted high-affinity binding sites in the IL6 promoter that are predicted to be abrogated by known single nucleotide polymorphisms (SNPs). Computational details of these analyses are provided in SI Text. Briefly, candidate ERTFs were identified through TELiS bioinformatics analysis (17) of TF-binding motifs (TFBMs) in promoters of genes that were found to be empirically differentially expressed in CNS prefrontal cortex tissue or CD11b+ splenocytes from animals that were subject to experimentally imposed social threat (described below) or in solid tissue samples from humans subject to chronic social adversity as previously described (42). The IL6 core promoter sequence was then computationally scanned for potential regulatory SNPs (rSNPs) predicted to affect the binding of candidate ERTFs. Analyses used a variant of the SNP_TRAST algorithm (18) to assess the impact of all dbSNP polymorphisms (29) on predicted TFBMs in the human genome sequence spanning −1,000 bp to + 200 bp relative to the RefSeq IL6 transcription start site (57). Polymorphisms predicted to inhibit ERTF binding to a high-affinity TFBM by more than 50% were considered biologically significant based on previous functional calibration studies (18). rs1800796 Modulation of GATA1 ERTF activity emerged as the sole candidate interaction meeting all analytic criteria.
Cell Culture and Gene Expression.
Monocyte-derived macrophages, Ramos B lymphoid cells, 3T3-L1 adipocytes, SKOV3ip ovarian carcinoma cells, and KS1 primary effusion lymphoma cells were cultured under standard conditions. Expression of IL6, GATA1, GATA2, GATA3, GATA4, GATA5, and GATA6 mRNA was assayed by quantitative real-time RT-PCR using established TaqMan Gene Expression Assays (Applied Biosystems) and standard threshold cycle analysis with normalization to ACTB mRNA. Additional details are available in SI Text.
Transcriptional Activity in Vitro.
Binding of nuclear proteins to the −174 bp region of the ancestral IL6 promoter sequence or the variant C allele (IL6 −187 to −163: 5′-AGT TGT GTC TTG C[G/C]A TGC TAA AGG A-3′) (Operon Technologies) was quantified by electrophoretic mobility shift assay (Cell Lytic NuCLEAR, Sigma) of cells treated 5 min with vehicle, 1 μM NE, 10 μM NE, or 2 ng/mL PMA (PKC-inducer as specificity control for PKA activation). Allele-specific chromatin immunoprecipitation (ChIP) was performed on rs1800795 heterozygous cells using ChIP-IT Express (Active Motif) and anti-GATA1 IgG (Active Motif) followed by real-time PCR detection of G vs. C allele DNA sequences (Applied Biosystems Custom Genotyping Assay 4331349). Additional details are provided in SI Text.
IL6 Promoter Activity.
Transcriptional response of rs1800795 G and C allele promoters was analyzed using luciferase reporter assays (QuantiGlo, Promega) of protein from 5 × 105 cells transfected with 1 μg of pIL6-174G or pIL6-174C (SI Text) and treated for 3–18 h with vehicle or NE. Role of the β-adrenoreceptor/cAMP/PKA signaling pathway in mediating NE effects was assessed by 15 min pretreatment with the β-adrenergic antagonist propranolol (10 μM) or protein kinase A (PKA) antagonist KT5720 (1 μM), or by replacing NE with 1 mM of the PKA activator db-cAMP (all from Sigma). Role of GATA1 in NE activation of the IL6 promoter was tested by siRNA knock-down of GATA1 mRNA beginning 24 h before stimulation and luciferase assay using GATA factor-specific reporter constructs (Panomics). SI Text provides additional details.
Transcriptional Activity in Vivo.
GATA1 activity in vivo was assayed by TELiS bioinformatics analysis (17) (http://www.telis.ucla.edu) of promoters showing empirical change in activity as a function of socio-environmental conditions. Sampling conditions, microarray assays, and bioinformatic analyses are detailed in SI Text. Briefly, adversity-sensitive genes were identified using microarray gene expression profiling, and promoters of those genes were tested for over-representation of GATA1 TFBMs as previously described (17) (TRANSFAC V$GATA1_01 position-specific weight matrix, −300 bp core promoter sequence, mat_sim ≥ 0.90). Differential gene transcription was assayed by Affymetrix high-density oligonucleotide arrays in (i) CNS prefrontal cortex and CD11b+ splenocytes from adult male C57BL/6 mice (Charles River Laboratories) subject to three daily cycles of 2-h exposure to social threat vs. nonthreatening control conditions (58), and (ii) human ovarian carcinoma tissues resected from patients confronting high vs. low levels of social adversity as previously described (42). Procedures were approved by Institutional Animal Care and Use Committees and study site Institutional Review Boards (42).
Molecular Epidemiology.
All available genomic DNA samples from the MacArthur Study of Successful Aging (43) were genotyped for rs1800795 using real-time PCR as described above. Primary analyses focused on Caucasian individuals at the North Carolina site (n = 184; Table S5) to mitigate ethnic stratification and site-related confounding. Primary results were confirmed in analyses of the pooled Caucasian sample from all three study sites (n = 381; SI Text). Depressive symptoms were measured by the Center for Epidemiologic Studies Depression scale (CES-D ≥ 16) (59) and the Hopkins Symptom Check List Depression scale (SCL-D >90th percentile) (60). Total and cause-specific mortality analyses were based on National Death Index (NDI) records (37% confirmed mortality), with lifespan censored at the date of last NDI search for living participants (all individuals ascertained >10 years). Mortality risk was analyzed using Cox proportional hazards regression (61) with sex-specific baseline hazard functions and regressors controlling for age at study entry, body mass index, household economic condition (satisfactory/unsatisfactory), smoking history (never, ex-, current) and alcohol consumption (never, occasional, moderate, heavy) (43). Genotype × Depression interaction was modeled as a product term in analyses containing Genotype and Depression main effects (62). Competing hazards analysis of cause-specific mortality (61) classified ICD-9/10 primary cause as inflammation related (cardiovascular disease, Alzheimer's disease, cancer) vs. not. Plasma C-reactive protein (CRP) was quantified by high-sensitivity ELISA (20) and classified as high at ≥ 3 mg/L. Additional details are given in SI Text. All research was approved by study site Institutional Review Boards (43).
Supplementary Material
Acknowledgments
This research was supported by the National Institute of Health Grants AG010415, AG028748, AG107265, AI052737, CA116778, CA110793, CA109298, and MH046801; the UCLA Norman Cousins Center; and the James L. Pendleton Charitable Trust.
Footnotes
* This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
Data deposition: Microarray data were deposited in Gene Expression Omnibus as series GEO Series GSE16661, GSE16660, and GSE9116.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911515107/DCSupplemental.
References
- 1.Robinson GE. Genomics. Beyond nature and nurture. Science. 2004;304:397–399. doi: 10.1126/science.1095766. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez LM, Blazer DG. Washington, DC: National Academies Press; 2006. [PubMed] [Google Scholar]
- 3.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: Joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 4.Finch CE. The Biology of Human Longevity: Inflammation, Nutrition, and Aging in the Evolution of Lifespans. Burlington, MA: Academic Press; 2007. [Google Scholar]
- 5.Meaney MJ, Szyf M, Seckl JR. Epigenetic mechanisms of perinatal programming of hypothalamic-pituitary-adrenal function and health. Trends Mol Med. 2007;13:269–277. doi: 10.1016/j.molmed.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Cole SW, et al. Social regulation of gene expression in human leukocytes. Genome Biol. 2007;8:R189. doi: 10.1186/gb-2007-8-9-r189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sloan EK, et al. Social stress enhances sympathetic innervation of primate lymph nodes: Mechanisms and implications for viral pathogenesis. J Neurosci. 2007;27:8857–8865. doi: 10.1523/JNEUROSCI.1247-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bierhaus A, et al. A mechanism converting psychosocial stress into mononuclear cell activation. Proc Natl Acad Sci USA. 2003;100:1920–1925. doi: 10.1073/pnas.0438019100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey M, Smale ST. Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 10.Buckland PR. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim Biophys Acta. 2006;1762:17–28. doi: 10.1016/j.bbadis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Hudson TJ. Wanted: Regulatory SNPs. Nat Genet. 2003;33:439–440. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- 12.Knight JC. Regulatory polymorphisms underlying complex disease traits. J Mol Med. 2005;83:97–109. doi: 10.1007/s00109-004-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prokunina L, Alarcón-Riquelme ME. Regulatory SNPs in complex diseases: Their identification and functional validation. Expert Rev Mol Med. 2004;6:1–15. doi: 10.1017/S1462399404007690. [DOI] [PubMed] [Google Scholar]
- 14.Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847–856. doi: 10.1038/35015718. [DOI] [PubMed] [Google Scholar]
- 15.Caspi A, et al. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 16.Cole SW. Social regulation of human gene expression. Curr Dir Psychol Sci. 2009;18:132–137. doi: 10.1111/j.1467-8721.2009.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole SW, Yan W, Galic Z, Arevalo J, Zack JA. Expression-based monitoring of transcription factor activity: The TELiS database. Bioinformatics. 2005;21:803–810. doi: 10.1093/bioinformatics/bti038. [DOI] [PubMed] [Google Scholar]
- 18.Stepanova M, Tiazhelova T, Skoblov M, Baranova A. Potential regulatory SNPs in promoters of human genes: A systematic approach. Mol Cell Probes. 2006;20:348–358. doi: 10.1016/j.mcp.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Ershler WB, Keller ET. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu Rev Med. 2000;51:245–270. doi: 10.1146/annurev.med.51.1.245. [DOI] [PubMed] [Google Scholar]
- 20.Gruenewald TL, Seeman TE, Ryff CD, Karlamangla AS, Singer BH. Combinations of biomarkers predictive of later life mortality. Proc Natl Acad Sci USA. 2006;103:14158–14163. doi: 10.1073/pnas.0606215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pepys MB, Hirschfield GM. C-reactive protein: A critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 23.Alley DE, et al. Socioeconomic status and C-reactive protein levels in the US population: NHANES IV. Brain Behav Immun. 2006;20:498–504. doi: 10.1016/j.bbi.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Banks J, Marmot M, Oldfield Z, Smith JP. Disease and disadvantage in the United States and in England. JAMA. 2006;295:2037–2045. doi: 10.1001/jama.295.17.2037. [DOI] [PubMed] [Google Scholar]
- 25.Danese A, et al. Elevated inflammation levels in depressed adults with a history of childhood maltreatment. Arch Gen Psychiatry. 2008;65:409–415. doi: 10.1001/archpsyc.65.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steptoe A, Willemsen G, Owen N, Flower L, Mohamed-Ali V. Acute mental stress elicits delayed increases in circulating inflammatory cytokine levels. Clin Sci (Lond) 2001;101:185–192. [PubMed] [Google Scholar]
- 27.Taylor SE, Lehman BJ, Kiefe CI, Seeman TE. Relationship of early life stress and psychological functioning to adult C-reactive protein in the coronary artery risk development in young adults study. Biol Psychiatry. 2006;60:819–824. doi: 10.1016/j.biopsych.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Fishman D, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. dbSNP 2009 rs1800795 dbSNP Database Record http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=1800795. Last accessed October 7, 2009.
- 31.Briest W, et al. Norepinephrine-induced interleukin-6 increase in rat hearts: Differential signal transduction in myocytes and non-myocytes. Pflugers Arch. 2003;446:437–446. doi: 10.1007/s00424-003-1043-x. [DOI] [PubMed] [Google Scholar]
- 32.Briest W, Elsner C, Hemker J, Müller-Strahl G, Zimmer HG. Norepinephrine-induced expression of cytokines in isolated biventricular working rat hearts. Mol Cell Biochem. 2003;245:69–76. doi: 10.1023/a:1022861609896. [DOI] [PubMed] [Google Scholar]
- 33.Bürger A, Benicke M, Deten A, Zimmer HG. Catecholamines stimulate interleukin-6 synthesis in rat cardiac fibroblasts. Am J Physiol Heart Circ Physiol. 2001;281:H14–H21. doi: 10.1152/ajpheart.2001.281.1.H14. [DOI] [PubMed] [Google Scholar]
- 34.Chrousos GP, Gold PW. The concepts of stress and stress system disorders. Overview of physical and behavioral homeostasis. JAMA. 1992;267:1244–1252. [PubMed] [Google Scholar]
- 35.Cohen S, Doyle WJ, Baum A. Socioeconomic status is associated with stress hormones. Psychosom Med. 2006;68:414–420. doi: 10.1097/01.psy.0000221236.37158.b9. [DOI] [PubMed] [Google Scholar]
- 36.Esler M, et al. The peripheral kinetics of norepinephrine in depressive illness. Arch Gen Psychiatry. 1982;39:295–300. doi: 10.1001/archpsyc.1982.04290030035006. [DOI] [PubMed] [Google Scholar]
- 37.Janicki-Deverts D, et al. Socioeconomic status is related to urinary catecholamines in the Coronary Artery Risk Development in Young Adults (CARDIA) study. Psychosom Med. 2007;69:514–520. doi: 10.1097/PSY.0b013e3180f60645. [DOI] [PubMed] [Google Scholar]
- 38.McEwen BS. Physiology and neurobiology of stress and adaptation: Central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- 39.Seeman T, Berkman LF, Blazer D. Social ties and support and neuroendocrine function: The MacArthur studies of successful aging. Ann Behav Med. 1994;16:95–106. [Google Scholar]
- 40.Sapolsky RM. Why Zebras Don't Get Ulcers: A Guide to Stress, Stress-Related Diseases, and Coping. New York: Freeman; 1994. [Google Scholar]
- 41.Weiner H. Perturbing the Organism: The Biology of Stressful Experience. Chicago: University of Chicago Press; 1992. [Google Scholar]
- 42.Lutgendorf SK, et al. Depression, social support, and beta-adrenergic transcription control in human ovarian cancer. Brain Behav Immun. 2009;23:176–183. doi: 10.1016/j.bbi.2008.04.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berkman LF, et al. High, usual and impaired functioning in community-dwelling older men and women: Findings from the MacArthur Foundation Research Network on Successful Aging. J Clin Epidemiol. 1993;46:1129–1140. doi: 10.1016/0895-4356(93)90112-e. [DOI] [PubMed] [Google Scholar]
- 44.Berkman LF, Kawachi I. Social Epidemiology. New York: Oxford University Press; 2000. [Google Scholar]
- 45.Seeman TE. Social ties and health: The benefits of social integration. Ann Epidemiol. 1996;6:442–451. doi: 10.1016/s1047-2797(96)00095-6. [DOI] [PubMed] [Google Scholar]
- 46.Panksepp J. Affective Neuroscience. New York: Oxford University Press; 1998. [Google Scholar]
- 47.Rivera-Chavez FA, Peters-Hybki DL, Barber RC, O'Keefe GE. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock. 2003;20:218–223. doi: 10.1097/01.shk.0000079425.52617.db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275:18138–18144. doi: 10.1074/jbc.M000379200. [DOI] [PubMed] [Google Scholar]
- 49.Chen CH, Zhang DH, LaPorte JM, Ray A. Cyclic AMP activates p38 mitogen-activated protein kinase in Th2 cells: Phosphorylation of GATA-3 and stimulation of Th2 cytokine gene expression. J Immunol. 2000;165:5597–5605. doi: 10.4049/jimmunol.165.10.5597. [DOI] [PubMed] [Google Scholar]
- 50.Klein-Hessling S, et al. Protein kinase A regulates GATA-3-dependent activation of IL-5 gene expression in Th2 cells. J Immunol. 2003;170:2956–2961. doi: 10.4049/jimmunol.170.6.2956. [DOI] [PubMed] [Google Scholar]
- 51.Lee HJ, et al. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel MD, Zhang DH, Ray P, Ray A. Activation of the interleukin-5 promoter by cAMP in murine EL-4 cells requires the GATA-3 and CLE0 elements. J Biol Chem. 1995;270:24548–24555. doi: 10.1074/jbc.270.41.24548. [DOI] [PubMed] [Google Scholar]
- 53.Tremblay JJ, Viger RS. Transcription factor GATA-4 is activated by phosphorylation of serine 261 via the cAMP/protein kinase a signaling pathway in gonadal cells. J Biol Chem. 2003;278:22128–22135. doi: 10.1074/jbc.M213149200. [DOI] [PubMed] [Google Scholar]
- 54.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 55.DeMichele A, et al. Interleukin-6 -174G→C polymorphism is associated with improved outcome in high-risk breast cancer. Cancer Res. 2003;63:8051–8056. [PubMed] [Google Scholar]
- 56.Moffitt TE, Caspi A, Rutter M. Strategy for investigating interactions between measured genes and measured environments. Arch Gen Psychiatry. 2005;62:473–481. doi: 10.1001/archpsyc.62.5.473. [DOI] [PubMed] [Google Scholar]
- 57.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stark JL, et al. Social stress induces glucocorticoid resistance in macrophages. Am J Physiol Regul Integr Comp Physiol. 2001;280:R1799–R1805. doi: 10.1152/ajpregu.2001.280.6.R1799. [DOI] [PubMed] [Google Scholar]
- 59.Radloff LS. The CES-D scale: A self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1:386–401. [Google Scholar]
- 60.Derogatis LR, Lipman RS, Rickels K, Uhlenhuth EH, Covi L. The Hopkins Symptom Checklist (HSCL). A measure of primary symptom dimensions. Mod Probl Pharmacopsychiatry. 1974;7:79–110. doi: 10.1159/000395070. [DOI] [PubMed] [Google Scholar]
- 61.Cox DR, Oakes D. Analysis of Survival Data. London: Chapman & Hall; 1984. [Google Scholar]
- 62.McCullagh P, Nelder JA. Generalized Linear Models. London: Chapman & Hall; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.