Abstract
Poxviruses are considered to be unique among all DNA viruses, because their infection cycle is carried out exclusively in the host cytoplasm. Such an infection strategy is of interest, because it necessitates generation of elaborate factories in which viral replication and assembly are promoted. By using diverse imaging techniques, we show that the infection cycle of the largest virus currently identified, the Acanthamoeba polyphaga Mimivirus, similarly occurs exclusively in the host cytoplasm. We further show that newly synthesized mRNAs accumulate at discrete cytoplasmic sites that are distinct from the sites where viral replication occurs, and this is observed in vaccinia infection. By revealing substantial physiologic similarity between poxviruses and Mimivirus and thus, implying that an entirely cytoplasmic viral replication might be more common than generally considered, these findings underscore the ability of DNA viruses to generate large and elaborate replication factories.
Keywords: electron tomography, nucleocytoplasmic large DNA viruses, poxviruses, viral factories
With the single exception of poxviruses, all currently known DNA viruses carry out replication and transcription either entirely or partially within host nuclei. The overwhelming bias to nucleus-centered viral transactions is rationalized by the fact that the nucleus provides the elaborate machinery required for these processes. Moreover, by concentrating essential factors into particular intranuclear sites as well as by enabling spatiotemporal regulation of viral replication and transcription, the nuclear environment considerably enhances the efficiency of these processes (1). Such underlying benefits render the entirely cytoplasmic infection cycle of poxviruses all of the more intriguing (2, 3). Nucleus-centered viral transactions present, however, remarkable hurdles associated with the prerequisite of transporting viral genomes from their entry sites at the cell periphery to and into the nucleus. Specifically, the cellular milieu is refractory to motion of DNA molecules because of the high viscosity of the cytosol and the dense molecular sieve generated by the cytoskeleton (4). Moreover, the nuclear envelope imposes a severe barrier for both entry and exit of long DNA molecules (1).
The hurdles associated with genome trafficking are particularly high for nucleocytoplasmic large DNA viruses (NCLDV), which include the eukaryote-infecting families Poxviridae, Phycodnaviridae, Iridoviridae, and Asfarviridae, all characterized by DNA genomes longer than 100 kbp (5). After entry, genomes of phycodnaviruses (6), iridoviruses (7), and the African swine fever virus (8) are released into the cytoplasm at the host cell periphery and then, are shuttled toward and transported into the nucleus where initial replication cycles occur. Viral DNA is subsequently delivered to specific cytoplasmic factories in which all transactions required for viral assembly take place. The factors that attenuate the barriers imposed by the constrained DNA motion and thus, enable the journey of large viral genomes to, into, and out of the nucleus remain unknown.
The amoeba-infecting giant virus Acanthamoeba polyphaga Mimivirus, which has recently been identified as a member of the NCLDV clade (5, 9), carries a dsDNA genome of 1.2 Mbp. With a genome considerably larger than all currently documented viral genomes, the hurdles inherent for DNA translocations during Mimivirus infection are unparalleled, thus rendering its replication cycle an appealing case study. It was previously shown that a large-scale opening of the Mimivirus capsid at a unique vertex, coined the stargate (10–12), enables rapid release of the genome-containing internal core into the host cytoplasm (10). Ensuing infection events remain, however, poorly understood. Initial structural studies were interpreted as implying that the Mimivirus genome is transported into the nucleus and shuttled back to the cytoplasm after a few rounds of replication (13), which has also been proposed for phycodnaviruses (6) and iridoviruses (7). Such a nuclear-dependent infection pathway was, however, contested by subsequent studies in which spherical inclusions similar in size to the Mimivirus internal core were detected in the host cytoplasm (14, 15).
By combining diverse imaging techniques, we show that, in contrast to all DNA viruses with the exception of poxviruses, the Mimivirus genome is not delivered to the nucleus and never crosses the nuclear membrane. Our studies further reveal that, like poxviruses, transcription is initiated within Mimivirus cores shortly after their delivery to the host cytoplasm. Ensuing genome release from the cores is accompanied by a burst of viral DNA replication within cytoplasmic factories. These observations indicate extensive physiologic similarity between Mimivirus and poxviruses and settle the quandary associated with DNA immobility.
Results
Mimivirus Cores.
At 2 h postinfection (PI) of Acanthamoeba polyphaga cells, Mimivirus particles are detected within host phagosomes. A large-scale opening of the viral icosahedral capsid at a unique vertex-centered structure, coined the stargate, results in the formation of a massive channel, which was previously proposed to enable delivery of the internal genome-containing Mimivirus core to the host cytoplasm (10). To appraise this conjecture, electron microscopy studies were conducted on Mimivirus-infected cells at early (2 h) PI time points. These studies revealed spherical structures that were regularly detected in the cytoplasm of infected amoeba but not in noninfected cells (Fig. 1 and Fig. S1). Notably, the frequency at which these particles are detected correlates with the titer of infecting viruses, which is further discussed below. Electron tomography analyses, performed on cryopreserved host cells at successive PI time points, indicated three distinct manifestations of the spherical particles. Initially, these structures are observed solely in Mimivirus virions enclosed within phagosomes (Fig. 2 A–C and Movie S1). At 2–3 h PI, the host cytoplasm contains free spherical particles (Fig. 2 D–F and Movie S2) as well as spheres surrounded by a dense substance that, as shown below, represents replicating DNA (Fig. 2 G–I and Movie S3). At 4 h PI, all spherical particles reveal the morphologic features depicted in Fig. 2 G–I.
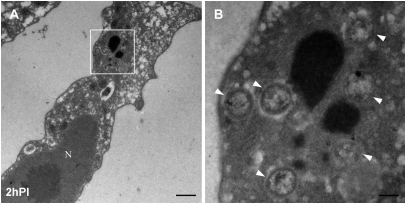
Fig. 1.
Mimivirus cores in the host cytoplasm at early infection stages. Acanthamoeba cells were fixed and processed for transmission electron microscope (TEM) cryosectioning at 2 h PI. (A) TEM image of infected cell. N, nucleus. (Scale bar, 1 μm.) (B) Magnification of the delineated area from A showing multiple cores (arrowheads) in the process of DNA release. (Scale bar, 200 nm.)
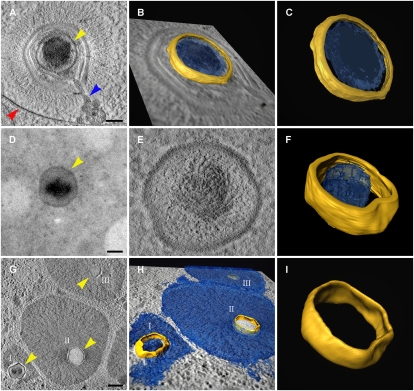
Fig. 2.
Mimivirus cores at successive infection stages. (A) Electron tomography slice of a Mimivirus particle enclosed within a phagosome (red arrowhead) and undergoing initial uncoating and inner membrane protrusion (blue arrowhead). The internal core is indicated by yellow arrowhead. (Scale bar, 100 nm.) (B and C) 3D surface rendering of the viral core shown in A. (D and E) Projection view and electron tomography slice of a free Mimivirus core (yellow arrowhead) in the host cytoplasm. (Scale bar, 100 nm.) (F) 3D surface rendering of the core shown in D and E. The nonuniform substance distribution within the core is consistent with the initiation of intracore transcription (Fig. 4). (G and H) Tomography slice and surface rendering of three early cytoplasmic viral factories surrounding three cores at various stages of genome release with partially (I) and completely (II and III) empty cores. (Scale bar, 300 nm.) (I) 3D surface rendering of core II in G and H. The structural resemblance of the core at the successive stages (C, F, and I) should be noted.
The width of the shell that defines the contours of the spherical particles at all infection phases as well as the overall diameter of these spheres are identical (10 nm and 320 nm, respectively). This finding implies that the spherical particles represent identical entities that correspond to viral cores on the basis of their resemblance to the Mimivirus internal core that was previously characterized by cryoelectron microscopy of whole extracellular Mimivirus particles (11). Yet, the extent of genome packaging within cores at various infection stages significantly differs. Whereas genomes in phagosome-enclosed virions seem to be evenly dispersed throughout the core (Fig. 2 A–C), free cytoplasmic cores reveal a nonuniform internal distribution with a dense substance at the core center (Fig. 2 D–F). This observation raises the possibility that the release of the cores to the cytoplasm coincides with initiation of intracore transactions, a conjecture corroborated by studies described below. In contrast, cores surrounded by replicating DNA are either partially or completely empty (Fig. 2 G–I), implying a temporal link between DNA release from the core and replication onset.
Early Replication and Transcription.
To get deeper insights into the processes that accompany and ensure the delivery of Mimivirus cores to the host cytoplasm, amoeba cells were infected with Mimivirus and incubated for 2 h in the presence of bromodeoxyuridine (BrdU) that is incorporated in actively replicating DNA. Labeling with anti-BrdU antibody along with DAPI counterstaining reveals two distinct cytoplasmic viral inclusions (Fig. 3 and Fig. S2). The first type exhibits an intense dotted DAPI staining with no BrdU labeling, whereas the second is characterized by relatively weak and disperse DAPI staining but extensive BrdU incorporation. After viewing the data presented in Fig. 1, we propose that the first category corresponds to free viral cores (Figs. 1 and 2 D–F), whereas the second type represents cores at a later stage (Fig. 2 G–I) from which genomes are released into the host cytoplasm. Genome release is accompanied with DNA decondensation resulting in attenuated DAPI staining as well as with initiation of massive DNA replication, which was indicated by BrdU incorporation. Thus, synthesis of viral DNA occurs in the host cytoplasm concomitantly with and after the exit of Mimivirus genomes from the cores, leading to the formation of cytoplasmic replication factories around the original viral cores.
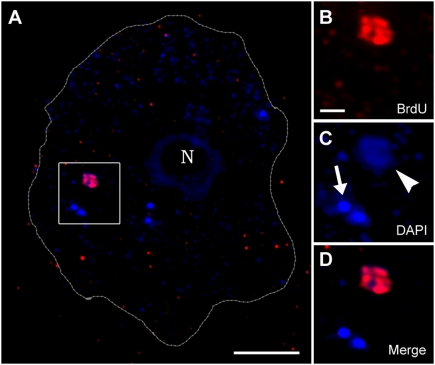
Fig. 3.
Mimivirus replication occurs in the host cytoplasm. Amoeba cells infected with Mimivirus in the presence of BrdU were labeled with anti-BrdU antibody and counterstained with DAPI at 2 h PI. (A) Infected cell containing an early cytoplasmic viral factory where extensive replication occurs; this is indicated by BrdU (red) and DAPI (blue) staining as well as five free viral cores that reveal only a dotted DAPI staining. The host cell contour obtained from a differential interference contrast (DIC) micrograph is delineated with a dashed line. N, host nucleus. (Scale bars, 5 μm.) (B and D) Magnified presentation of the area delineated by a white square in A depicting the BrdU (B), DAPI (C), and merged (D) channels. The weaker DAPI staining revealed by the factory compared with free cores (arrowhead and arrow in C, respectively) is assigned to DNA decondensation occurring on DNA release from the core. (Scale bar, 1 μm.)
Infected amoeba cells were treated with bromouridine (BrU) that is incorporated in transcribed RNA. After cryosectioning and anti-BrU labeling, cells at 4 h PI reveal BrU incorporation that is localized at discrete sites within cytoplasmic viral cores (Fig. 4A and Fig. S3). In addition, hybridization of 4 h PI cells with poly(dT)-Cy5 and costaining with DAPI revealed accumulation of newly transcribed mRNAs at foci adjacent to regions where viral DNA replication occurs (Fig. 4 B–D). These results imply that early transcription is initiated in the cores during DNA release and that the newly synthesized mRNAs subsequently accumulate at discrete cytoplasmic sites that are adjacent to, but not colocalized with, DNA replication sites. Notably, transcription within viral cores is consistent with the nonuniform appearance of free, cytoplasmic viral cores (Fig. 2 D–F).
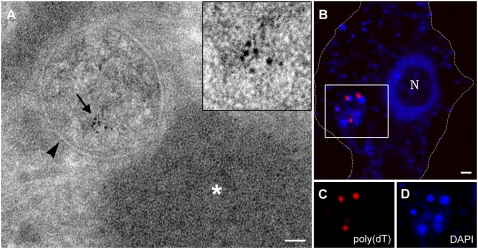
Fig. 4.
Intracore transcription and fate of early mRNAs. (A) Mimivirus-infected amoeba cells transfected with BrU at 2 h PI for 30 min were cryosectioned, and thin sections were treated with anti-BrU at 4 h PI. Exclusive BrU localization at a discrete site (arrow) within the viral core (arrowhead) indicates active transcription in the core. The dense region adjacent to the core (star) represents viral DNA. (Scale bar, 50 nm.) (B) Four hour PI cells were hybridized with poly(dT)-Cy5 (red) and costained with DAPI (blue). Early mRNAs synthesized in the cores accumulate in cytoplasmic sites adjacent to DNA replication sites. The cell contour is delineated on the basis of DIC images. (Scale bar, 1 μm.) (C and D) Single channel magnifications of the delineated square in B.
Cytoplasmic Factories.
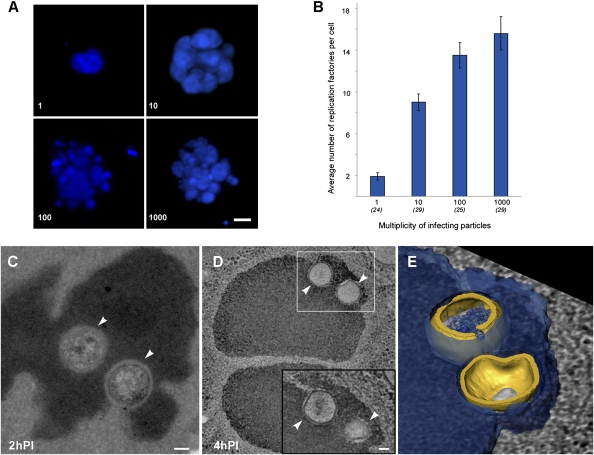
The results presented above indicate that early transcription of the Mimivirus genome occurs within viral cores shortly after their exit from Mimivirus particles and that replication of viral DNA is initiated during genome release into the host cytoplasm. This sequence of events implies that the Mimivirus infection cycle occurs entirely in the cytoplasm. To further assess this notion, amoeba cells were infected with increasing titers of Mimivirus particles and stained with DAPI at 5 h PI (Fig. 5 A and B). A linear correlation is detected between viral titers and the number of replication factories generated within host cells. This observation indicates that each Mimivirus core delivered to the host cytoplasm forms a replication factory at the site where the genome was released from the core. The lower ratio between factories and viral titers observed at high titers can be accounted for by the fusion of several expanding factories that are generated within a discrete site of the cytoplasm and remain localized at this site (Fig. 5 C–E). Notably, fusion of early factories is consistent with previous studies that showed the presence of a single viral factory at late infection stages (10).
Fig. 5.
Correlation between infecting viral particles and viral factories. Amoeba cells were infected with Mimivirus at the indicated multiplicity of infective particles (MOIP) where a MOIP value of 1 corresponds to a viral titer that results in the infection of 90–100% of a given population of host cells. (A) Whole volume view of DAPI-stained cells at 5 h PI. (Scale bar, 1 μm.) (B) Average number of cytoplasmic viral factories as a function of MOIP obtained from the indicated number of infected cells per each MOIP value. Bars are standard errors of the mean. (C–E) Fusion of replication factories at early times PI. Amoeba cells were infected with a viral titer corresponding to MOIP = 100 and processed for cryosectioning. (C) A TEM image of a replication factory seeded from two neighboring viral cores (white arrowheads) in an infected cell at 2 h PI. (Scale bar, 100 nm.) (D) Electron tomography slice of two replication factories at 4 h PI, each generated by the fusion of viral genomes released from multiple viral cores (arrowheads). (Inset) A different tomographic slice of the delineated region. (Scale bar, 100 nm.) (E) Surface rendering of the delineated region in D showing two viral cores (gold) engulfed by viral DNA (blue). Collectively, these observations support the notion that early viral factories, which are generated in the host cytoplasm at the site of genome release from incoming viral particles, eventually fuse to form a single large factory.
Discussion
Cytoplasmic Infection Cycle of Mimivirus.
A main incentive for the current studies was the apparent contradiction between the inherent immobility of free DNA molecules in the cytoplasm (4, 16) and previously reported observations implying that the huge Mimivirus genome is extensively translocated in the host cell during the infection cycle (13). Our findings resolve this quandary by showing very limited genome motility throughout the infection cycle.
In keeping with previous observations (14), our results point to an exclusively cytoplasmic Mimivirus infection cycle. Thus, after a phagosome-mediated entry, the genome-containing viral cores are released into the host cytoplasm (Figs. 1 and 2 A–F). Transcription is initiated within the cores from which newly transcribed mRNAs are delivered to discrete cytoplasmic sites (Fig. 4). These observations are consistent with the previously reported presence of all five RNA polymerase subunits in the Mimivirus core (17). The subsequent release of viral genomes into the cytoplasm is accompanied by a burst of DNA replication that takes place around the viral cores where replication factories are generated (Figs. 2 G–I and 3) in close opposition to the sites where mRNAs accumulated (Fig. 4). Whereas mRNA extrusion in vaccinia was proposed to occur through pores located at the core wall (18) and genome release was proposed to proceed through a major opening of the core (19), the mechanisms responsible for these transactions in Mimivirus remain to be resolved. The correlation between the number of infecting Mimivirus particles and replication factories (Fig. 5 A and B) indicates that each core seeds its own factory and that factories remain around their seeding cores, which is consistent with the results presented in Figs. 1 and 2. When factories expand because of extensive replication, they are not transported to the nucleus but eventually fuse to form a large single factory (Fig. 5) within the host cytoplasm (10).
Poxviruses Paradigm.
In contrast to all other DNA viruses, the infection cycles of poxviruses (2, 3, 20–22) and Mimivirus are entirely cytoplasmic. Early transcription in both poxviruses (20, 23) and Mimivirus is initiated within intact cores after their delivery to the host cytoplasm through the previously described (10–12) stargate portal. As is the case for poxviruses (2, 20), Mimivirus mRNAs accumulate at sites that were distinct from those where viral replication is initiated. Moreover, early Mimivirus factories (Fig. 5), as well as poxvirus factories (22), eventually fuse to form a large cytoplasmic factory. Finally, recent studies indicated that packaging of Mimivirus and poxviruses genomes into preformed capsids proceeds through a similar pathway (10, 24). Thus, whereas structural and phylogenetic analyses position the Mimivirus in between Iridoviridae and Phycodnaviridae (15), the Mimivirus infection cycle implies extensive Poxviridae-like physiology.
Notably, an entirely cytoplasmic Mimivirus infection cycle, which was previously suspected (14, 15) and established here, does not imply a nucleus-independent process, because nuclear factors are likely to be leaked or actively delivered from the host nucleus to the cytoplasmic factory. Indeed, the involvement of nuclear enzymes in the cytoplasmic replication of the Vaccinia virus has recently been shown (22, 25) and suggested to represent leakage of these factors from the host nucleus (25). Nevertheless, a cytoplasmic cycle raises intriguing questions, because the nucleus provides an optimal, ready-made site for viral replication (1). Thus, an exclusive cytoplasmic infection necessitates de novo generation of a platform that confers the machinery, ingredients, and most significantly, elaborate architecture required for replication of large DNA viruses (26, 27). The recently highlighted structural complexity of vaccinia cytoplasmic factories (22) renders the spatiotemporal features that characterize the factory of the larger and more complex Mimivirus into a particularly attractive case study in self-assembly. Moreover, the findings reported here call into question our current understanding of infection strategies of other large DNA viruses of the NCLDV clade for which a nucleus-dependent replication cycle was proposed but not explicitly shown or mechanistically elucidated.
Materials and Methods
Electron Tomography.
Acanthamoeba polyphaga were infected with Mimivirus, processed, and studied by electron tomography as previously described (10). Immunolabeling methods are described in SI Materials and Methods.
Immunofluorescence and BrdU Labeling.
Achanthamoeba cells were grown on glass coverslips in PYG growth medium (2% Proteose Peptone, 0.1% yeast extract, 100 mM glucose) and infected with Mimivirus at MOI of 10. Incorporation of BrdU to visualize viral replication was performed as previously described (28). Briefly, after labeling, cells were washed in PBS, fixed with 4% paraformaldehyde for 15 min, washed, permeabilized with 0.1% Triton X-100, and incubated at room temperature for 1 h in 2% BSA in PBS. Cells were incubated with mouse anti-BrdU antibody (Sigma) for 2 h at room temperature, washed, incubated again with goat anti-mouse conjugated to Cy3 (Jackson) for 45 min, and stained with 0.5 ng/mL DAPI for 15 min at room temperature in the dark. Coverslips were mounted with Mowiol. For multiplicity of infective particles (MOIP) experiments, cells were grown as described above and infected in increasing Mimivirus titers as indicated in Fig. 5.
Fluorescent in Situ Hybridization of Poly(dT) Probes.
Acanthamoeba cells were grown on glass coverslips, infected, and fixed at 4 h PI with 4% paraformaldehyde. Cells were subsequently washed, permeabilized, treated with formamide (40% in 1× SSC buffer at 55 °C) for 5 min and immediately placed on a drop of hybridization buffer (30% Dextran Sulfate, 10 mg/mL BSA, 20 mM Ribonucleoside Vanadyl Complex, and 10 mM sodium phosphate buffer in 2× SSC) containing 500 ng of poly(dT)-Cy5 probe (GeneLink) and 100 ng of carrier DNA (salmon sperm DNA; Sigma). Before the hybridization, probe and carrier were resuspended in 5 volumes of 80% formamide and heated to 85 °C for 5 min. Samples were incubated for 5 h at 55 °C in humid chamber, equilibrated in 40% formamide in 1× SSC for 15 min, washed with 1× SSC twice for 5 min, stained with DAPI, and mounted on slides as described above.
Fluorescence Microscopy and Image Processing.
For all fluorescence experiments, cells were visualized and photographed using a Deltavision system (Applied Precision). Images were deconvoluted with the conservative SoftWorx package. Whole volume images were obtained using the volume-viewer application.
Supplementary Material
Acknowledgments
We thank Dr. Vlad Brumfeld and Vladimir Kiss for their assistance in fluorescence microscopy studies. The electron microscopy and fluorescence studies were conducted at the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging at the Weizmann Institute of Science. This study was supported by the Minerva Foundation, Germany.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912737107/DCSupplemental.
References
- 1.Whittaker GR, Kann M, Helenius A. Viral entry into the nucleus. Annu Rev Cell Dev Biol. 2000;16:627–651. doi: 10.1146/annurev.cellbio.16.1.627. [DOI] [PubMed] [Google Scholar]
- 2.Schramm B, Locker JK. Cytoplasmic organization of Poxvirus DNA replication. Traffic. 2005;6:839–846. doi: 10.1111/j.1600-0854.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 3.Moss B. In: Fields Virology. Knipe DM, editor. Vol. 74. Philadelphia: Lippicott Williams & Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- 4.Dauty E, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm. J Biol Chem. 2005;280:7823–7828. doi: 10.1074/jbc.M412374200. [DOI] [PubMed] [Google Scholar]
- 5.Iyer LA, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Yamada T, Onimatsu H, Van Etten JL. Chlorella viruses. Adv Virus Res. 2006;66:293–336. doi: 10.1016/S0065-3527(06)66006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams T, Barbosa-Solomieu V, Chinchar VG. A decade of advances in iridovirus research. Adv Virus Res. 2005;65:173–248. doi: 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- 8.Eulalio A, et al. African swine fever virus p37 structural protein is localized in nuclear foci containing the viral DNA at early post-infection times. Virus Res. 2007;130:18–27. doi: 10.1016/j.virusres.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 10.Zauberman N, et al. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga Mimivirus. PLoS Biol. 2008;6:1104–1114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao CA, et al. Cryo-electron microscopy of the giant Mimivirus. J Mol Biol. 2005;353:493–496. doi: 10.1016/j.jmb.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 12.Xiao C, et al. Structural studies of the giant Mimivirus. PLoS Biol. 2009;7:958–966. doi: 10.1371/journal.pbio.1000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzan-Monti M, La Scola B, Barrassi L, Espinosa L, Raoult D. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One. 2007;2:e328. doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claverie JM, et al. Mimivirus and Mimiviridae: Giant viruses with an increasing number of potential hosts, including corals and sponges. J Invertebr Pathol. 2009;101:172–180. doi: 10.1016/j.jip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Claverie JM, Abergel C. Mimivirus and its virosome. Annu Rev Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 16.Weiner A, Zauberman N, Minsky A. Recombinational DNA repair in a cellular context: A search for the homology search. Nat Rev Microbiol. 2009;7:748–755. doi: 10.1038/nrmicro2206. [DOI] [PubMed] [Google Scholar]
- 17.Renesto P, et al. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol. 2006;80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cyrklaff M, et al. Cryo-electron tomography of vaccinia virus. Proc Natl Acad Sci USA. 2005;102:2772–2777. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cyrklaff M, et al. Whole cell cryo-electron tomography reveals distinct disassembly intermediates of vaccinia virus. PLoS One. 2007;2:e420. doi: 10.1371/journal.pone.0000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallardo M, et al. Relationship between vaccinia virus intracellular cores, early mRNAs, and DNA replication sites. J Virol. 2002;76:5167–5183. doi: 10.1128/JVI.76.10.5167-5183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broyles SS. Vaccinia virus transcription. J Gen Virol. 2003;84:2293–2303. doi: 10.1099/vir.0.18942-0. [DOI] [PubMed] [Google Scholar]
- 22.Katsafanas GC, Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallardo M, Schleich S, Locker JK. Microtubule-dependent organization of vaccinia virus core-derived early mRNAs into distinct cytoplasmic structures. Mol Biol Cell. 2001;12:3875–3891. doi: 10.1091/mbc.12.12.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Locker JK. Membrane rupture generates single open membrane sheets during Vaccinia Virus assembly. Cell Host Microbe. 2009;6:81–90. doi: 10.1016/j.chom.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Oh J, Broyles SS. Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J Virol. 2005;79:12852–12860. doi: 10.1128/JVI.79.20.12852-12860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novoa RR, et al. Virus factories: Associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traktman P, Boyle K. Methods for analysis of poxvirus DNA replication. Methods Mol Biol. 2004;269:169–186. doi: 10.1385/1-59259-789-0:169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.