Abstract
The peptidoglycan layer is a vital component of the bacterial cell wall. The existing paradigm describes the peptidoglycan network as a static structure that is cross-linked predominantly by 4→3 transpeptide linkages. However, the non-classical 3→3 linkages predominate the transpeptide networking of the peptidoglycan layer of non-replicating M. tuberculosis 1,2. The molecular basis of these linkages, their significance to the physiology of the peptidoglycan layer, virulence and susceptibility of M. tuberculosis to drugs remain undefined. Here, we identify MT2594 (Rv2518c) as an L,D-transpeptidase that generates 3→3 linkages in M. tuberculosis. We show that the loss of this gene leads to altered colony morphology, loss of virulence and increased susceptibility to amoxicillin–clavulanate during the chronic phase of infection. This suggests that 3→3 cross-linking is vital to the physiology of the peptidoglycan layer. Although a functional homolog exists, expression of MT2594 is dominant throughout the growth phases of M. tuberculosis. The 4→3 transpeptide linkages are targeted by one of the most widely used classes of antibacterial drugs in human clinical use today, namely β-lactams. Recently, meropenem–clavulanate was shown to be effective against drug resistant M. tuberculosis 3. Our study suggests that a combination of an L,D-transpeptidase and a β-lactamase inhibitors could effectively target persisters during chronic phase of TB.
Keywords: M. tuberculosis; persister; L,D-transpeptidation; peptidoglycan remodeling
Tuberculosis (TB) continues to be a major public health threat around the world. The estimate that more lives may have been lost in 2009 due to TB than in any year in history is alarming 4. An increasing number of cases reporting infection with multi-(MDR) and extensively drug-resistant (XDR) strains of M. tuberculosis has diminished our capability to respond effectively against this threat. A recent study reporting high mortality rates of patients co-infected with HIV and XDR-TB illustrates the need for new drugs to treat TB 5. A major reason for emergence of drug resistance is thought to be poor compliance to treatment regimens as the current therapy requires a combination of drugs to be taken daily for 6 months or more 4. While >99% of M. tuberculosis bacilli are killed within two weeks, it takes the remainder of the therapy to effectively kill the surviving population 6. These bacilli, broadly defined as ‘persisters’, are able to transiently tolerate drugs. The phenomenon of persistence is poorly understood. In vitro models designed to mimic the physiology of persisters are based on exposure to nitric oxide and depletion of oxygen and nutrients as these conditions are thought to prevail in a persisting infection in vivo 7,8,9.
A higher percentage of E. coli bacilli are able to survive exposure to drugs at stationary phase and persist compared to exponential phase of growth 10. Little is known about changes in the cell wall during chronic phase of infection and whether it regulates persistence of M. tuberculosis in the host. Understanding the regulation of cell wall physiology and its consequences may enable us to effectively target and kill persisters by interfering with this process. The cell wall of M. tuberculosis accounts for up to 40% of cell’s dry mass compared to 5% and 10% in gram-positive and negative bacteria 11. It is approximated that the degree of peptidoglycan cross–linking is ~50% in E. coli 12 whereas it is ~70–80% in Mycobacterium spp. 13. Mycobacterial peptidoglycan layer is cross–linked with both 4→3 and 3→3 linkages 1. Recently it was shown that 80% of the transpeptide linkages in the peptidoglycan of M. tuberculosis at stationary phase are of the non–classical 3→3 type 2. In this study, we report identification of a gene MT2594(Rv2518c) that encodes an L,D–transpeptidase for synthesis of non–classical 3→3 cross-linkages and show that inactivation of the gene results in altered colony morphology, attenuation of persistence, and increased susceptibility to amoxicillin/clavulanate both in vitro and in the mouse model of TB.
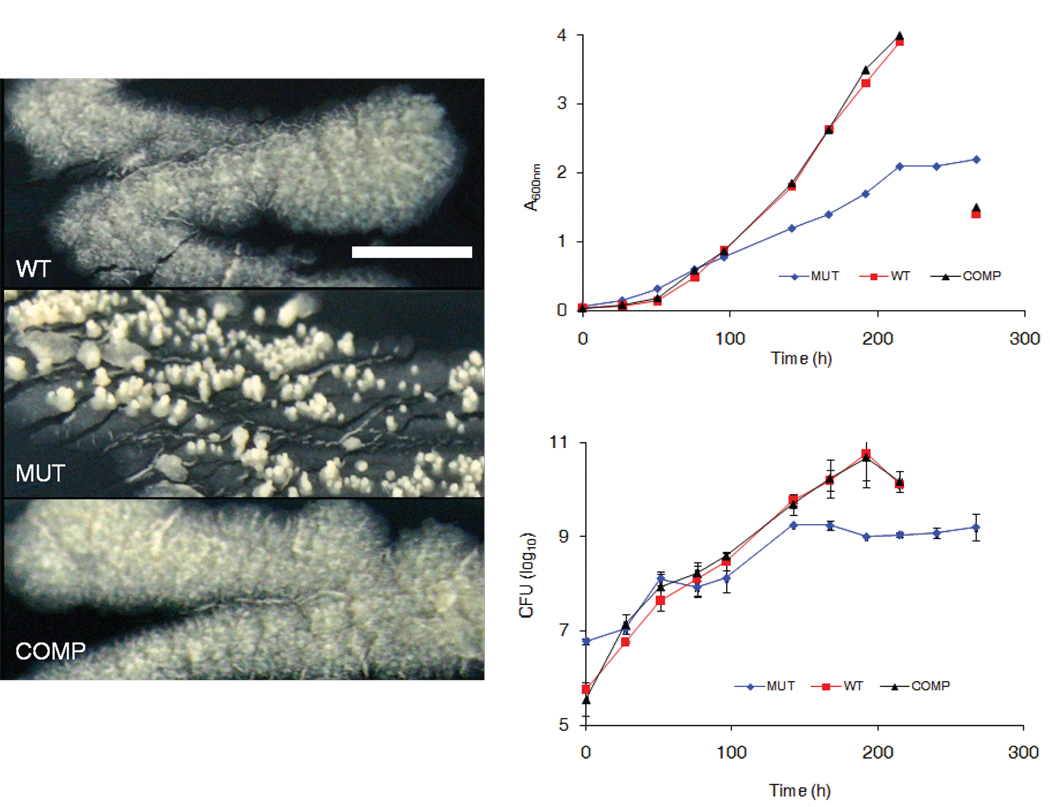
A mutant M. tuberculosis resulting from inactivation of gene MT2594 (Rv2518c), hereafter referred to ldtMt2, was isolated by screening a collection of 5,100 unique transposon insertion mutants for growth attenuation 14. Colonies of this mutant (ldtMt2::Tn) were smaller, smooth and had punctuated aerial growth rather than the typical large, rough and laterally diffuse morphology observed in the parent strain (Fig. 1a). In liquid broth, culture of the mutant strain reached lower optical and cell densities compared with the parental strain (Fig. 1b). The wild–type phenotypes were restored upon complementation of the mutant with a single copy of the gene. The ratio of colony forming units (CFU) of the mutant to WT at the beginning of the growth assay was 10:1 whereas after 192 h it was 1:100. This data shows that there was ~1,000 fold larger increase in the wild-type population over 192 h compared to the mutant strain. The doubling times of 18.1, 14.5 and 14.8 h were derived from CFU data for the mutant, wild-type and the complemented strains, respectively.
Figure 1.
Morphology and growth in vitro. (a) Morphologies of wild-type M. tuberculosis (WT), LdtMt2 mutant (MUT) and the complemented strain (COMP) on solid media after 21 days of growth at 37 °C. (scale = 1 cm).(b) Growth of wild-type M. tuberculosis (WT), LdtMt2 mutant (MUT) and the complemented strain (COMP) in Middlebrook 7H9 liquid medium at 37 °C. The decrease in optical density and CFU at the final time point for WT and COMP is due to clumped cultures.
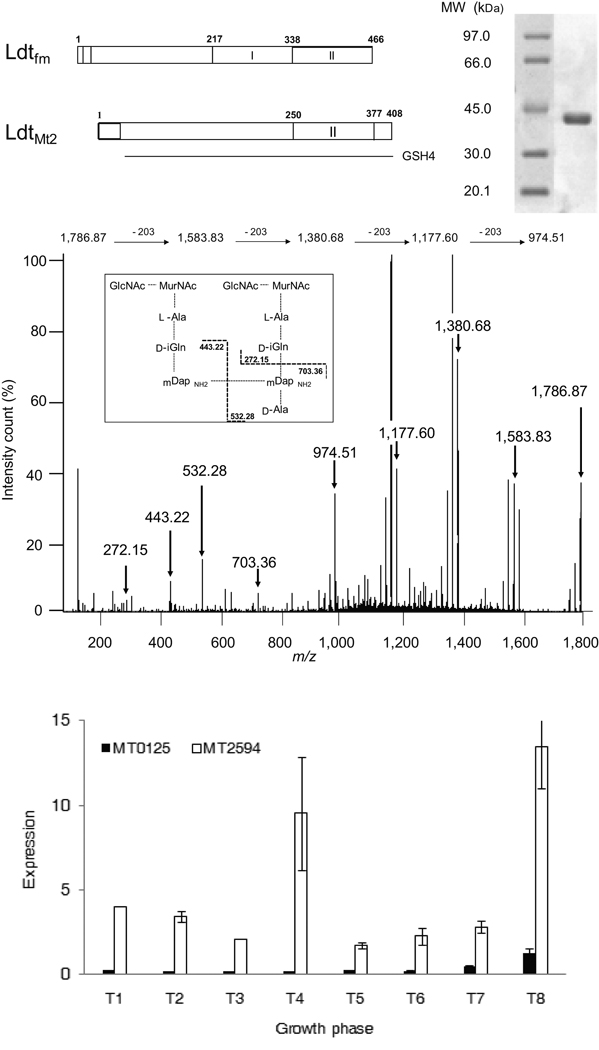
MT2594 (LdtMt2) is annotated as a hypothetical protein with an unknown function. The N–terminus contains a putative single transmembrane domain spanning residues (positions 20–42) anchoring the remainder of the protein that is predicted to protrude outside the cell membrane into the cell wall (www.ch.embnet.org). The C–terminal region of MT2594 is similar to the catalytic domain of the prototypic peptidoglycan L,D–transpeptidase from E. faecium (29% identity) including the active site cysteine residue within the invariant SHGC motif 15 (Fig. 2a). To determine if ldtMt2 encodes a functional L,D–transpeptidase, we produced a soluble fragment of the protein in E. coli that was purified and assayed for cross-linking activity (Fig. 2b). The substrate was a disaccharide–tetrapeptide monomer isolated from the peptidoglycan of C. jeikeium, which has the same structure as the predominant monomer of M. tuberculosis 16. Electrospray mass spectrometry analysis of the reaction products revealed the formation of a peptidoglycan dimer (m/z = 1,786.87 [M+H]1+, Fig. 2c) from two disaccharide–tetrapeptide monomers (m/z = 938.44 [M+H]1+). Tandem mass spectrometry of the dimer confirmed the presence of a 3→3 cross-link connecting two meso–diaminopimelic acids at the third position of the stem peptides (Fig. 2c). LdtMt2 did not catalyze formation of dimers with disaccharide–peptide containing a stem pentapeptide. Thus, LdtMt2 is specific for stem tetrapeptides as LdtMt1 2 and the prototypic L,D–transpeptidase from E. faecium 15. These results show that MT2594 is an L,D–transpeptidase that catalyzes formation of 3→3 peptidoglycan cross–links.
Figure 2.
Characterization of LdtMt2 from M. tuberculosis. (a) Domain composition of L,D-transpeptidases from E. faecium (Ldtfm) and M. tuberculosis (LdtMt2). Residues 250–377 of LdtMt2 share homology with the catalytic domain of Ldtfm (Domain II, 338–466). (b) Purification of a soluble fragment of LdtMt2 produced in E. coli. (c) Structure and inferred fragmentation pattern of the peptidoglycan dimer formed in vitro by LdtMt2. The ion at m/z 974.51 was generated by losses of the two GlcNAc-MurNAc residues following fragmentations of the ether links connecting the lactoyl group to the C–3 position of MurNAc. Loss of each sugar decreased the m/z by 203. Cleavage of additional peptide bonds from the ion at m/z 974.51 gave ions at 703.36, 532.28, 433.22 and 272.15 as indicated in the inset. (d) Transcription profile of LdtMt2 (MT2594) and LdtMt1 (MT0125). RNA isolated from wild-type Mtb cultures at growth phases T1 (A600nm=0.2), T2 (A600nm=0.5), T3 (A600nm=0.8), T4 (A600nm=0.9), T5 (A600nm=01.9), T6 (A600nm=3.0), T7 (2 days post A600nm =3.0) and T8 (3 days post clumping).
Further investigation revealed four putative paralogs of ldtMt2 in the genome of M. tuberculosis (MT0125, MT0202, MT0501 and MT1477). To gain an insight into the level of expression of the five paralogs, we performed quantitative RT–PCR analyses on eight RNA samples prepared from exponential and stationary phases of growth (Supplementary Fig. 2). The ldtMt2 mRNA was at least five fold more abundant than the combined expression of the four paralogs. Next, we assessed their functional relevance to L,D–transpeptidation. Mutants lacking MT0202, MT0501 and MT1477 have morphologies and growth phenotypes similar to that of the parent wild-type strain (Supplementary Fig. 3). We purified MT0202, MT0501 and MT1477 but did not detect any L,D–transpeptidase activity using the peptidoglycan precursor as the substrate. MT0125 of M. tuberculosis strain CDC1551 is identical to Rv0116c of strain H37Rv, a gene designated ldtMt1 that was recently shown to also encode a peptidoglycan L,D–transpeptidase 2. Between the two L,D–transpeptidases, MT2594 was expressed at a level at least 10 fold higher than that of ldtMt1 at all phases of growth (Fig. 2d). The morphology, growth and virulence deficient phenotype of our mutant indicates that low-level expression of ldtMt1 did not compensate for the loss of the L,D–transpeptidase activity of MT2594 although analysis of the peptidoglycan structure showed that 3→3 cross–linkages were synthesized by LdtMt1 in stationary phase culture of the mutant (Supplementary Fig. 4).
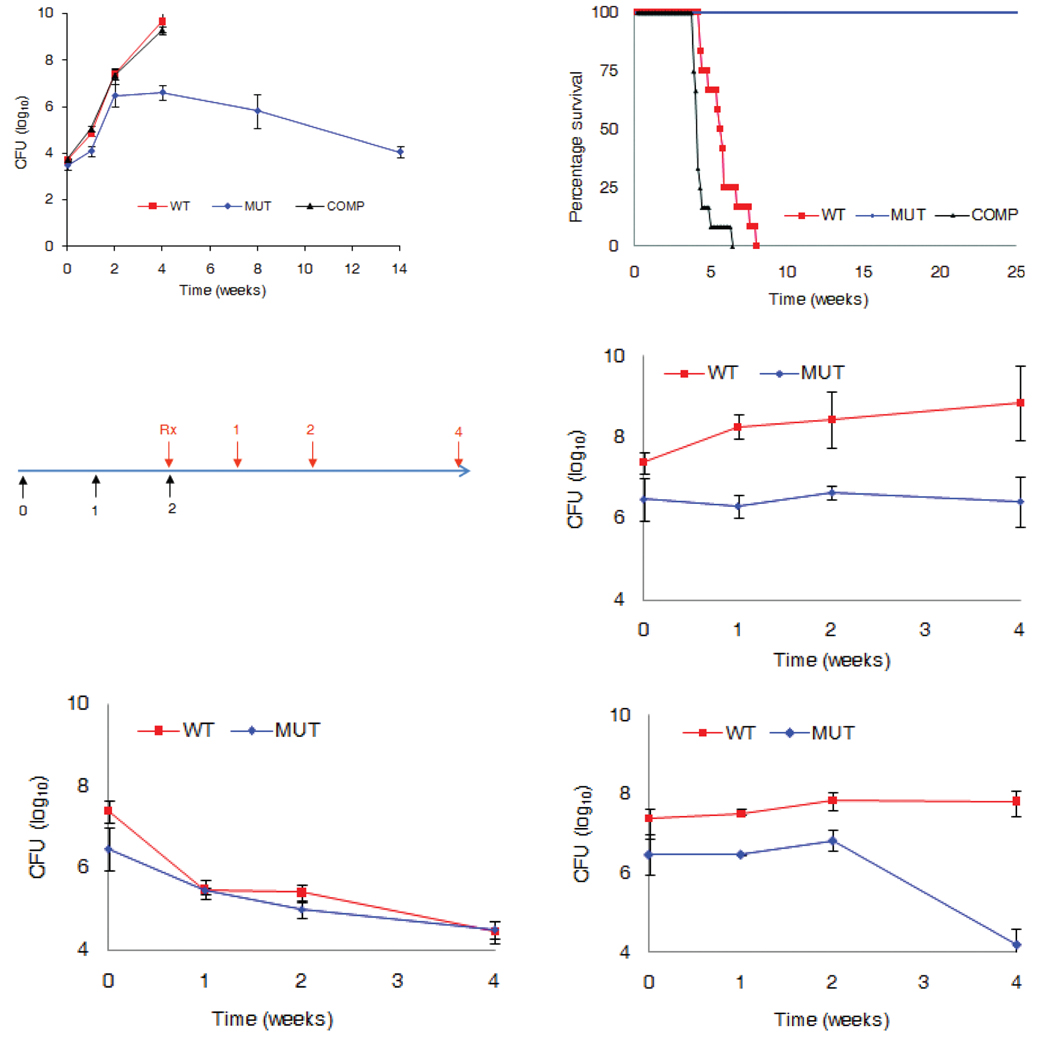
Next, we assessed if inactivation of ldtMt2 affected in vivo growth and virulence of M. tuberculosis. Approximately 3.5 log10 of CFUs of each strain was implanted in the lungs of three groups of immunocompetent BALB/c mice. The mutant established infection and exhibited a normal growth pattern during the first two weeks but discontinued proliferation beyond this stage of infection (Fig. 3a). The wild–type and the complemented strains proliferated rapidly until four weeks of infection, at which stage a heavy bacterial burden led to death of the animals. The median survival time for the wild–type and complement infected group were 38 and 30 days, respectively (Fig. 3b). The mice infected with the mutant strain did not die and signs of morbidity were not observed despite the presence of ca. 104 CFU in the lungs during the persistent stage of infection.
Figure 3.
Assessment of growth, virulence and susceptibility to amoxicillin in vivo. (a) Bacterial burden in the lungs of mice infected with wild-type M. tuberculosis (WT), LdtMt2 mutant (MUT) or the complemented (COMP) strain are shown. (b) Virulence of each strain was assessed by determining time-to-death following infection of mice, 12 per group, with the three strains. (c) Mice were infected with either WT or the MUT strain. Following two weeks of infection, mice were treated daily with either no drug placebo (d), or 25 mg kg−1 isoniazid (e), or 200 mg kg−1 amoxicillin and 50 mg kg−1 clavulanate (f). Bacterial burden was determined by enumerating CFU from the lungs of mice.
The non-classical 3→3 linkages comprise the majority of the transpeptide linkages in non–replicating M. tuberculosis 2. We hypothesized that the loss of LdtMt2 may compromise the mutant’s ability to adapt during the chronic phase of infection, a critical stage in the pathogenesis of TB. If the failure in adaptation was the result of a defect in peptidoglycan cross-linking by LdtMt2, another consequence could be an increased susceptibility to β–lactams. We tested this hypothesis by assessing susceptibility of the LdtMt2 mutant to amoxicillin. M. tuberculosis produces a β–lactamase that is inactivated by clavulanic acid 17. The MIC of the commercial clavulanic acid–amoxicillin combination (Augmentin®) was 1.2 and 0.14 µg ml−1 for wild-type and the mutant strain respectively. Loss of MT2594 did not alter susceptibility to isoniazid (MIC = 0.03 µg ml−1) and D–cycloserine (5 µg ml−1). Thus, loss of LdtMt2 was associated with increased susceptibility to amoxicillin in the presence of clavulanic acid that inhibited the β–lactamase. In a recent study authors assessed susceptibility of drug sensitive laboratory strain H37Rv and 13 XDR strains of M. tuberculosis to amoxicillin 3. While H37Rv was found to be resistant to amoxicillin–clavulanate, all 13 XDR strains were highly susceptible with an MIC ranging between 0.32 and 10 ug ml−1. It has been reported that amoxicillin–clavulanate lacks early bactericidal activity, a measure of effectiveness of the drugs during the first two days of treatment 18. However, amoxicillin–clavulanate has also been used to treat MDR–TB patients 19. An explanation for these observations is that amoxicillin–clavulanate lacks potency during the early phase but shows activity during the extended phase of treatment.
Next we assessed susceptibility of the mutant strain to amoxicillin in the mouse. Mice infected with either the wild–type M. tuberculosis or the mutant strain were treated daily with either phosphate buffered saline (PBS) as a placebo or 25mg kg−1 of isoniazid or 200 mg kg−1 amoxicillin in combination with 50 mg kg−1 clavulanate (Fig. 3c). Isoniazid was similarly effective against both wild-type and the mutant strain (Fig. 3e). Bacterial burden in mice infected with the wild–type strain treated with amoxicillin–clavulanate was similar to the group that received no treatment placebo (Fig. 3d, f). Amoxicillin–clavulanate was ineffective during the first two weeks of treatment in mice infected with the mutant as the bacterial burden remained unchanged. However, a decrease of more than 2 log10 in CFU was observed between two and four weeks of treatment illustrating that the mutant is selectively susceptible to amoxicillin–clavulanate during the chronic phase of infection (Fig. 3f).
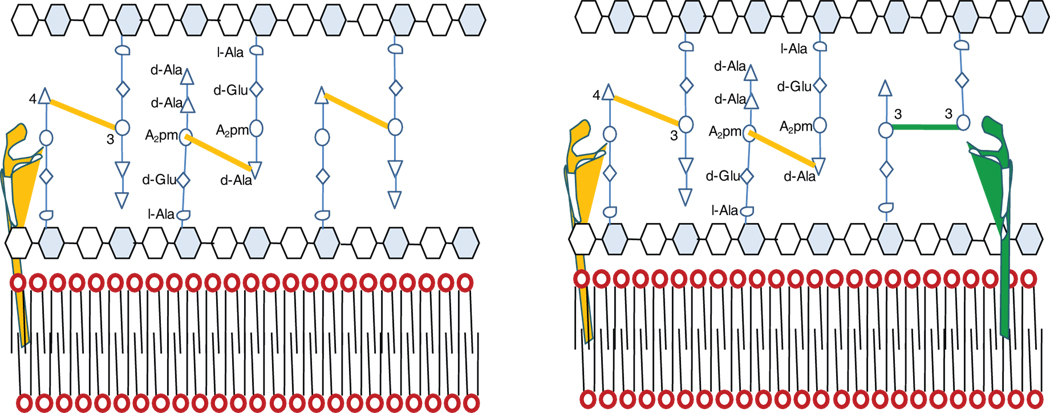
All bacteria possess an elaborate peptidoglycan layer in their cell wall. In E. coli the main inter–peptide cross-linking occurs between the penultimate D–alanine (D–ala) at the fourth position of the donor and mDAP at the third position of the acceptor. Formation of these 4→3 bond is catalyzed by the D,D-transpeptidases that are the essential target of β–lactam antibiotics 20. The drugs are structural analogs of the D–Ala–D–Ala extremity of the peptidoglycan precursors and act as suicide substrate in an acylation reaction 21. E. coli has served as the model organism for studying peptidoglycan metabolism but the existing paradigm built on this organism is incomplete. In this classical model the peptidoglycan layer is regarded as a static network involving polymerization of glycan chains and cross–linking of adjacent chains by formation of 4→3 peptide bonds (Fig. 4a) 22. Based on the data presented in this report and our recent findings 2 we propose a model describing the peptidoglycan layer as a dynamic structure whose inter–peptide linkages are altered as an adaptive response to a change in the environment and growth phase (Fig. 4b). The peptide chains of the peptidoglycan layer are linked with both 4→3 and 3→3 bonds.
Figure 4.
Proposed model for physiology of the peptidoglycan layer in M. tuberculosis. (a) classical model of peptidoglycan crosslinking containing 4→3 inter peptide bonds. (b) model based on recent data: the peptidoglycan is cross-linked with classical 4→3 and non-classical 3→3 inter peptide bonds. Both 4→3 (orange) and 3→3 (green) transpeptidases are involved in maintenance and remodeling of the peptidoglycan layer in the proposed model.
In addition to transpeptidases, endo– and carboxypeptidases are also present in bacteria and function to modify the peptidoglycan network 23. These enzymes have yet to be definitively identified in M. tuberculosis. A recent report showed gene pgdA to encode a N–deacetylase that is involved in modification of the peptidoglycan layer in L. monocytogenes 24. A putative homolog of pgdA exists in M. tuberculosis. This gene, MT1128 (Rv1096), has yet to be characterized and its in vivo function identified. Although MT2594 is a 3→3 transpeptidase, its loss and accompanying changes in the cross-linking may have pleiotropic effects on the metabolism of the peptidoglycan layer.
In this report we have shown a novel molecular basis of 3→3 linkages and their physiological role for viability and virulence of M. tuberculosis. We have also shown that L,D–transpeptidation is required to resist killing by amoxicillin–clavulanate and that inhibition of LdtMt2 alone may be sufficient to target the 3→3 linkages despite presence of redundancy. It may be inferred from our findings that both 3→3 and 4→3 trans-peptide linkages need to be destroyed to effectively kill M. tuberculosis. We have presented an unexploited enzyme in the pathway that has been targeted by one the most successful antibiotics in human clinical use, namely the β–lactams. Therefore, a regimen containing a combination of inhibitors of L,D–transpeptidase and β–lactamase, and a β–lactam may be able to kill M. tuberculosis by comprehensively destroying the peptidoglycan layer. As peptidoglycan layer is a vital structure of the bacterial cell wall, insight and applications resulting from studies in M. tuberculosis is likely broadly applicable to other bacteria.
METHODS
Bacterial strains and Culture Conditions
We used M. tuberculosis CDC1551, a clinical isolate, as the host strain to generate a transposon insertion mutant in MT2594 (ldtMt2::Tn) as described 14. This mutant carries a Himar1 transposon insertion at +872 base from the putative translation start site of the gene. Next, we generated a complemented strain by transforming the mutant with pGS202_2594. This is a single copy integrating plasmid based on pMH94 backbone 25, which we modified into a GATEWAY compatible destination vector (Invitrogen). We cloned a wild-type copy of MT2594 along with its promoter into this destination vector pGS202 to generate pGS202_2594. We verified genotypes of the strains by Southern blotting. We used Middlebrook 7H9 liquid medium supplemented with 0.2% glycerol, 0.05% Tween–80, 10% vol/vol oleic acid–albumin–dextrose–catalase (OADC) and 50 ug ml−1 cycloheximide for in vitro growth, and Middlebrook 7H11 solid medium (Becton-Dickinson) for enumerating colony forming units (CFU) in in vitro and in vivo growth studies.
Production and Purification of Recombinant LdtMt2
We amplified a portion of ltdMt2 with primers 5’-TTTTCATGATCGCCGATCTGCTGGTGC-3’ and 5’-TTGGATCCCGCCTTGGCGTTACCGGC-3’, digested with BspHI–BamHI (underlined) and cloned into pET2818 15. The resulting plasmid, encodes a fusion protein consisting of a methionine specified by the ATG initiation codon of pET2818, residues 55 to 408 of LdtMt2, and a C-terminal polyhistidine tag with the sequence GSH6. We grew E. coli BL21(DE3) harboring pREP4GroESL 26 and pET2818ΩldtMt2 at 37 °C in 3 L of brain heart infusion broth containing 150 µg ml−1 ampicillin, induced expression using Isopropyl–D–thiogalactopyranoside. We purified LdtMt2 from a clarified lysate by affinity chromatography on Ni2+-nitrilotriacetate-agarose resin (Qiagen GmbH, Germany) followed by anion exchange chromatography (MonoQ HR5/5, Amersham Pharmacia) with a NaCl gradient in 50 mM Tris–HCl pH 8.5. We performed an additional size exclusion chromatography on a Superdex HR10/30 column equilibrated with 50 mM Tris–HCl (pH 7.5) containing 300 mM NaCl. Finally, we concentrated the protein by ultrafiltration (Amicon Ultra–4 centrifugal filter devices, Millipore) and stored at −20 °C in the same buffer supplemented with 20% glycerol.
L,D-transpeptidase assays
We purified disaccharide-tetrapeptide containing amidated meso-diaminopimelic acid (GlcNAc–MurNAc–L–Ala1–D–iGln2–mesoDapNH23–D–Ala4) from C. jeikeium strain CIP103337 and determined the concentration after acid hydrolysis 27,28. Next, we tested in vitro formation of muropeptide dimers in 10 µL of 50 mM Tris–HCl (pH 7.5) containing 300 mM NaCl, 5 µM LdtMt2, and 280 µM disaccharide–tetrapeptide. We incubated he reaction mixture for two hours at 37 °C and analyzed the resulting muropeptides by nanoelectrospray tandem mass spectrometry using N2 as the collision gas 28.
Growth and Virulence Analysis in Mice
We used 4–5 week old, female BALB/c mice (Charles River Laboratories) to study in vivo virulence of the strains and their susceptibility to drugs. We infected mice with a log phase culture of wild–type M. tuberculosis, or ldtMt2::Tn or the complemented strain in an aerosol chamber. For assessing in vivo growth of each strain, we sacrificed four mice per group at days 1, 7, 14, 28, 56 and 98 following infection, obtained lungs and spleen, homogenized and cultured appropriate dilutions on Middlebrook 7H11 medium to determine CFU. We allocated 12 mice for each infection group to assess virulence of each strain, for which we determined median-survival-time that mice from each group survived following infection. Protocols for experiments involving mice were approved by Johns Hopkins University Animal Care and Use Committee.
Drug Susceptibility Testing in Mice
We determined minimum inhibitory concentrations for amoxicillin–clavulanate, imipenem (Merck), isoniazid (Sigma) and cycloserine (Sigma) using the broth dilution method 29. We used Augmentin (GlaxoSmithKline), a preparation containing amoxicillin and calvulanate as M. tuberculosis contains β–lactamases 17. For this we inoculated 105 M. tuberculosis bacilli in 2.5 ml of 7H9 broth and added drugs at different concentrations. We incubated these cultures at 37 °C and evaluated for growth by visual inspection at 7 and 14 days. For those samples with diminished growth compared to no-drug control, we determined CFU. We used four-five week old, female BALB/c mice for in vivo assessment of susceptibility of M. tuberculosis lacking LdtMt2 to amoxicillin. We infected two groups of mice, 36 per group, with aerosolized cultures of either wild-type M. tuberculosis or ldtMt2::Tn. We allocated 12 mice from each group into 3 sub–groups and initiated daily treatment at 2 weeks following infection. We provided each sub–group either 25 mg kg−1 isoniazid, or 200 mg kg−1 amoxicillin and 50 mg kg−1 clavulanate, or no drug at all by oral gavage. For analysis, we sacrificed four mice from each treatment sub-group at 1, 2 and 4 weeks following initiation of therapy, obtained lungs, homogenized in 1 ml of PBS and determined CFU in each organ by plating appropriate dilutions of the homogenates on Middlebrook 7H11 selective plates.
Supplementary Material
ACKNOWLEDGMENT
The support of NIH award AI30036 is gratefully acknowledged. This work was also supported by the Foundation pour la Recherche Médicale (Equipe FRM 2006 (DEQ200661107918). M. Lavollay is the recipient of INSERM PhD fellowship (Poste d’Accueil pour Pharmacien, Médecin, et Vétérinaire).
Footnotes
AUTHOR CONTRIBUTIONS: R. Gupta, W. R. Bishai, and G. Lamichhane designed the project. J-L. Mainardi, and M. Arthur, designed the biochemical characterization of MT2594. M. Lavollay and J-L. Mainardi performed biochemistry and analyzed data. R. Gupta and G. Lamichhane conducted genetics, microbiology and mouse experiments. G. Lamichhane wrote the manuscript with contributions from co-authors.
BIBILOGRAPHY
- 1.Wietzerbin J, et al. Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry. 1974;13:3471–3476. doi: 10.1021/bi00714a008. [DOI] [PubMed] [Google Scholar]
- 2.Lavollay M, et al. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L,D-transpeptidation. J Bacteriol. 2008;190:4360–4366. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science (New York, N.Y. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fauci AS. Multidrug-resistant and extensively drug-resistant tuberculosis: the National Institute of Allergy and Infectious Diseases Research agenda and recommendations for priority research. J Infect Dis. 2008;197:1493–1498. doi: 10.1086/587904. [DOI] [PubMed] [Google Scholar]
- 5.Gandhi NR, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co- infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 6.Jindani A, Dore CJ, Mitchison DA. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 days. Am J Respir Crit Care Med. 2003;167:1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- 7.Wayne LG, Sohaskey CD. Nonreplicating persistence of mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 8.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 9.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goren MB, Brennan PJ. In: Tuberculosis. Youmans GP, editor. Philadelphia: W. B. Saunders; 1979. p. 63. [Google Scholar]
- 12.Vollmer W, Holtje JV. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuhashi M. Biosynthesis in the bacterial cell wall. Tanpakushitsu kakusan koso. 1966;11:875–886. [PubMed] [Google Scholar]
- 14.Lamichhane G, et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2003;100:7213–7218. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mainardi JL, et al. A novel peptidoglycan cross-linking enzyme for a beta-lactam-resistant. J Biol Chem. 2005;280:38146–38152. doi: 10.1074/jbc.M507384200. [DOI] [PubMed] [Google Scholar]
- 16.Lavollay M, et al. The β-lactam-sensitive D,D-carboxypeptidase activity of Pbp4 controls the L,D and D,D transpeptidation pathways in Corynebacterium jeikeium. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06887.x. In press. [DOI] [PubMed] [Google Scholar]
- 17.Hugonnet JE, Blanchard JS. Irreversible inhibition of the Mycobacterium tuberculosis beta- lactamase by clavulanate. Biochemistry. 2007;46:11998–12004. doi: 10.1021/bi701506h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donald PR, et al. Early bactericidal activity of amoxicillin in combination with clavulanic acid in patients with sputum smear-positive pulmonary tuberculosis. Scand J Infect Dis. 2001;33:466–469. doi: 10.1080/00365540152029954. [DOI] [PubMed] [Google Scholar]
- 19.Nadler JP, Berger J, Nord JA, Cofsky R, Saxena M. Amoxicillin-clavulanic acid for treating drug-resistant Mycobacterium tuberculosis. Chest. 1991;99:1025–1026. doi: 10.1378/chest.99.4.1025. [DOI] [PubMed] [Google Scholar]
- 20.Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 21.Waxman DJ, Strominger JL. Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 22.Crick DC, Brennan PJ. Biosynthesis of the arabinogalactan-peptidoglycan complex. In: Daffe M, Reyrat J, editors. The Mycobacterial Cell Envelope. Washington DC: ASM; 2008. pp. 25–40. [Google Scholar]
- 23.Templin MF, Ursinus A, Holtje JV. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 1999;18:4108–4117. doi: 10.1093/emboj/18.15.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boneca IG, et al. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc Natl Acad Sci USA. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amrein KE, et al. Purification and characterization of recombinant human p50csk protein- tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc Natl Acad Sci USA. 1995;92:1048–1052. doi: 10.1073/pnas.92.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auger G, van Heijenoort J, Mengin-Lecreulx D, Blanot D. A MurG assay which utilises a synthetic analogue of lipid I. FEMS Microbiol Lett. 2003;219:115–119. doi: 10.1016/S0378-1097(02)01203-X. [DOI] [PubMed] [Google Scholar]
- 28.Arbeloa A, et al. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J Biol Chem. 2004;279:41546–41556. doi: 10.1074/jbc.M407149200. [DOI] [PubMed] [Google Scholar]
- 29.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.