Abstract
Repair of DNA double-strand-breaks (DSBs) by homologous recombination is crucial for cell proliferation and tumor suppression. However, despite its importance, the molecular intermediates of mitotic DSB-repair remain undefined. The double Holliday Junction (dHJ), presupposed to be the central intermediate for more than 25 years1, has only been identified during meiotic recombination2. Moreover, evidence has accumulated for alternative, dHJ-independent mechanisms3–6, raising the possibility that dHJs are not formed during DSB-repair in mitotically cycling cells. Here we identify intermediates of DSB-repair using a budding yeast assay system designed to mimic physiological DSB repair. This system utilizes diploid cells and provides the possibility for allelic recombination either between sister-chromatids or between homologs, as well as direct comparison with meiotic recombination at the same locus. In mitotically cycling cells, we detect inter-homolog Joint Molecule (JM) intermediates whose size and strand-composition are identical to the canonical dHJ structures observed in meiosis2. However, in contrast to meiosis, JMs between sister chromatids form in preference to those between homologs. Moreover, JMs appear to represent a minor pathway of DSB repair in mitotic cells, being detected at ~10-fold lower levels (per DSB) than during meiotic recombination. Thus, although dHJs are identified as intermediates of DSB-promoted recombination in both mitotic and meiotic cells, their formation is distinctly regulated according to the specific dictates of the two cellular programs.
To identify intermediates of mitotic recombination, an inducible site-specific DSB assay system was constructed by modifying the HIS4LEU2 locus, which has been used extensively to characterize meiotic recombination intermediates in Saccharomyces cerevisiae (Fig. 1a)7. To better model physiological DSB-repair in mammalian cells, three features were incorporated. First, we used diploid strains, rather than the haploids typically utilized in yeast DSB-repair studies. Second, we specified that chromosome breakage should be induced relatively inefficiently such that an intact sister-chromatid repair template is available in addition to the uncut homolog templates. The commonly employed HO-endonuclease normally cleaves with high efficiency, effectively precluding inter-sister recombination8. In our hands, the I-Sce I endonuclease9 cleaves much less efficiently so that an intact sister-chromatid will generally be available for repair (Fig. 1b,c). A single I-Sce I recognition site was introduced into one homolog of a diploid strain at the location where meiotic DSBs occur (Fig. 1a); and an SCEI gene under the control of the galactose-inducible GAL1 promoter10 was integrated elsewhere in the genome. Third, as described below, we monitored DSB-repair intermediates during a period when both homolog and sister templates are available for repair. Intermediates were monitored using a series of gel electrophoresis and Southern hybridization assays analogous to those used to study meiotic recombination (Fig. 1b and Fig. 2)11,2.
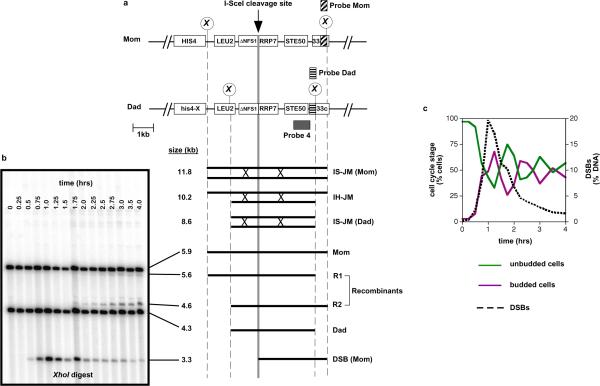
Figure. 1. Inducible DSB-repair system.
a, Map of the HIS4LEU2-SceI locus showing diagnostic restriction sites and probe positions. DNA species detected following Southern hybridization are shown below. Lollipops indicate XhoI restriction site polymorphisms. Diploid strains contain the I-Sce I recognition site on either the Mom or the Dad homolog. IS-JM, inter-sister joint molecule; IH-JM, inter-homolog joint molecule.
b, Image of one-dimensional gel hybridized with Probe 4 showing the DNA species detailed in (a). JMs are detected using two-dimensional gel analysis (see Fig. 2).
c, Time course analysis of DSB formation and repair in cycling diploid cells following DSB-induction. Sampling intervals are 15 minutes. Cell cycle stage was assessed microscopically.
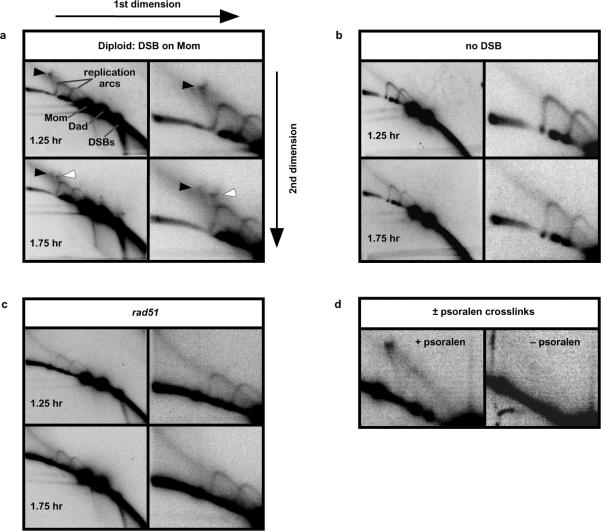
Fig. 2. Detection of Joint Molecules during DSB-repair.
a, Images of native/native 2D gels hybridized with Probe 4. Panels on the right-hand side show magnifications of the JM region. Black and white arrows highlight the two JM species.
b–d, JM signals are dependent on DSB-induction, the Rad51 protein and stabilization by psoralen crosslinking.
Cell cultures were synchronized in G1 and then transferred to fresh media to reinitiate growth, with addition of galactose to induce I-Sce I expression. After 45 minutes, I-Sce I expression was repressed by the addition of glucose. Under these conditions, DSBs appear within 30 minutes of I-Sce I induction and reach maximum levels after one hour, coincident with a period where a majority of cells are in the S and G2/M phases of the cell cycle (Fig. 1c). Thus, many DSBs are occurring in cells containing both homolog and sister-chromatid repair templates. Moreover, the maximum DSB level, ~20% of all chromatids, implies that breakage of both sister-chromatids in S/G2 cells should be rare (~4% of cells), i.e. an intact sister-chromatid will nearly always be available for repair. Most DSBs are repaired within 2–3 hrs of induction and their disappearance is concurrent with the appearance of inter-homolog recombinant products (Fig. 1b and 4c. Inter-homolog recombinants are further analyzed in Supplemental Figs. S2 and S3).
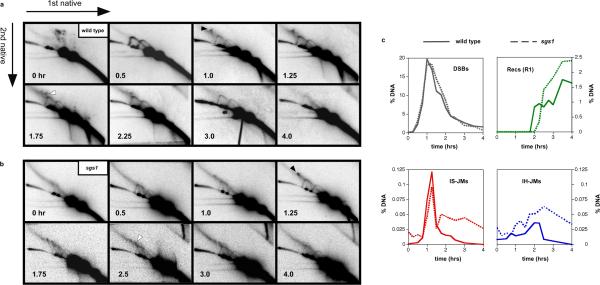
Fig. 4. Temporal analysis of JMs in wild-type and sgs1 cells.
a and b, 2D analysis of JM formation over the time-course of DSB-repair in wild-type and sgs1 strains. Intersister JMs and interhomolog JMs are highlighted by black and white arrows, respectively.
c, Quantitation of DSBs, JMs and recombinants (Recs). DSBs and Recs were quantitated from 1D gel analysis. JMs were quantitated from 2D gels. See Supplemental Figs. S2 and S3 for additional analysis and interpretation of recombinants in this system.
To detect JM intermediates, cell samples were first treated with psoralen to produce DNA interstrand crosslinks that stabilize such structures12,2. DNA was then extracted and analyzed using native/native two-dimensional (2D) electrophoresis, which identifies JMs by virtue of their branched structure (Fig. 2a)12. For diploid cells carrying an I-Sce I cutting site on the “Mom” homolog (Fig. 1a), this analysis reveals transient formation of two distinct branched molecules, in addition to the Y-arcs formed by replicating DNA (Fig. 2a). Formation of these branched species is dependent on both DSB-induction (Fig. 2b) and the DNA strand-exchange protein, Rad51 (Fig. 2c), indicating that they are intermediates of DSB-repair by homologous recombination. Moreover, recovery of these intermediates is dependent on psoralen cross-linking indicating the presence of migratable strand-exchange junctions (Fig. 2d).
The gel migration patterns of the two intermediates are consistent with their being, respectively, inter-sister and inter-homolog JMs. Inter-sister strand-exchange is expected to form a “Mom+Mom” JM of ~11.8 kb (Fig. 1a and indicated by the black arrowhead in Fig. 2a), whereas inter-homolog strand-exchange will form a “Dad+Mom” JM of ~10.2 kb (white arrowhead in Fig. 2a,). Since DSBs occur only on the Mom chromosome, no intersister “Dad+Dad” JMs are expected and none were observed.
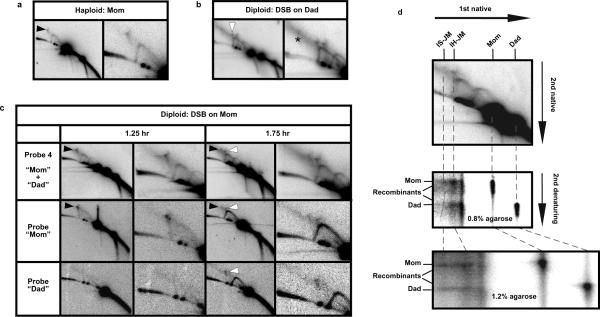
These JM assignments were confirmed in three ways. (1) JM formation was monitored in cycling haploid cells, where Mom+Mom inter-sister JMs are the only possible intermediates. As expected, only the larger of the two JM species seen in diploid cells was detected (Fig. 3a). (2) JMs were also monitored in diploid cells carrying the I-Sce I recognition site on the Dad homolog instead of on Mom (Fig. 3b). In this case, the Mom+Mom inter-sister JM signal was absent. However, the putative interhomolog-JM signal was seen and a smaller JM species was also detected, consistent with the formation of JMs between Dad sister-chromatids (indicated by a caret in Fig. 3b; also see Fig. 1a). (3) The chromatid composition of detected JMs was analyzed by sequentially hybridizing 2D Southern blots with probes specific to either the Mom or Dad homologs (Fig. 3c; probe positions shown in Fig. 1a). This analysis was performed on samples from diploid cells carrying the I-Sce I site on the Mom homolog. At an early time point (1.25 hrs), only the larger JM is detected, and it hybridizes exclusively to the Mom-specific probe indicating that it contains only Mom chromatids and thus is an inter-sister Mom+Mom JM, as inferred above (Fig. 3c; “Probe Mom”). After 1.75 hrs, the smaller JM (seen in Fig. 2a, above) is also detected and it hybridizes to both Mom- and Dad-specific probes, as expected for a bi-parental inter-homolog JM (Fig. 3c).
Fig. 3. Analysis of Joint Molecule composition.
a, 2D gel analysis of JMs during DSB-repair in cycling haploid cells.
b, 2D gel analysis of JMs in diploid cells carrying the I-Sce I recognition site on the Dad homolog. The smaller Dad+Dad intersister JM is highlighted by a caret. The asterisk highlights a high molecular weight species of unknown identity.
c, Successive hybridizations of 2D gels with Probe 4 and homolog-specific probes. Mom+Mom intersister JMs and Mom+Dad interhomolog JMs are highlighted by black and white arrows, respectively.
d, Native/denaturing 2D gel analysis of the strand composition of JMs. The corresponding native/native 2D gel is shown in the top panel to align JM species with their component strands in the lower panels. The same JM sample was analyzed using two different agarose concentrations in the second dimension. Component strands are more sharply resolved in the 1.2% gel and show that interhomolog JMs comprise primarily parental length strands. The lower detection limit for this experiment is estimated to be 28% recombinant strands, i.e. 28% of interhomolog-JMs could be sHJs and go undetected by this assay.
The structure of mitotic JMs was further investigated using native/denaturing 2D gel analysis in which psoralen cross-links are removed following electrophoresis in the first dimension gel, and component single-strands are then separated by denaturing electrophoresis in the second dimension (Fig. 3d)2. For inter-homolog JMs, in which parental duplexes differ in size, this analysis can distinguish between JMs involving odd or even numbers of Holliday junctions: JMs with even numbers of junctions, such as dHJs, contain only parental-length strands; in contrast, single Holliday junctions (sHJs) (or JMs with odd numbers of junctions) are comprised of equal numbers of parental and crossover-length strands (Supplemental Fig. S4). This analysis reveals that interhomolog-JMs comprise primarily parental length strands implying that most molecules contain an even number of HJs, presumably two (Fig. 3d). We infer that mitotic inter-homolog-JMs are most likely dHJs. The fact that recovery of mitotic interhomolog-JMs is dependent on psoralen cross-linking (Fig. 2d) further supports this inference because, while dHJs are dependent on psoralen for stabilization, analogous sHJ structures are not13. Because inter-sister-JMs contain four identical length strands, component strand analysis cannot distinguish whether they contain odd or even numbers of junctions.
As noted above, the two JMs species detected in diploids form at different times during the course of DSB-repair. Inter-sister JMs appear first and levels peak ~1.25 hrs after DSB-induction, i.e. just 15 minutes after the peak of DSB formation (Fig 4a,c; for normalized data see Supplemental Fig S5). We infer that homology search and DNA strand-exchange between sister-chromatids occurs rapidly following chromosome breakage. In contrast, inter-homolog JMs peak almost an hour later, between 2.0 to 2.25 hrs (a time when the ensuing cell division would normally have occurred; Fig. 4c and SUPPLEMENT 4d). This contrasts with meiotic recombination in which inter-sister and inter-homolog-JMs form with essentially identical timing, ~1.5 hrs after DSB formation (see Supplemental Fig. S6)7.
A straightforward explanation for the later appearance of inter-homolog JMs is that they arise from DSBs induced in the early stages (G1 or early-S) of the second cell cycle, when a sister-chromatid repair template is no longer present. This scenario implies that inter-homolog JMs are essentially never formed if a sister-template is available. An alternative, explanation is that late arising inter-homolog JMs arise from DSBs induced during the G1 or early-S stages of the first cell cycle, but are not repaired until some later event(s) occur(s). In support of this idea, studies of spontaneous and X-ray induced gene conversion indicate that DSBs incurred in G1 are associated with crossing-over that occurs in the ensuing G2 phase14,15. Under this scenario, late appearance of inter-homolog JMs could be due to the fact that resection of DSB-ends, which is a prerequisite for homologous recombination, requires cyclin-dependent kinase activity and, thus, progression beyond G1 into the cell cycle16,17. It is also likely that homology search and stable DNA strand-exchange between homologs occurs more slowly than between sister chromatids because they are not already connected by cohesion18. Alternatively, or in addition, inter-homolog recombination could be actively suppressed until replication of the homolog template is completed. Such negative regulation of inter-homolog recombination could allow time for the homolog template to be replicated without interference from ongoing recombination.
Quantitative analysis reveals two important features regarding the levels of inter-sister and inter-homolog JMs (Fig. 4b). First, steady-state levels of inter-sister JMs peak at ~0.12% of hybridizing DNA compared to ~0.03% for inter-homolog-JMs, pointing to a preference for use of the sister template. In actuality, if inter-homolog-JMs arise from DSBs formed in G1/early S (above), then the sister template may always be utilized for repair during late-S/G2. The inter-sister bias revealed here at the level of JM intermediates is consonant with that previously inferred by measuring recombination products19,20,21. Furthermore, it is the opposite of the bias observed during meiotic recombination at the HIS4LEU2 locus, where inter-homolog JMs are favored over inter-sister JMs by ~5:111 (see Supplemental Fig. S6).
Second, although DSB levels induced in our mitotic assay occur at similar levels to those observed during meiosis at the same locus (~20%), the total level of detected JMs (inter-sister and inter-homolog combined) is ≥10-fold lower during mitotic DSB-repair than during meiotic recombination (Supplemental Fig. S6)11. During meiosis, about one half of DSBs at HIS4LEU2 are processed via JMs, all (or most) of which are specifically resolved into crossover products11,7,22. The remaining half are processed via a non-JM pathway, most likely synthesis-dependent strand annealing (SDSA)5,23, which yields intact duplexes that have not undergone crossing-over (“non-crossovers”). Thus, taken at face value, the relative paucity of JMs per DSB during mitotic repair implies that as many as 90% of events proceed via a non-JM pathway which, presumptively, yields non-crossover products.
The alternative possibility, that dHJs are the major intermediates of mitotic DSB-repair, could be true if we failed to detect the majority of JMs, e.g. if they migrate out of the assayed region, or if they have a much shorter lifespan relative to meiotic dHJs. If the former possibility were true, JM yield should increase when longer fragments are analyzed. However, we find that JMs are recovered at very similar levels for the 4.3 and 5.9 kb XhoI fragments (shown in Fig. 1a) as for 13.6 kb SacI fragments that span the HIS4LEU2-SceI locus (Supplemental Fig. S7), i.e. JM yield for the XhoI fragments is efficient. If the latter possibility were true, JM levels should increase when turnover-mediating factors are eliminated. In vitro, dHJs can be rapidly dissociated by helicase/topoisomerase ensembles: RecQ DNA helicase, BLM, type-I topoisomerase, TOPIIIα, and the specificity factor, RMI (RMI1+RMI2) in human23,24; and the orthologous Sgs1-Top3-Rmi1 in budding yeast (S. Kowalczykowski, personal communication). Rapid dissociation of mitotic JMs by Sgs1-Top3-Rmi1 could explain why detected levels are so much lower than during meiosis. To address this possibility, we analyzed JM formation in an sgs1 mutant diploid (Fig. 4b, c and Supplemental Figs. S2 and S5). Importantly, peak JM levels in sgs1 cells are only slightly higher than those measured in wild-type cells indicating that rapid turnover of JMs by Sgs1-Top3-Rmi1 cannot account for the low levels detected during mitotic DSB repair (Fig. 4c). However, JM resolution does appear to be defective in the absence of Sgs1 helicase activity, as indicated by the persistence of both inter-sister- and inter-homolog-JM signals. Taken together, these data support the inference that JMs represent a minor pathway of DSB-repair in mitotically cycling cells and provide compelling in vivo evidence for the “dHJ-dissolution” activity of the Sgs1-Top3-Rmi1 complex.
The stark differences between mitotic and meiotic JMs reflect the distinct logic for homologous recombination in the two cell types. During mitotic DSB-repair, use of the sister template minimizes alteration of genetic content (loss-of-heterozygosity); and preferential occurrence of non-crossovers minimizes deleterious effects that can result from crossing-over, such as chromosome rearrangements and missegregation21,24. In contrast, during meiosis, most DSBs are directed to homolog templates in order to facilitate pairing of parental homologs. Moreover, formation of dHJs and their resolution into crossovers is essential to direct segregation of homologs to opposite poles at the first meiotic division25.
Methods Summary
All strains are derived from SK1 strains NHY53 and NHY56 and are described in Supplemental Information. To synchronize cells in G1, cultures were diluted 1/100 and grown in YP-lactate (1% Bacto yeast extract, 2% Bacto peptone, 3% lactic acid, pH 5.5) for ~18 hrs at 30°C. Cells were then harvested and resuspended in fresh YP containing 2% galactose to induce I-Sce I expression. After 45 mins, I-Sce I expression was repressed by addition of 3% glucose. Cell samples were treated with psoralen and DNA was purified as previously described2. 1D and 2D gel analyses were as described2,7,11.
Methods
Strain Construction
The linker 5'-CGCGCGGCCTAGGGATAACAGGGTAATGGCGC-3', containing a single I-Sce I cleavage site (highlighted in bold), was inserted into the MluI sites of plasmids pNH16 (to create pMB34) and pNH93 (to create pMB7), which carry the “Mom” and “Dad” HIS4LEU2 loci, respectively7. The linker 5'-CGCGCGGCCGAGGGATAACAGGGTAATGGCGC-3', which contains a mutated I-Sce I recognition site that is not cleaved by I-Sce I in vivo, was inserted into the MluI site of plasmid pNH93 (to create pMB8). Underlined sequences in the linkers indicate the polymorphic AvrII restriction site analyzed in Supplemental Fig. S3. The resulting plasmids were cut with SacI and PstI, to release the HIS4LEU2 constructs from the vector backbone, and then transformed into precursor strains NHY53 and NHY56 (Supplemental Table S1) using standard methods (http://home.cc.umanitoba.ca/~gietz/). Correct integration at the native HIS4 locus was conformed by Southern analysis with Probe 4. Plasmid pWY20310 contains a pGAL1 driven SCEI gene marked with URA3 inserted into the LYS2 gene. To target integration of this construct to the native LYS2 locus, pWY203 was cleaved with AflII prior to transformation.
The sgs1 allele used in this study was sgs1-ΔC795::hphMX4 and has been described previously11. sgs1-ΔC795 encodes a truncated Sgs1 protein26 lacking the conserved helicase and HRDC domains, which are essential for dHJ dissociation in vitro27,28. The sgs1-ΔC795 strain, which grows normally, was used in preference to an sgs1Δ strain, which grows slowly and synchronizes poorly (data not shown). rad51Δ::hisG is a complete deletion of RAD51 and has been described29.
Analysis of Cell Cycle Stage
To asses the synchrony of cultures and follow progression through the cell cycle, cell samples (0.5 mL) were fixed in 40% ethanol 0.1M sorbitol, stained with 4',6-diamidino-2-phenylindole and visualized microscopically with both fluorescence and bright-field illumination. Cell cycle stage (G1, S, G2 or M) was scored according to standard criteria. ~200 cells were analyzed for each time point.
DNA Analysis
Detailed methods for psoralen cross-linking, DNA extraction, gel electrophoresis and Southern analysis methods are described in Oh et al.30. For 1D gels, 2μg of genomic DNA was digested and analyzed for each time point; 6 μg of DNA was used for 2D gels. JM signals were quantified using a phosphorimager. For the JM component-strand analysis experiment in Fig. 3d, given that mitotic JMs are relatively rare, we first used preparative gel electrophoresis to obtain a sample enriched for these molecules. DNA fragments of the appropriate size were recovered from the preparative gel and then analyzed by native/denaturing 2D gel electrophoresis as described2,30.
Supplementary Material
Acknowledgements
We thank Robert Schiestl (UCLA) for plasmid pWY203 and Wolf Heyer for critical reading of the manuscript. This work was supported by NIH NIGMS grants GM025326 to N.K. and GM074223 to N.H.
References
- 1.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 2.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83(5):783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 3.Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 4.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Molecular and cellular biology. 1994;14(3):1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5(11):e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyer WD, Ehmsen KT, Solinger JA. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem Sci. 2003;28(10):548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106(1):59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 8.Haber JE. Uses and abuses of HO endonuclease. Methods Enzymol. 2002;350:141–164. doi: 10.1016/s0076-6879(02)50961-7. [DOI] [PubMed] [Google Scholar]
- 9.Monteilhet C, Perrin A, Thierry A, Colleaux L, Dujon B. Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990;18(6):1407–1413. doi: 10.1093/nar/18.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galli A, Schiestl RH. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics. 1998;149(3):1235–1250. doi: 10.1093/genetics/149.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh SD, et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130(2):259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell L, Byers B. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Analyt Biochem. 1983;130:527–535. doi: 10.1016/0003-2697(83)90628-0. [DOI] [PubMed] [Google Scholar]
- 13.Cromie GA, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127(6):1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roman H, Fabre F. Gene conversion and associated reciprocal recombination are separable events in vegetative cells of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America; 1983. pp. 6912–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee PS, et al. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 2009;5(3):e1000410. doi: 10.1371/journal.pgen.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431(7011):1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. The EMBO journal. 2004;23(24):4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11(12):991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 19.Fabre F, Boulet A, Roman H. Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae. Mol Gen Genet. 1984;195(1–2):139–143. doi: 10.1007/BF00332736. [DOI] [PubMed] [Google Scholar]
- 20.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132(2):387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29(Pt 2):196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 22.Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117(1):29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 23.Jessop L, Allers T, Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169(3):1353–1367. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beumer KJ, Pimpinelli S, Golic KG. Induced chromosomal exchange directs the segregation of recombinant chromatids in mitosis of Drosophila. Genetics. 1998;150(1):173–188. doi: 10.1093/genetics/150.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter N. Meiotic Recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Springer-Verlag; Heidelberg: 2006. pp. 381–442. [Google Scholar]
- 26.Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154(3):1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426(6968):870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, et al. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. The EMBO journal. 2005;24(14):2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69(3):457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 30.Oh SD, Jessop L, Lao JP, Lichten MJ, Hunter NH. Stabilization and Electrophoretic Analysis of Meiotic Recombination Intermediates. In: Keeney S, editor. Methods in Molecular Biology, Meiosis: Molecular and Genetic Methods. Humana Press; New York: 2009. pp. 209–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.