Abstract
Intramuscular administration of DNA vaccines can lead to the generation of antigen-specific immune responses through cross-priming mechanisms. We propose a strategy that is capable of leading to local inflammation and enhancing cross-priming, thus resulting in improved antigen-specific immune responses. Therefore, in the current study, we evaluated immunologic responses elicited through electroporation mediated intramuscular administration of a DNA vaccine encoding calreticulin (CRT) linked to HPV-16 E7 (CRT/E7) in combination with DNA expressing HLA-A2 as compared to CRT/E7 DNA vaccination alone. We found that the co-administration of a DNA vaccine in conjunction with a DNA encoding an xenogenic MHC molecule could significantly enhance the E7-specific CD8+ T cell immune responses as well an antitumor effects against an E7-expressing tumor, TC-1 in C57BL/6 tumor-bearing mice. Furthermore, a similar enhancement in E7-specific immune responses was observed by co-administration of CRT/E7 DNA with DNA encoding other types of xenogenic MHC class I molecules. This strategy was also applicable to another antigenic system, ovalbumin. Further characterization of the injection site revealed that co-administration of HLA-A2 DNA led to a significant increase in the number of infiltrating CD8+ T lymphocytes as well as CD11b/c+ antigen presenting cells. Furthermore, the E7-specific immune responses generated by intramuscular co-administration of CRT/E7 with HLA-A2 DNA were reduced in HLA-A2 transgenic mice. Thus, our data suggest that intramuscular co-administration of DNA encoding xenogenic MHC class I can further improve the antigen-specific immune responses as well as antitumor effects generated by DNA vaccines through enhancement of cross-priming mechanisms.
Keywords: DNA vaccine, xenogenic MHC class I, electroporation
Introduction
DNA vaccines have emerged as a potentially promising approach for cancer immunotherapy based on their safety, stability and ease of preparation (for review, see 1,2). However, naked DNA vaccines suffer from limited immunogenicity and therefore require additional strategies to improve their ability to generate strong immune responses. We have developed a number of innovative strategies to enhance DNA vaccine potency (for review, see 3,4). One approach to enhance DNA vaccine potency is the employment of a molecule that is capable of binding with molecules expressed on dendritic cells. For example, we have previously developed a DNA vaccine encoding the human papillomavirus type 16 (HPV-16) E7 antigen linked to a chaperone protein, calreticulin (CRT) (CRT/E7 DNA) 5,6. We have employed the CRT/E7 DNA vaccine in several recent studies using the intradermal administration via gene gun 7-9. We have also shown that intramuscular administration of DNA vaccines encoding CRT/E7 induced strong E7-specific CD8+ T cell immune responses 10.
Intramuscular administration of DNA vaccines is a commonly used method for administration of DNA vaccines (for review see 11-13). However, this approach can result in suboptimal immunogenicity despite the introduction of high concentrations of injected plasmid 14. Intramuscular administration of DNA vaccines may be enhanced by the employment of electroporation (for reviews see 15,16). Electroporation involves the administration of a naked DNA plasmid into muscle by needle injection, followed by a brief electrical pulse to the surrounding tissue. A transient increase in the permeability of the plasma membrane induced by the electrical current allows increased uptake and, thus, expression of the DNA plasmid 17,18. In addition to the increased uptake of DNA by myocytes resulting in increased protein expression levels of the target antigen, pro-inflammatory cytokines and monocytes/macrophages are also recruited to the site of vaccination, which can enhance antigen presentation to the immune system 19. In fact, electroporation has been shown to significantly enhance the potency of therapeutic HPV DNA vaccines compared to other forms of administration 20.
Intramuscular administration of DNA vaccines via electroporation has been shown to lead to the generation of antigen-specific immune responses through cross-priming mechanisms 21. In general, myocytes do not serve as efficient antigen-presenting cells (APCs) since they lack co-stimulatory molecules. Furthermore, since myocytes are not normally surrounded by lymphocytes, this restricts their ability to prime naïve T cells. Thus, DNA vaccines administered intramuscularly will most likely require the release of antigen expressed by myocytes, which can then be uptaken by professional APCs in order to prime antigen-specific T cells (so-called cross-priming). Thus, we reasoned that a strategy that is capable of leading to local inflammation and release of antigen from the myocytes may potentially enhance the cross-priming of antigen-specific T cells, resulting in improved antigen-specific T cell immune responses generated by DNA vaccination.
Therefore, in the current study, we evaluated immunologic responses elicited through administration of CRT/E7 DNA vaccine in combination with DNA encoding a xenogenic MHC molecule followed by electroporation. We found that the co-administration of a DNA vaccine in conjunction with HLA-A2 DNA could significantly enhance the E7-specific CD8+ T cell immune responses as well an antitumor effects against an E7-expressing tumor, TC-1 in C57BL/6 tumor-bearing mice. Furthermore, a similar enhancement in E7-specific immune responses was observed by co-administration of CRT/E7 DNA with DNA encoding other types of xenogenic MHC class I molecules. This strategy was also applicable to other antigenic systems, such as ovalbumin. Furthermore, the E7-specific immune responses generated by intramuscular co-administration of CRT/E7 with HLA-A2 DNA were reduced in HLA-A2 transgenic mice. Further characterization of the injection site revealed that co-administration of HLA-A2 DNA led to a significant increase in the number of infiltrating CD8+ T lymphocytes as well as CD11b/c+ antigen presenting cells. This strategy may potentially be used in other antigenic systems for the control of infectious diseases and/or cancer.
Materials and Methods
Mice
Six- to eight-week-old female C57BL/6 mice were purchased from the National Cancer Institute (Frederick, MD). HLA-A2 transgenic mice (C57BL/6-Tg(HLA-A2.1)1Enge/J) were purchased from The Jackson Laboratory (Bar Harbor, ME). All animal procedures were performed according to approved protocols and in accordance with recommendations for the proper use and care of laboratory animals.
Generation and characterization of plasmid DNA constructs
The generation of pcDNA3-CRT/E7 5, pcDNA3-OVA 22 and pcDNA3-Luc 22 has been described previously. pcDNA3-HLA A2, HLA A3, and HLA A24 were kindly provided by Dr. Elizabeth M. Jaffee at Johns Hopkins University. For the generation of pcDNA3-HLA-A2/Luc, HLA A2 DNA fragments was amplified by PCR using primers 5'-AAATCTAGAATGGCCGTCATGGCGCCCCG-3' and 5'-AAAGAATTCCACTTTACAAGCTGTGAG-3' and previously described pcDNA3-HLA A2 vector as the template. The amplified DNA products were cloned into the Xba I/EcoR I sites of pcDNA3 vector (No stop). And then the luciferase DNA fragment was isolated from pcDNA3-Luc by PCR using primers 5’-AAAGAATTCATGGAAGACGCCAAAAAC-3’ and 5’-AAAGGATCCTTACACGGCGATCTTTCC-3’ and cloned into EcoR I/BamH I sites of pcDNA3-HLA-A2(no stop). The target gene HLA-A2 in recombinant pcDNA3-HLA-A2/Luc was then confirmed by sequencing and the expression of the target protein HLA-A2/LUC was demonstrated (See Supplementary Figure 1).
For the identification of HLA-A2 expression in cells transfected with pcDNA3-HLA-A2/Luc plasmid, BHK-21 cells (4×104 per well) were seeded into 24-well round-bottomed plate and after overnight incubation, BHK-21 cells were transfected with 1ug of pcDNA3-HLA-A2, pcDNA3-HLA-A2/Luc or pcDNA3-No insert plasmid DNA using lipofectamin 2000 reagent (Invitrogen, Carlsbad, CA). After one day, the expression of HLA-A2 was measured by flow cytometry analysis after staining using PE- labeled mouse anti HLA-A2 antibody (BD Biosciences, San Jose, CA). Analysis was performed on a Becton-Dickinson FACScan with CELLQuest software (Becton Dickinson Immunocytometry System, Mountain View, USA).
Cells
The HPV-16 E7-expressing murine tumor model, TC-1, has been described previously 23. In brief, HPV-16 E6, E7, and the ras oncogene were used to transform primary C57BL/6 mice lung epithelial cells to generate the TC-1 cell line. The DC-1 cells 24 were derived from a well-characterized dendritic cell line and were kindly provided by Dr. Kenneth Rock (University of Massachusetts, Worcester, MA, USA) 25. These cell lines were cultured in vitro in RPMI 1640 supplemented with 10% fetal bovine serum, 50units/ml of penicillin/streptomycin, 2mM L-glutamine, 1mM sodium pyruvate, and 2mM non-essential amino acids, and grown at 37°C with 5% CO2.
In vivo electroporation and gene delivery
For the immunization of mice, control plasmid (pcDNA3-no insert), CRT/E7, HLA-A2, HLA-A3, HLA-A24, or OVA plasmid vectors were delivered intramuscularly followed by electroporation twice with a 1-week interval. Hair around the quadriceps femoris muscle of mice was removed. The muscle was then injected with 2ug (1ug of pcDNA3- CRT/E7 was mixed with 1μg of pcDNA3-No insert, HLA-A2, HLA-A3, HLA-A24, or OVA) of plasmid DNA in 50μl using the TriGrid Electroporation Delivery System (Ichor Medical Systems). The electroporation protocol involved rectangular wave, direct-current pulses applied at 220V/cm peak amplitude and 8% dusty cycle over 0.5 s. The TriGrid electroporation system consists of an electric pulse generator and a TriGrid array comprising four electrodes 2mm in length with 2.5mm interelectrode spacing.
Intracellular cytokine staining and flow cytometry analysis
Mice were immunized with 2 μg of the various DNA vaccines and received a booster with the same regimen 1 week later. Splenocytes were harvested 1 week after the last vaccination. Before intracellular cytokine staining, 6×105 pooled splenocytes from each vaccination group were incubated for 16 hours with either 1 μg/ml of E7 peptide (RAHYNIVTF) or OVA peptide (SIINFEKL) containing an MHC class I epitope 22 for detecting E7 or OVA-specific CD8+ T cells. Cell surface marker staining of CD8 and intracellular cytokine staining for IFN-γ using Phycoerythrin-conjugate monoclonal rat anti-mouse CD8 antibody (BD Pharmingen, San Diego, CA) and FITC-conjugated anti-mouse IFN-γ antibodies (BD Pharmingen, San Diego, CA) as well as flow cytometry analysis, were performed using conditions described previously 26. Analyses were done on a Becton-Dickinson FACScan with CELLQuest software (Becton Dickinson Immunocytometry System, Mountain View, CA).
Immunohistochemistry
For the immunohistochemistry, 1ug of pcDNA-no insert or HLA-A2 plasmid DNA were injected in vivo by electroporation-mediated intramuscle injection into the mice. Muscle from the site of plasmid electroporation were fixed with 5% neutralized formalin and embedded in paraffin. The formalin-fixed and paraffin-embedded sections were deparaffinized and hydrated in xylene and serial alcohol solutions, respectively. And then sections were stained with H&E for visualization of cellular inflammation. For the antibody staining, the antigen retrieval was performed in a pressure cooker with antigen retrieval buffer pH 6 (Dako, Glostrup, Denmark) at 124°C, for 30 sec. Endogenous peroxidase was blocked by incubation in 3% H2O2 for 10 min. LSAB®+ (DakoCytomation; CD8, and CD11b/c) was used for immunohistochemical labeling. Nonspecific binding of antibodies were blocked with serum-free protein block (DakoCytomation) for 15 min and then incubated with primary antibodies at 4°C overnight. We used the following antibodies obtained from commercial suupliers: rat anti-F4/80 (CI:A3-1, 1:50, Abcam), rabbit anti-CD8 (EP1150Y, 1:500, Abcam), and rabbit anti-CD11b/c (1:200, Abcam) antibodies. The stain visualized using 3,3’-diaminobenzidine plus (DakoCytomation) and was lightly counterstained with hematoxylin, dehydrated in ethanol, and cleared in xylene. The slides were covered and observed under a light microscope (Axioplot, Carl Zeiss, Jena, Germany). The counting of each antibody positive cells was performed as previously described {Peng, 2008 #75. The data were generated using 200 × HPF (high power field) from 3 different fields for each muscle tissue.
Luciferase-based bioluminescence imaging
For the in vivo bioluminescence imaging, 1ug of pcDNA3-HLA-A2/Luc or pcDNA3-Luc plasmid DNA were injected in vivo by electroporation-mediated intramuscle injection into the mice. The intensity of bioluminescence was checked on days 0 and 3 after injection. Image on day 0 was taken from 4 h later after injection. D-Luciferin was dissolved to 7.8 mg/mL in PBS, filter sterilized, and stored at −80°C. Mice were given D-luciferin by i.p. injection (200 uL/mouse, 75 mg/kg) and anesthetized with isoflurane. In vivo bioluminescence imaging for LUC was conducted on a cryogenically cooled IVIS system (Xenogen) using Living Image acquisition and analysis software (Xenogen). Mice were then placed onto the warmed stage inside the light-tight camera box with continuous exposure to 1% to 2% isoflurane. Images were acquired 10 min after D-luciferin administration and imaged for 1min. The levels of light from the bioluminescent cells were detected by IVIS camera system, integrated, and digitized. Region of interest from displayed images was designated around the injection sites and quantified as total photon counts using Living Image 2.50 software (Xenogen).
In vivo tumor treatment experiments
For the tumor treatment experiment, mice were challenged with 2×l05 TC-1 tumor cells/mouse in the tail vein to simulate hematogenous spread of tumors {Ji, 1998 #73}. Four days after tumor challenge, mice were vaccinated and boosted 1 week later with pcDNA-No insert, HLA-A2, CRT/E7 or CRT/E7 and HLA-A2 via electroporation. Mice were monitored twice a week and sacrificed on day 35 after the last vaccination. The mean number of pulmonary nodules in each mouse was determined by experimenters blinded to the sample identity.
Long-term in vivo tumor protection experiments
C57BL/6 mice (five per group) were vaccinated and received a booster with pcDNA-No insert, HLA-A2, CRT/E7 or CRT/E7 and HLA-A2 via electroporation. Seven weeks after the last vaccination, mice were subcutaneously challenged with 1 × l05 TC-1 cells/mouse in the right leg and evaluate long-term survival. Mice were monitored for evidence of survival by palpation and inspection twice a week. The mice were killed at any time during the experiment if tumor size was over than 2 cm in the longest dimension.
Statistical analysis
All data are expressed as means ± standard deviation (S.D.) and are representative of at least two separate experiments. Results for intracellular cytokine staining with flow cytometry analysis and tumor treatment experiments were evaluated by analysis of variance (ANOVA). Comparisons between individual data points were made using Student's t-test. In the tumor protection experiments, the principal outcome measure was time to tumor development. The event time distributions for different mice were compared using the Kaplan and Meier method and the log-rank statistic. Graphs were done using SigmaPlot program file (Systat Software, Inc.). All p values < 0.05 were considered significant.
Results
Intramuscular vaccination with CRT/E7 DNA in conjunction with HLA-A2 DNA followed by electroporation generates significantly higher frequency of E7-specific CD8+ T cells in vaccinated mice
In order to determine the E7-specific CD8+ T cell immune response in mice vaccinated with CRT/E7 DNA with HLA-A2 DNA, we vaccinated C57BL/6 mice (5 per group) intramuscularly with CRT/E7 DNA mixed with no insert or HLA-A2 DNA followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and characterized the presence of E7-specific CD8+ T cells in treated mice using intracellular cytokine staining for IFN-γ followed by flow cytometry analysis. As shown in Figure 1, mice vaccinated with CRT/E7 DNA with HLA-A2 DNA generated a significantly higher number of E7-specific CD8+ T cells compared to mice vaccinated with CRT/E7 DNA with backbone DNA (no insert). These results indicate that co-administration of HLA-A2 DNA can significantly enhance the E7-specific CD8+ T cell immune responses generated by CRT/E7 DNA vaccination.
Figure 1. Intracellular cytokine staining followed by flow cytometry analysis to characterize the E7-specific CD8+ T cell immune response in vaccinated mice.
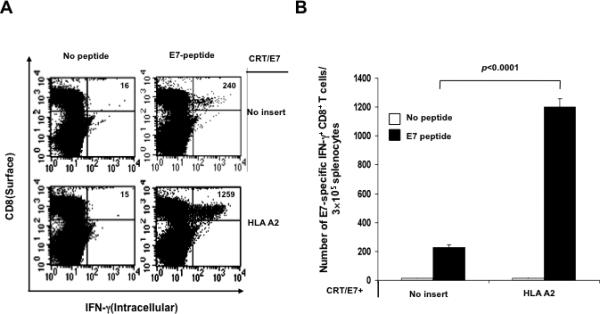
C57BL/6 mice (5 per group) were immunized with CRT/E7 DNA mixed with no insert or HLA-A2 DNA intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and stimulated with the E7 peptide. Cells were characterized for E7-specific CD8+ T cells using intracellular IFN-γ staining followed by flow cytometry analysis. Splenocytes without peptide stimulation were used as negative control. (A) Representative data of intracellular cytokine staining followed by flow cytometry analysis showing the number of E7-specific IFNγ+ CD8+ T cells in the various groups (right upper quadrant). (B) Bar graph depicting the numbers of E7-specific IFN-γ-secreting CD8+ T cells per 3×105 pooled splenocytes (mean± s.d.).
We also characterized the HLA-A2-specific CD8+ T cell immune responses in vaccinated mice. We observed that mice vaccinated with CRT/E7 DNA with HLA-A2 DNA generated a significantly higher number of HLA-A2-specific CD8+ T cells compared to mice vaccinated with CRT/E7 DNA with backbone DNA (no insert) (See Supplementary Figure 2).
TC-1 tumor-bearing mice treated with CRT/E7 DNA vaccine in conjunction with HLA-A2 DNA generate significantly better therapeutic anti-tumor effects
To determine the therapeutic antitumor effects generated by HLA-A2 DNA in conjunction with CRT/E7 DNA vaccination, we first challenged groups of C57BL/6 mice (5 per group) via tail vein with TC-1 tumor cells and then treated them with CRT/E7 DNA with or without HLA-A2 DNA intramuscularly followed by electroporation. Tumor-bearing mice treated with pcDNA3-no insert were used as negative controls. As shown in Figure 2A, tumor-bearing mice treated with HLA-A2 DNA with CRT/E7 DNA showed significantly reduced number of pulmonary tumor nodules as compared to tumor-bearing mice treated with DNA vaccine alone (p<0.002).
Figure 2. In vivo tumor treatment and prevention experiments.
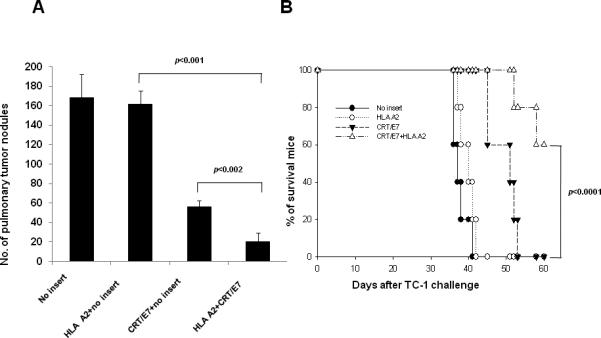
(A) Bar graph depicting the number of pulmonary tumor nodules in TC-1 tumor bearing mice treated with CRT/E7 and/or HLA-A2 DNA. C57BL/6 mice (5 per group) were challenged via tail vein with 2×105 /mouse of TC-1 tumor cells. Four days later, the mice were immunized with 2ug of pcDNA-No insert, 1ug of HLA-A2 mixed 1ug of pcDNA3-no insert, 1ug of CRT/E7 mixed 1ug of pcDNA3-no insert or 1ug of CRT/E7 mixed 1ug of HLA-A2 intramuscularly followed by electroporation twice with a 1-week interval. Tumor growth was monitored over time and survival of tumor-bearing mice was analyzed by Kaplan & Meier analysis. The data are expressed as mean number of pulmonary lung nodules ± SD. twice with a 1-week interval. (B) Kaplan & Meier survival analysis of TC-1 tumor challenged mice treated with mice treated with CRT/E7 and/or HLA-A2 DNA. C57BL/6 mice (5 per group) were vaccinated with 2ug of pcDNA-No insert, 1ug of HLA-A2 mixed 1ug of pcDNA3-no insert, 1ug of CRT/E7 mixed 1ug of pcDNA3-no insert or 1ug of CRT/E7 mixed 1ug of HLA-A2 intramuscularly followed by electroporation twice with a 1-week interval. Seven weeks after the last vaccination, mice were subcutaneously challenged with 1 × l05 TC-1 cells/mouse in the right leg and long-term survival was evaluated. Mice were monitored for evidence of survival by palpation and inspection twice a week. Data shown are representative of two experiments performed.
We also tested the long-term antitumor effects of vaccinated mice using a tumor prevention experiment. C57BL/6 mice (5 per group) were vaccinated with CRT/E7 DNA with or without HLA-A2 DNA intramuscularly followed by electroporation twice with a 1-week interval. Seven weeks after the last vaccination, mice were subcutaneously challenged with 1 × l05/mouse of TC-1 cells in the right leg and the long-term survival was evaluated. As shown in Figure 2B, mice vaccinated with HLA-A2 DNA with CRT/E7 DNA showed improved long-term survival compared to mice vaccinated with HLA-A2 DNA alone or the DNA vaccine alone (p < 0.0001). Thus, our data indicate that the treatment with HLA-A2 DNA can significantly enhance the therapeutic and preventive antitumor effects generated by CRT/E7 DNA vaccination in TC-1 tumor-bearing mice.
Vaccination with CRT/E7 DNA in conjunction with DNA encoding different xenogenic MHC class I is capable of generating a significantly higher frequency of E7-specific CD8+ T cells in vaccinated mice
In order to determine if the observed enhancement in E7-specific CD8+ T cell immune response in vaccinated mice by HLA-A2 DNA is applicable to other xenogenic MHC class I molecules, we vaccinated C57BL/6 mice (5 per group) with CRT/E7 DNA mixed with DNA encoding other xenogenic MHC class I molecules including HLA-A3, HLA-A24 and HLA-A2 intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and characterized the presence of E7-specific CD8+ T cells in treated mice using intracellular cytokine staining for IFN-γ followed by flow cytometry analysis. As shown in Figure 3, mice vaccinated with CRT/E7 DNA in conjunction with DNA encoding any of the xenogenic MHC class I molecules generated a significantly higher number of E7-specific CD8+ T cells compared to mice vaccinated with CRT/E7 DNA with backbone DNA (no insert). Among the various xenogenic MHC class I molecules tested, HLA-A2 DNA generated the highest E7-specific immune responses. These results indicate that the observed enhancement in E7-specific CD8+ T cell immune response in mice vaccinated with CRT/E7 DNA in conjunction with HLA-A2 DNA is also applicable to other xenogenic MHC class I molecules.
Figure 3. Intracellular cytokine staining and flow cytometry analysis to characterize the antigen specific CD8+ T cell response by vaccination with various xenogenic MHC class I DNA.
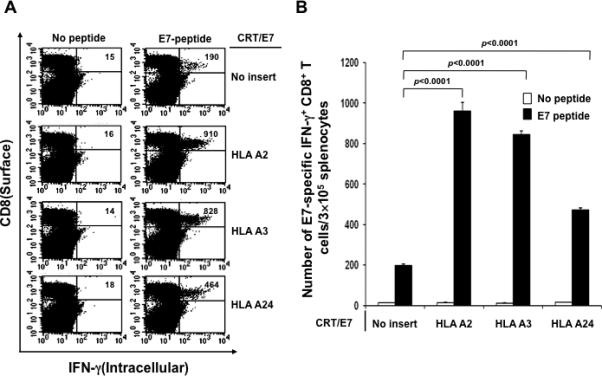
C57BL/6 mice (5 per group) were immunized with CRT/E7 DNA mixed with no insert or HLA-A2, HLA-A3, HLA-A24 DNA intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and stimulated with the E7 peptide. Cells were characterized for E7-specific CD8+ T cells using intracellular IFN-γ staining followed by flow cytometry analysis. Splenocytes without peptide stimulation were used as negative control. (A) Representative data of intracellular cytokine staining followed by flow cytometry analysis showing the number of E7-specific IFNγ+ CD8+ T cells in the various groups (right upper quadrant). (B) Bar graph depicting the numbers of E7-specific IFN-γ-secreting CD8+ T cells per 3×105 pooled splenocytes (mean± s.d.).
Vaccination with OVA-specific DNA vaccine in conjunction with HLA-A2 DNA generates a significantly higher frequency of OVA-specific CD8+ T cells in vaccinated mice
In order to determine if the observed enhancement in E7-specific CD8+ T cell immune response in vaccinated mice can be applied to other antigenic systems, we vaccinated C57BL/6 mice (5 per group) with DNA encoding ovalbumin (OVA) mixed with no insert or HLA-A2 DNA intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and characterized the presence of OVA-specific CD8+ T cells in treated mice using intracellular cytokine staining for IFN-γ followed by flow cytometry analysis. As shown in Figure 4, mice vaccinated with OVA DNA mixed with HLA-A2 DNA generated a significantly higher number of OVA-specific CD8+ T cells compared to mice vaccinated with OVA DNA with backbone DNA (no insert). These results indicate that ability of DNA encoding xenogenic MHC molecules to enhance the antigen-specific CD8+ T cell immune responses generated by DNA vaccination can be applied to other antigenic systems, such as ovalbumin.
Figure 4. Intracellular cytokine staining followed by flow cytometry analysis to characterize the OVA-specific CD8+ T cell immune response in vaccinated mice.
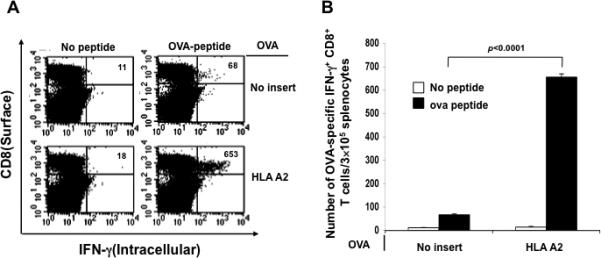
C57BL/6 mice (5 per group) were immunized with pcDNA3 OVA mixed with pcDNA3 HLA-A2 or no insert intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and stimulated with the OVA peptide. Cells were characterized for OVA-specific CD8+ T cells using intracellular IFN-γ staining followed by flow cytometry analysis. Splenocytes without peptide stimulation were used as negative control. (A) Representative data of intracellular cytokine staining followed by flow cytometry analysis showing the number of OVA-specific IFNγ+ CD8+ T cells in the various groups (right upper quadrant). (B) Bar graph depicting the numbers of OVA-specific IFN-γ-secreting CD8+ T cells per 3×105 pooled splenocytes (mean± s.d.).
Vaccination with CRT/E7 DNA in conjunction with HLA-A2 DNA fails to generates significantly higher frequency of E7-specific CD8+ T cells in vaccinated HLA-A2 transgenic mice
In order to determine if the observed enhancement in E7-specific CD8+ T cell immune response by co-administration of DNA encoding HLA-A2 will occur in HLA-A2 transgenic mice, we vaccinated HLA-A2 transgenic mice (5 per group) with CRT/E7 DNA mixed with no insert or HLA-A2 DNA intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and characterized the presence of E7-specific CD8+ T cells in treated mice using intracellular cytokine staining for IFN-γ followed by flow cytometry analysis. As shown in Figure 5, HLA-A2 transgenic mice vaccinated with CRT/E7 DNA mixed with HLA-A2 DNA generated comparable number of E7-specific CD8+ T cells compared to mice vaccinated with CRT/E7 DNA with backbone DNA (no insert). These results indicate that co-administration of DNA encoding xenogenic MHC molecules is required to enhance the antigen-specific CD8+ T cell immune responses generated by DNA vaccination.
Figure 5. Intracellular cytokine staining followed by flow cytometry analysis to characterize the E7-specific CD8+ T cell immune response in vaccinated HLA-A2 transgenic mice.
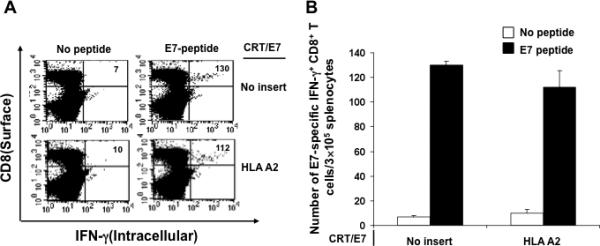
HLA-A2 transgenic mice (5 per group) were immunized with CRT/E7 DNA mixed with no insert or HLA-A2 DNA intramuscularly followed by electroporation twice with a 1-week interval. One week after the last vaccination, splenocytes from vaccinated mice were harvested and stimulated with the E7 peptide. Cells were characterized for E7-specific CD8+ T cells using intracellular IFN-γ staining followed by flow cytometry analysis. Splenocytes without peptide stimulation were used as negative control. (A) Representative data of intracellular cytokine staining followed by flow cytometry analysis showing the number of E7-specific IFNγ+ CD8+ T cells in the various groups (right upper quadrant). (B) Bar graph depicting the numbers of E7-specific IFN-γ-secreting CD8+ T cells per 3×105 pooled splenocytes (mean± s.d.).
Co-administration of HLA-A2 DNA leads to a significant increase in the number of infiltrating CD8+ T lymphocytes and CD11b/c+ antigen presenting cells as well as a reduction of protein expressing cells in the injection site
In order to investigate the mechanisms of the observed enhancement in E7-specific CD8+ T cell immune response by co-administration of DNA encoding HLA-A2, C57BL/6 mice were injected with pcDNA-no insert or HLA-A2 DNA by intramuscular injection followed by electroporation. Muscle sections from the site of injection was removed on day 3 and 5 and characterized for the presence of CD8 and CD11b/c immune cells. As shown in Figure 6A, muscle sections from mice injected with HLA-A2 demonstrated a significant increase in the number of F4/80+ inflammatory cells, the number of infiltrating CD8+ T lymphocytes as well as CD11b/c+ antigen presenting cells compared to sections from mice injected with pcDNA-no insert. In addition, we found that the number of F4/80+ inflammatory cells and CD11b/c+ cells decreased over a period of time. The number of CD8+ T lymphocytes were slightly increased on day 5 compared to day 3, but significantly decreased on day 7 (Figure 6B). We further characterized the luciferase expression in C57BL/6 mice injected intramuscularly with DNA encoding luciferase in conjunction with HLA-A2 DNA or luciferase DNA alone followed by electroporation. The bioluminescence intensity was measured on days 0 and 3 after injection. As shown in Figure 6C and D, mice injected with luciferase DNA in conjunction with HLA-A2 DNA demonstrated a significant reduction in luminescent intensity compared to mice injected with luciferase DNA alone 3 days after injection. Taken together, our data suggest that co-administration of HLA-A2 DNA leads to significant local inflammation and infiltration of immune cells, resulting in the reduction of antigen-expressing cells at the injection site.
Figure 6. Characterization of the infiltrating CD8+ T lymphocytes as well as CD11b/c+ antigen presenting cells and kinetics of protein expression in the injection site following administration of HLA-A2 DNA.
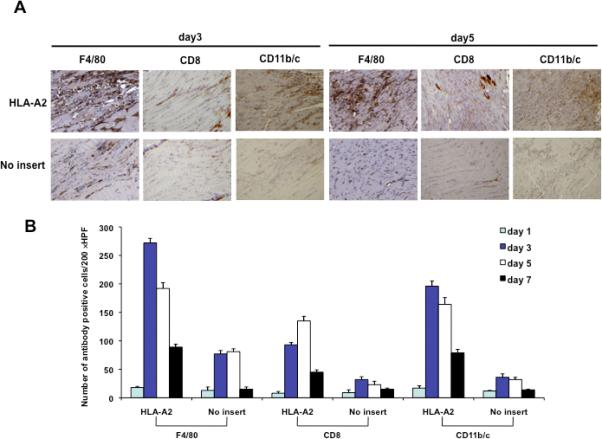
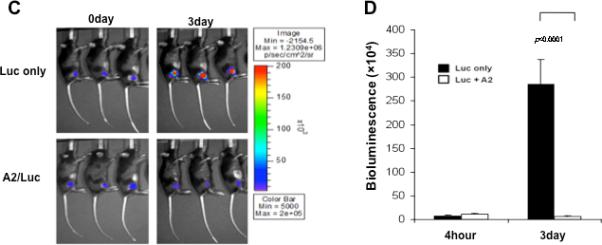
(A) Representative immunocytochemistry images of muscle sections from mice injected with or without HLA-A2 DNA. C57BL/6 mice were injected with 1ug of pcDNA-no insert or HLA-A2 plasmid DNA by electroporation-mediated i.m. injection. Muscle sections from the site of injection was removed on day 3 and 5. Sections were stained with hematoxylin-eosin, F4/80, CD8, and CD11b/c antibody. (B) 5-(B) Graphical representation of the number of F4/80, CD8, and CD11b/c positive cells per 200× HPF. The data were generated using 200 × HPF from 3 different fields for each muscle tissue and shown as mean±s.d. The data presented in this figure are from one representative experiment of two performed. (C) Representative luminescence images of mice injected with pcDNA3-HLA-A2/Luc or Luc plasmid DNA on day 0 and day 4. C57BL/6 mice (3 per group) were injected with 1ug of pcDNA3-HLA-A2/Luc or Luc plasmid DNA intramuscularly followed by electroporation. The intensity of bioluminescence was checked on days 0 and 3 after injection. Image on day 0 was taken 4 hours after injection. (D) Bar graph depicting quantification of luminescence intensity in mice injected with pcDNA3-HLA-A2/Luc or Luc plasmid DNA (mean ± S.D.). Data shown are representative of two experiments performed.
Discussion
In the current study, we observed that the co-administration of a DNA vaccine in conjunction with DNA encoding xenogenic MHC class I molecules could significantly enhance the E7-specific CD8+ T cell immune responses as well an antitumor effects against TC-1 in C57BL/6 tumor-bearing mice. This strategy was also applicable to other antigenic systems, such as ovalbumin. Furthermore, the ability to enhance the antigen-specific immune responses is clearly due to the co-administration of xenogenic MHC class I DNA since co-administration of CRT/E7 with HLA-A2 DNA failed to enhance the immune responses in HLA-A2 transgenic mice. Further characterization of the injection site revealed that co-administration of HLA-A2 DNA led to a significant increase in the number of infiltrating CD8+ T lymphocytes as well as CD11b/c+ immune cells as well as a reduction of protein expressing cells in the injection site. Thus, our data suggest that intramuscular co-administration of DNA encoding xenogenic MHC class I can further improve the antigen-specific immune responses as well as antitumor effects generated by DNA vaccines possibly through enhancement of cross-priming mechanisms.
The encouraging results from the current study suggest the possibility that other pro-inflammatory molecules and cytokines may potentially be used to enhance the immune responses generated by DNA vaccination administered intramuscularly. It has previously been shown that co-administration of DNA vaccines with DNA encoding pro-inflammatory cytokines such as GM-CSF 27 and IL-12 28 intramuscularly results in enhancement of antigen-specific immune responses and antitumor effects in vaccinated animals (for reviews see 4,29). It is quite likely that these molecules may lead to a local inflammatory response, resulting in the increased release of antigen from muscle cells.
Our data demonstrate that co-administration of HLA-A2 DNA leads to an increase in the number of F4/80+ inflammatory cells, infiltrating CD8+ T lymphocytes and CD11b/c+ antigen presenting cells to the injection site (Figure 6). The presence of antigen-presenting cells in the injection site raises the possibility that cross-priming may occur in the local environment in the vicinity of Ag-expressing cells in addition to the draining lymph nodes. It has been shown that dendritic cells can be recruited to the local tissues for CD8+ T cell cross priming 30. Thus, it is conceivable that the inflammation generated by co-administration of xenogenic MHC class I DNA may lead to the recruitment of antigen-presenting cells, resulting in cross priming in the local muscle tissue in addition to the draining lymph nodes.
For clinical translation it is important to consider issues related to the safety of the vaccination. Since the current approach leads to local inflammation in the injection site, it may raise concerns for myositis. Although our data demonstrate that the mice injected with HLA-A2 DNA demonstrated significant local inflammation in the muscle on day 3 following injection, inflammation gradually reduced 5 days after the injection (Figure 6) and eventually the mice completely recover from the injection. Thus, we expect that DNA vaccination with this approach may not create issues related to myositis based on our preclinical data.
In summary, our results demonstrate that intramuscular co-administration of DNA encoding xenogenic MHC class I followed by electroporation can improve the antigen-specific immune responses as well as antitumor effects generated by DNA vaccines through enhancement of cross-priming mechanisms. Our study represents a novel approach that may potentially be used in other antigenic systems for the control of infectious diseases and/or cancer.
Supplementary Material
Supplementary Figure 1. Characterization of HLA-A2 or Luc expression in cells transfected with pcDNA3-HLA-A2/Luc plasmid. BHK-21 cells (4×104 per well) were transfected with pcDNA3-HLA-A2, pcDNA3-HLA-A2/Luc, pcDNA3-Luc or pcDNA3-No insert plasmid DNA. (A) Flow cytometry analysis to demonstrate expression of HLA-A2 in transfected cells. One day after transfection, transfected cells were analyzed by flow cytometry using HLA-A2 specific antibody. Bold line (1) represents untransfected cells, scattered line (2) represents pcDNA3-HLA-A2 or pcDNA3-HLA-A2/Luc transfected cells, and line (3) represents pcDNA3-No insert transfected cells. The data presented is a representation of two independent experiments. (B) Representative luminescence imaging depicting the luciferase expression level from pcDNA3-HLA-A2/Luc, pcDNA3-Luc or pcDNA3-No insert plasmid DNA transfected cells. (C) Bar graph depicting the quantification of luminescence intensity in each transfected cells. The data presented is a representation of two independent experiments.
Supplementary Figure 2. Intracellular cytokine staining followed by flow cytometry analysis to characterize the HLA-A2-specific CD8+ T cell immune response in vaccinated mice. C57BL/6 mice (5 per group) were immunized with CRT/E7 DNA mixed with no insert or HLA-A2 DNA. One week after the last vaccination, splenocytes from vaccinated mice were harvested. 1×107 pfu of wild type (wild type VV) or HLA-A2 expressed vaccinia virus (HLA-A2 VV) were infected into 1×106 DC-1 cells. One day later, these cells were incubated with splenocytes from vaccinated mice for 16 hours. Cells were characterized for HLA-A2-specific CD8+ T cells using intracellular IFN-γ staining followed by flow cytometry analysis. (A) Flow cytometry analysis to characterize HLA-A2 expression on DC-1 cells infected with wild type VV or HLA-A2 VV. PE-conjugated HLA-A2 antibody was used to detect HLA-A2 expression. The isotype antibody was used as the negative control (grey profile). (B) Representative data of intracellular cytokine staining followed by flow cytometry analysis showing the number of HLA-A2-specific IFNγ+ CD8+ T cells in the various groups (right upper quadrant). (C) Bar graph depicting the numbers of HLA-A2-specific IFN-γ-secreting CD8+ T cells per 3×105 pooled splenocytes (mean± s.d.).
Acknowledgements
This work was supported by the American Cancer Society (C.F. Hung) and National Cancer Institute SPORE in Cervical Cancer P50 CA098252 and the 1 RO1 CA114425-01 (T.-C. Wu).
References
- 1.Donnelly JJ, Ulmer JB, Shiver JW, Liu MA. DNA vaccines. Annu Rev Immunol. 1997;15:617–648. doi: 10.1146/annurev.immunol.15.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Gurunathan S, Klinman DM, Seder RA. DNA vaccines: immunology, application, and optimization*. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 3.Hung CF, Wu TC. Improving DNA vaccine potency via modification of professional antigen presenting cells. Curr Opin Mol Ther. 2003;5:20–24. [PubMed] [Google Scholar]
- 4.Tsen SW, Paik AH, Hung CF, Wu TC. Enhancing DNA vaccine potency by modifying the properties of antigen-presenting cells. Expert Rev Vaccines. 2007;6:227–239. doi: 10.1586/14760584.6.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng WF, Hung CF, Chai CY, Hsu KF, He L, Ling M, et al. Tumor-specific immunity and antiangiogenesis generated by a DNA vaccine encoding calreticulin linked to a tumor antigen. J Clin Invest. 2001;108:669–678. doi: 10.1172/JCI12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng S, Tomson TT, Trimble C, He L, Hung CF, Wu TC. A combination of DNA vaccines targeting human papillomavirus type 16 E6 and E7 generates potent antitumor effects. Gene Ther. 2006;13:257–265. doi: 10.1038/sj.gt.3302646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng CW, Monie A, Trimble C, Alvarez RD, Huh WK, Buchsbaum DJ, et al. Combination of treatment with death receptor 5-specific antibody with therapeutic HPV DNA vaccination generates enhanced therapeutic anti-tumor effects. Vaccine. 2008;26:4314–4319. doi: 10.1016/j.vaccine.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tseng CW, Monie A, Wu CY, Huang B, Wang MC, Hung CF, et al. Treatment with proteasome inhibitor bortezomib enhances antigen-specific CD8+ T-cell-mediated antitumor immunity induced by DNA vaccination. J Mol Med. 2008;86:899–908. doi: 10.1007/s00109-008-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng S, Trimble C, Alvarez RD, Huh WK, Lin Z, Monie A, et al. Cluster intradermal DNA vaccination rapidly induces E7-specific CD8+ T-cell immune responses leading to therapeutic antitumor effects. Gene Ther. 2008;15:1156–1166. doi: 10.1038/gt.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JW, Hung CF, Juang J, He L, Kim TW, Armstrong DK, et al. Comparison of HPV DNA vaccines employing intracellular targeting strategies. Gene Ther. 2004;11:1011–1018. doi: 10.1038/sj.gt.3302252. [DOI] [PubMed] [Google Scholar]
- 11.Bodles-Brakhop AM, Draghia-Akli R. DNA vaccination and gene therapy: optimization and delivery for cancer therapy. Expert Rev Vaccines. 2008;7:1085–1101. doi: 10.1586/14760584.7.7.1085. [DOI] [PubMed] [Google Scholar]
- 12.Hung CF, Ma B, Monie A, Tsen SW, Wu TC. Therapeutic human papillomavirus vaccines: current clinical trials and future directions. Expert Opin Biol Ther. 2008;8:421–439. doi: 10.1517/14712598.8.4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung CF, Monie A, Alvarez RD, Wu TC. DNA vaccines for cervical cancer: from bench to bedside. Exp Mol Med. 2007;39:679–689. doi: 10.1038/emm.2007.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bansal A, Jackson B, West K, Wang S, Lu S, Kennedy JS, et al. Multifunctional T-cell characteristics induced by a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine regimen given to healthy adults are dependent on the route and dose of administration. J Virol. 2008;82:6458–6469. doi: 10.1128/JVI.00068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luxembourg A, Evans CF, Hannaman D. Electroporation-based DNA immunisation: translation to the clinic. Expert Opin Biol Ther. 2007;7:1647–1664. doi: 10.1517/14712598.7.11.1647. [DOI] [PubMed] [Google Scholar]
- 16.Rols MP. Mechanism by which electroporation mediates DNA migration and entry into cells and targeted tissues. Methods Mol Biol. 2008;423:19–33. doi: 10.1007/978-1-59745-194-9_2. [DOI] [PubMed] [Google Scholar]
- 17.Aihara H, Miyazaki J. Gene transfer into muscle by electroporation in vivo. Nat Biotechnol. 1998;16:867–870. doi: 10.1038/nbt0998-867. [DOI] [PubMed] [Google Scholar]
- 18.Mir LM, Bureau MF, Gehl J, Rangara R, Rouy D, Caillaud JM, et al. High-efficiency gene transfer into skeletal muscle mediated by electric pulses. Proc Natl Acad Sci U S A. 1999;96:4262–4267. doi: 10.1073/pnas.96.8.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng B, Zhao Y, Xu L, Xu Y. Electric pulses applied prior to intramuscular DNA vaccination greatly improve the vaccine immunogenicity. Vaccine. 2007;25:2064–2073. doi: 10.1016/j.vaccine.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 20.Best SR, Peng S, Juang CM, Hung CF, Hannaman D, Saunders JR, et al. Administration of HPV DNA vaccine via electroporation elicits the strongest CD8+ T cell immune responses compared to intramuscular injection and intradermal gene gun delivery. Vaccine. 2009;27:5450–5459. doi: 10.1016/j.vaccine.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu TM, Ulmer JB, Caulfield MJ, Deck RR, Friedman A, Wang S, et al. Priming of cytotoxic T lymphocytes by DNA vaccines: requirement for professional antigen presenting cells and evidence for antigen transfer from myocytes. Mol Med. 1997;3:362–371. [PMC free article] [PubMed] [Google Scholar]
- 22.Kim TW, Hung CF, Ling M, Juang J, He L, Hardwick JM, et al. Enhancing DNA vaccine potency by coadministration of DNA encoding antiapoptotic proteins. J Clin Invest. 2003;112:109–117. doi: 10.1172/JCI17293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin KY, Guarnieri FG, Staveley-O'Carroll KF, Levitsky HI, August JT, Pardoll DM, et al. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56:21–26. [PubMed] [Google Scholar]
- 24.Kim TW, Hung CF, Boyd DA, He L, Lin CT, Kaiserman D, et al. Enhancement of DNA vaccine potency by coadministration of a tumor antigen gene and DNA encoding serine protease inhibitor-6. Cancer Res. 2004;64:400–405. doi: 10.1158/0008-5472.can-03-1475. [DOI] [PubMed] [Google Scholar]
- 25.Shen Z, Reznikoff G, Dranoff G, Rock KL. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J Immunol. 1997;158:2723–2730. [PubMed] [Google Scholar]
- 26.Chen CH, Wang TL, Hung CF, Yang Y, Young RA, Pardoll DM, et al. Enhancement of DNA vaccine potency by linkage of antigen gene to an HSP70 gene. Cancer Res. 2000;60:1035–1042. [PubMed] [Google Scholar]
- 27.Leachman SA, Tigelaar RE, Shlyankevich M, Slade MD, Irwin M, Chang E, et al. Granulocyte-macrophage colony-stimulating factor priming plus papillomavirus E6 DNA vaccination: effects on papilloma formation and regression in the cottontail rabbit papillomavirus--rabbit model. J Virol. 2000;74:8700–8708. doi: 10.1128/jvi.74.18.8700-8708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MS, Sin JI. Both antigen optimization and lysosomal targeting are required for enhanced anti-tumour protective immunity in a human papillomavirus E7-expressing animal tumour model. Immunology. 2005;116:255–266. doi: 10.1111/j.1365-2567.2005.02219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lori F, Weiner DB, Calarota SA, Kelly LM, Lisziewicz J. Cytokine-adjuvanted HIV-DNA vaccination strategies. Springer Semin Immunopathol. 2006;28:231–238. doi: 10.1007/s00281-006-0047-y. [DOI] [PubMed] [Google Scholar]
- 30.Le Borgne M, Etchart N, Goubier A, Lira SA, Sirard JC, van Rooijen N, et al. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity. 2006;24:191–201. doi: 10.1016/j.immuni.2006.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Characterization of HLA-A2 or Luc expression in cells transfected with pcDNA3-HLA-A2/Luc plasmid. BHK-21 cells (4×104 per well) were transfected with pcDNA3-HLA-A2, pcDNA3-HLA-A2/Luc, pcDNA3-Luc or pcDNA3-No insert plasmid DNA. (A) Flow cytometry analysis to demonstrate expression of HLA-A2 in transfected cells. One day after transfection, transfected cells were analyzed by flow cytometry using HLA-A2 specific antibody. Bold line (1) represents untransfected cells, scattered line (2) represents pcDNA3-HLA-A2 or pcDNA3-HLA-A2/Luc transfected cells, and line (3) represents pcDNA3-No insert transfected cells. The data presented is a representation of two independent experiments. (B) Representative luminescence imaging depicting the luciferase expression level from pcDNA3-HLA-A2/Luc, pcDNA3-Luc or pcDNA3-No insert plasmid DNA transfected cells. (C) Bar graph depicting the quantification of luminescence intensity in each transfected cells. The data presented is a representation of two independent experiments.
Supplementary Figure 2. Intracellular cytokine staining followed by flow cytometry analysis to characterize the HLA-A2-specific CD8+ T cell immune response in vaccinated mice. C57BL/6 mice (5 per group) were immunized with CRT/E7 DNA mixed with no insert or HLA-A2 DNA. One week after the last vaccination, splenocytes from vaccinated mice were harvested. 1×107 pfu of wild type (wild type VV) or HLA-A2 expressed vaccinia virus (HLA-A2 VV) were infected into 1×106 DC-1 cells. One day later, these cells were incubated with splenocytes from vaccinated mice for 16 hours. Cells were characterized for HLA-A2-specific CD8+ T cells using intracellular IFN-γ staining followed by flow cytometry analysis. (A) Flow cytometry analysis to characterize HLA-A2 expression on DC-1 cells infected with wild type VV or HLA-A2 VV. PE-conjugated HLA-A2 antibody was used to detect HLA-A2 expression. The isotype antibody was used as the negative control (grey profile). (B) Representative data of intracellular cytokine staining followed by flow cytometry analysis showing the number of HLA-A2-specific IFNγ+ CD8+ T cells in the various groups (right upper quadrant). (C) Bar graph depicting the numbers of HLA-A2-specific IFN-γ-secreting CD8+ T cells per 3×105 pooled splenocytes (mean± s.d.).


