Abstract
Ducks and wild waterfowl perpetuate all strains of influenza viruses in nature. In their natural host, influenza viruses typically cause asymptomatic infection and little pathology. Ducks are often resistant to influenza viruses capable of killing chickens. Here, we show that the influenza virus sensor, RIG-I, is present in ducks and plays a role in clearing an influenza infection. We show evidence suggesting that RIG-I may be absent in chickens, providing a plausible explanation for their increased susceptibility to influenza viruses compared with ducks. RIG-I detects RNA ligands derived from uncapped viral transcripts and initiates the IFN response. In this study, we show that the chicken embryonic fibroblast cell line, DF-1, cannot respond to a RIG-I ligand. However, transfection of duck RIG-I into DF-1 cells rescues the detection of ligand and induces IFN-β promoter activity. Additionally, DF-1 cells expressing duck RIG-I have an augmented IFN response resulting in decreased influenza replication after challenge with either low or highly pathogenic avian influenza virus. Implicating RIG-I in the antiviral response to an infection in vivo, we found that RIG-I expression is induced 200 fold, early in an innate immune response in ducks challenged with the H5N1 virus A/Vietnam/1203/04. Finding this natural disease resistance gene in ducks opens the possibility of increasing influenza resistance through creation of a transgenic chicken.
Keywords: comparative immunology, evolution, H5N1 avian influenza, animal reservoir, chicken
All strains of influenza A virus are perpetuated in the duck reservoir (1). Although rare, when influenza viruses cross over from the avian reservoir into humans they may evolve into pandemic strains. After emerging in 1996, the highly pathogenic avian influenza (HPAI) H5N1 viruses have evolved into multiple clades and subclades and have spread to Europe and Africa, probably by wild birds (2). Although H5N1 viruses do not frequently infect humans they are often highly pathogenic and cause death in otherwise healthy individuals. Among 445 laboratory-confirmed human H5N1 infections, the mortality rate is 60% (3).
In 2002, H5N1 viruses began killing wild waterfowl (4, 5), including ducks, which is highly unusual and a disruption of the normal ecology (1). Typically, ducks do not show signs of disease upon infection with influenza. H5N1 viruses may rapidly adapt, becoming less pathogenic to ducks (6, 7), suggesting that virus evolution helps maintain the balance between virus and host. Many strains of highly pathogenic H5N1 now cause asymptomatic infection in ducks, making them a “Trojan horse” in the spread of these influenza viruses (6–9). In contrast, many of these same H5N1 viruses cause 100% mortality in chickens within hours or days. We therefore reasoned that superior innate immunity might protect the duck during this critical period.
The molecular basis of the natural resistance of ducks to influenza infection is unresolved. A successful innate immune response to influenza infection involves a robust, yet transient induction of IFN-stimulated antiviral genes. RIG-I is a cytoplasmic RNA sensor (10), and triggering by influenza virus leads to production of IFN-β and expression of downstream IFN-stimulated antiviral genes (11). A hallmark of lethal influenza virus infection is interference with expression of RIG-I and downstream genes (12). Further, RNA viruses have been shown to be more virulent and replicate to higher levels in mice lacking RIG-I (13). Considering this, we speculated that the outcome of influenza infection in birds may also be determined by RIG-I and the downstream subset of IFN-stimulated genes. Here we show that the duck, the natural host of influenza, has an intact and functional RIG-I that is induced upon infection with H5N1 A/Vietnam/1203/04 (VN1203), whereas chickens appear to lack RIG-I. However, we demonstrate that transfection of duck RIG-I into a chicken embryonic fibroblast cell line can restore recognition of RIG-I ligand and initiate an antiviral IFN response.
Results
RIG-I Is Present in Ducks and Apparently Absent in Chickens.
Given its role in antiviral defense in mammals, we searched for avian homologues of RIG-I. We identified a duck (Anas platyrhynchos) RIG-I homologue with 53% amino acid identity to human and 78% identity to zebra finch RIG-I (Fig. 1). Remarkably, we are unable to identify a chicken homologue of RIG-I, even though we used a variety of approaches to identify one. Searches of the chicken (Gallus gallus) genome (14) with the duck or finch RIG-I sequence do not reveal a match. In addition, a phylogenetic analysis of RIG-like receptors also noted that RIG-I was absent in chickens (15). However, the related melanoma differentiation associated gene–5 (MDA5) is present in the chicken genome (16). RIG-I and MDA5 initiate signaling cascades that converge on the same pathway at IPS-1 and lead to induction of IFN-β and expression of downstream IFN-stimulated antiviral genes (Fig. S1). MDA5 is a detector of long double-stranded RNA, polyinosinic–polycytidylic acid [poly (I:C)], and picornaviruses (13). To ensure that the helicase we isolated was RIG-I and distinct from MDA5, we amplified a large fragment of duck MDA5. The fragment of MDA5 shared 91% amino acid identity with chicken MDA5 (Fig. S2), yet only 33% identity with duck RIG-I.
Fig. 1.
Amino acid alignment of duck, zebra finch, and human RIG-I. Alignment of duck RIG-I (accession no. EU363349), zebra finch (accession no. XM_002194524), and human RIG-I (accession no. AF038963) was performed using the ClustalW program and edited with Boxshade. Black shading indicates amino acid identity and gray shading indicates similarity (50% threshold). Plus signs indicate human residues involved in polyubiquitination and asterisks indicate residues involved in ligand binding. The ATP binding motif is boxed.
We identified the RIG-I syntenic region on the chicken Z chromosome where acontinase I is encoded; this is a gene flanking the mammalian RIG-I homologue. A local BLAST search of the adjacent 4 Mb reveals no match to RIG-I, although there are sequence ambiquities in this region. Confirming the syntenic region is conserved in other birds, we identified a RIG-I homologue in the recently released draft genome for zebra finch (Taeniopygia guttata) on chromosome Z and flanked by acontinase I. A search of the finch expressed sequence tag (EST) database revealed two RIG-I transcripts among the 92,000 sequences. In contrast, no RIG-I sequences are present among the 600,000 ESTs from chicken. Notably, MDA5 transcripts are present. Thus, RIG-I appears to be absent from the chicken genome sequence derived from the Red Jungle Fowl, which resembles the ancestral chicken, as well as the modern chicken lines represented in the EST sequences. Combined, these data suggest that chickens may have lost RIG-I before their domestication.
Avian RIG-I proteins have features in common with mammalian RIG-I. Duck RIG-I is 933 amino acids, and zebra finch RIG-I is 927 amino acids. Domain prediction reveals the expected tandem N-terminal CARD domains, a helicase domain and a DeXD/H box helicase domain, consistent with the mammalian structure (10, 17). RIG-I is a ligand-dependent ATPase, and the Walker A ATP-binding motif is conserved. The hydrophobic core and the four lysine residues implicated in ligand-binding, K858/861/888/907 (17), are completely conserved within the C-terminal regulatory domain. However, residues T55 and K172, critical for interaction and polyubiquitination by TRIM25 needed for IPS-1 binding and signal induction (18, 19), are not conserved, suggesting this pathway does not function or involves different residues in birds.
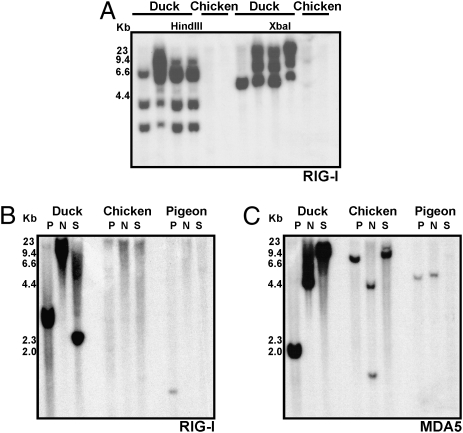
To provide further evidence for the absence of RIG-I in chickens, we hybridized a Southern blot of genomic DNA from ducks and chickens with a probe amplified from the helicase region of duck RIG-I (Fig. 2A). Although we observed a polymorphic pattern of hybridization to duck DNA, we did not detect cross-hybridization to chicken DNA. However, the duck probe cross-hybridizes with RIG-I from the more phylogenetically distant pigeon (Fig. 2B), providing evidence that the probe can recognize RIG-I from other avian species. The duck RIG-I probe hybridized to pigeon DNA in only the PstI digestion, although this may be because the NdeI and SpeI digestions produced hybridizing fragments of DNA too small to be visualized. Hybridization of our duck probe to pigeon DNA was observed on several other blots, whereas hybridization to chicken DNA was never observed. It is noteworthy that pigeons are remarkably resistant to influenza viruses, including HPAI H5N1 (20). In comparison, strong cross-hybridization of a duck MDA5 probe with DNA from pigeon and chicken (Fig. 2C) suggests it has diverged considerably less than RIG-I and is presumably under less selective pressure in avian species. Demonstrating that our duck RIG-I probe hybridizes to pigeon but not to chicken genomic DNA further supports the absence of RIG-I in chickens. However, we cannot rule out that chicken RIG-I has diverged to an extent that it is not detectable through bioinformatics or hybridization approaches, which may preclude function in any case.
Fig. 2.
RIG-I is present in ducks and pigeons, but apparently absent in chickens. (A) Hybridization of a multiple exon duck RIG-I probe to HindIII and XbaI-digested genomic DNA from four White Pekin ducks and two White Leghorn chickens. (B) Hybridization of a single exon duck RIG-I probe to PstI, NdeI, and SacI digested genomic DNA from duck, chicken, and pigeon. (C) Hybridization of a single exon duck MDA5 probe to same blot.
Duck RIG-I Detects in Vitro Transcribed RNA and Activates the Chicken IFN-β Promoter.
The apparent absence of RIG-I in chickens led us to investigate whether the DF-1 chicken cell line (chicken embryonic fibroblasts) can respond to a RIG-I ligand. RIG-I signaling is activated by 5′triphosphate RNA (5′ppp RNA) containing short regions with double-stranded conformation, such as that derived from viral RNA with panhandle structures (21, 22), and from in vitro transcribed products (23). With this in mind, we challenged DF-1 cells with an in vitro transcribed 21mer 5′ppp RNA shown to activate mammalian RIG-I (23). Transfection of this ligand failed to activate a chicken IFN-β promoter-luciferase reporter (Fig. 3A). Poly (I:C), known to drive the chicken IFN-β promoter in this cell line (24), showed a 2.4-fold increase in IFN-β promoter activity compared with unstimulated cells. Poly (I:C) triggers the cytoplasmic receptor MDA5 and the endosomal dsRNA receptor TLR3. However, the response to poly (I:C) in DF-1 cells is largely through MDA5, as viral inhibition of MDA5 signaling nearly abrogates IFN-β promoter activity (23). Thus DF-1 cells respond to poly (I:C) mostly through MDA5, demonstrating that they possess the downstream components of the shared RIG-I/MDA5 pathway.
Fig. 3.
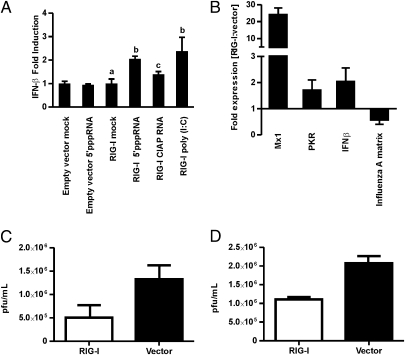
Duck RIG-I rescues detection of 5′ppp RNA and induces an antiviral response in DF-1 chicken embryonic fibroblast cells. (A) IFN-β promoter activity in RIG-I or empty vector–transfected DF-1 cells following 15 h of ligand stimulation compared to mock-treated cells, shown as mean fold induction (±SD). Results are representative of three independent experiments and were analyzed using a single-factor ANOVA and Tukey post hoc test (different letters, P < 0.05). (B) RIG-I–transfected DF-1 cells respond to BC500 infection (MOI, 1) with increased expression of chicken IFN-β and the IFN-stimulated genes Mx1 and PKR, and decreased influenza matrix gene expression, relative to empty vector–transfected cells. RNA was extracted from cells for qRT-PCR 15 h PI, and fold difference in gene expression calculated for RIG-I and vector-only–transfected DF-1 cells. Results are representative of three independent experiments and error bars show RQMin/Max at a 95% confidence level and represent SE (n = 3). (C and D) RIG-I–transfected DF-1 cells had significantly lower influenza virus titers compared with empty vector–transfected cells for BC500 (C) or VN1203 (D). Both infections were performed 24 h after transfection at an MOI of 1. After 15 h, titer was determined by plaque assay from triplicate wells and results were analyzed with the two-tailed Student‘s t test (n = 3; P = 0.002).
We next investigated whether duck RIG-I could confer recognition of a RIG-I ligand and signal through these downstream components. DF-1 cells were transfected with duck RIG-I before stimulation with 5′ppp RNA. DF-1 cells expressing duck RIG-I responded to 5′ppp RNA with a twofold induction of the IFN-β promoter compared with mock-transfected cells. However, phosphatase removal of the 5′ triphosphate abrogated the response, similar to the previous report in mammalian cells (23). Thus duck RIG-I is functional and can induce IFN-β promoter activity in the DF-1 chicken cell line. Although the twofold induction of the IFN-β promoter is modest, this is consistent with the up-regulation by poly (I:C) stimulation downstream of chicken MDA5. Given the estimated 90 My of divergence between chicken and duck (25), it is remarkable that the duck RIG-I even binds to chicken IPS-I to connect to downstream signaling components.
Transfected Duck RIG-I Detects Influenza and Induces an Antiviral Response in Chicken Cells.
To determine if duck RIG-I could detect influenza virus and induce an antiviral response in the chicken DF-1 cells, we transfected the cells with duck RIG-I or vector only, followed by infection with influenza viruses. We chose H5N2 A/mallard/British Columbia/500/2005 (BC500), a low pathogenic avian influenza (LPAI) isolated from wild ducks that causes no pathology in its natural host and VN1203, an HPAI isolated from a fatal human infection and known to be lethal to ducks and chickens (7). After a 15 h infection with BC500, there was increased expression of IFN-β as well as the antiviral IFN-stimulated genes Mx1 and PKR, known to be RIG-I responsive in mouse fibroblasts (11) (Fig. 3B). Although IFN-β and PKR were only slightly up-regulated, the IFN-stimulated gene Mx1 was induced approximately 30 fold. Influenza A matrix gene expression was significantly reduced in RIG-I–transfected DF-1 cells compared with vector-transfected control cells (Fig. 3B). Furthermore, the IFN response initiated by duck RIG-I resulted in significant reduction in viral titer for both BC500 (Fig. 3C) and VN1203 (Fig. 3D) infection, indicating that duck RIG-I is capable of reducing influenza replication in chicken cells.
RIG-I Is Highly Up-Regulated in Ducks Infected with VN1203.
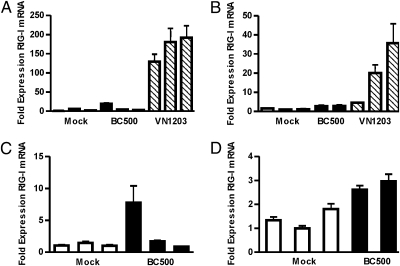
To determine whether RIG-I contributes to the antiviral response to influenza infection in ducks, we measured the expression of RIG-I in 6-week-old White Pekin duck tissues following infections with BC500 (H5N2) or VN1203 (H5N1). Infection with VN1203 induced significant up-regulation of RIG-I gene expression in the infected lung (Fig. 4A). RIG-I expression was induced more than 200-fold by d 1 postinfection (PI), whereas by d 3 PI, RIG-I was only modestly expressed, suggesting the induction is early and transient (Fig. 4B). In comparison, infection with BC500 induced only slight up-regulation of RIG-I in lung tissue. Because LPAI strains predominately replicate within the intestinal epithelium of ducks (26), we also assessed RIG-I expression in BC500 infected duck intestine (Fig. 4 C and D). RIG-I expression was not significantly induced by BC500 infection in duck intestine on d 1 or d 3 PI. Duck RIG-I is thus expressed early during an innate immune response to a highly pathogenic influenza virus. However, it is not clear why VN1203 infection results in tremendous up-regulation of duck RIG-I whereas BC500 does not. Considering that RIG-I expression is itself regulated by IFN, high expression in the lungs following VN1203 infection may reflect IFN levels. Additionally, influenza viruses vary in their ability to actively inhibit IFN induction in birds and mammals (27, 28), a function that is dependent on the viral protein NS1 (29, 30) that directly targets the RIG-I pathway preventing activation of IFN-β (31). One possibility is that the NS1 protein of BC500 interferes with viral activation of the IFN pathway more efficiently than the NS1 of VN1203. Although VN1203 is potentially lethal in ducks (7), none of the ducks showed severe symptoms or died within 3 d of the experiment. We show duck RIG-I is functional and initiates antiviral gene expression in chicken cells. Thus, we speculate that the robust and transient up-regulation of RIG-I contributes to the successful innate response of ducks.
Fig. 4.
RIG-I is dramatically up-regulated in duck lung infected with VN1203 but not BC500. Lung and intestinal RNA was extracted d 1 or d 3 PI and analyzed by qRT-PCR for RIG-I expression compared with a mock-infected animal. Fold expression of RIG-I mRNA in duck lung tissue infected with BC500 or VN1203 at d 1 PI (A) or d 3 PI (B). Fold expression of RIG-I mRNA in duck intestine infected with BC500 at d 1 PI (C) or d 3 PI (D). Error bars show RQMin/Max at a 95% confidence level and represent SE (n = 4).
Discussion
This study establishes the presence of RIG-I in ducks—the natural reservoir of influenza viruses—but its apparent absence in chickens. This may reflect the differential susceptibility of ducks and chickens to influenza-induced pathology. We found that RIG-I is expressed during the innate immune response to influenza infection in ducks, providing evidence for the antiviral relevance of RIG-I in the natural host of the virus. RIG-I is highly up-regulated early in an innate immune response to VN1203 infection in ducks, likely contributing to antiviral defense against this potentially lethal virus. In contrast, RIG-I appears to be absent in chickens and a chicken embryonic fibroblast cell line fails to respond to 5′ppp RNA, a function that can be conferred to the cells by transfection with duck RIG-I. Expression of duck RIG-I augments the antiviral IFN response and reduces influenza replication in chicken cells.
Despite the lack of RIG-I, chicken cells do produce IFNs. Indeed, IFNs were initially discovered in chicken cells treated with heat-inactivated influenza virus (32). Although other pathways can produce IFN-α, IFN-β expression upon influenza infection is largely dependent on RIG-I. Embryonic fibroblasts from a RIG-I–KO mouse fail to induce IFN-β and a subset of genes involved in innate immunity following influenza infection (11). siRNA knockdown of RIG-I (33) or introduction of a dominant-negative RIG-I (34) has been demonstrated to significantly reduce the influenza-induced IFN-β production in human cell lines. Additionally, IFN-β–KO mice show reduced survival and enhanced influenza viral titers in the lung (35). Thus, IFN-β appears to be protective during an influenza infection and cannot be compensated for by IFN-α. Chicken embryonic fibroblasts infected with influenza produce IFN and inhibit an IFN-sensitive GFP-tagged vesicular stomatitis virus (27); however, this response is 80% IFN-α (36). Infection of primary chicken embryo fibroblasts with H5N1 strains induced low Mx1 and IFN-α gene expression, whereas IFN-β was not significantly induced (37). Similar to influenza infection, Newcastle Disease Virus, also detected by RIG-I, causes substantially more pathology in chickens than ducks. In accord with our findings, others have found that Newcastle Disease Virus infection does not induce IFN-β promoter activity in chicken embryonic fibroblasts (38), also consistent with lack of RIG-I.
Here we provide evidence for the antiviral function of RIG-I in ducks, and its apparent absence in chickens. RIG-I independent pathways, such as TLR7, also contribute to influenza detection and IFN production in chickens (39) and ducks (40). However, chickens lack a key regulator of the antiviral response, so it is not surprising that they suffer remarkable pathology from HPAI infection compared with ducks. Chickens lacking RIG-I would be without a first line of defense at the lung epithelial cell layer during influenza infection, and antiviral genes downstream of RIG-I may not be expressed (11). Considering that RIG-I may also inhibit influenza replication directly and independent of IFN (10), loss of this undefined pathway may also contribute to unchecked virus replication in chickens. RIG-I loss-of-function mutants in humans (41, 42) may also be predicted to cause increased viral pathogenesis. The presence of a functional RIG-I in ducks eliciting an early antiviral response may contribute to survival of what may otherwise be a lethal influenza infection.
Materials and Methods
Identification and Cloning of Duck RIG-I.
A PCR fragment of RIG-I was obtained from duck (A. platyrhynchos) splenic cDNA using primers 5′-GAT CCC AGC AAT GAG AAT CCT AAA CT-3′ and 5′-CAA TGT CAA TGC CTT CAT CAG C-3′ based on a conserved region of the human RIG-I sequence. The complete cDNA sequence was obtained via 5′ and 3′ RACE using the SMART RACE cDNA Amplification Kit (Clontech). The complete coding region of duck RIG-I was amplified using primers in the 5′UTR 5′-CGG CCG GCA GAG CCC AGC C-3′ and 3′ UTR 5′-GTG TAG GAG AGT AAT AGA TGC ACT A-3′ using Phusion High-Fidelity DNA Polymerase (New England Biolabs), cloned into pCR 2.1-TOPO (Invitrogen) and completely sequenced.
Plasmids.
pcDNA-RIG was obtained by cloning duck RIG-I into the mammalian expression vector pcDNA 3.1 hygro+ (Invitrogen). Duck RIG-I was digested out of pCR 2.1-TOPO (Invitrogen) using SpeI and NotI. RIG-I was then inserted between NheI and NotI sites of pcDNA 3.1 hygro+ (Invitrogen). The chicken IFN-β promoter luciferase reporter (pGL3-chIFNβ) was constructed from White Leghorn chicken genomic DNA using primers with incorporated BglII and MluI sites that amplified −158 to +14 of the chicken IFN-2 promoter, as previously done (24, 38). The promoter fragment was then inserted between BglII and MluI sites of the pGL3-basic luciferase reporter vector (Promega).
In Vitro Transcription and RNAs.
The 21-mer RNA, 5′-pppGGGGCUGACCCUGAAGUUCCC-3′ (23) was transcribed from annealed DNA oligonucleotides 5′-TAA TAC GAC TCA CTA TAG GG-3′ and 5′-GGG AAC TTC AGG GTC AGC CCC TAT AGT GAG TCG TAT TA-3′ containing a T7 promoter site, using the T7 Megashortscript kit (Ambion). In vitro transcription was carried out overnight, followed by DNaseI digestion and precipitation. Calf intestinal alkaline phosphatase (CIAP) treatment was carried out for 3 h using 30 μg of in vitro transcribed RNA, 30 U of CIAP (Invitrogen), 1× CIAP buffer, and 200 U RNase Out (Invitrogen). CIAP-treated 5′ppp RNA was purified by phenol-chloroform extraction and precipitation. Poly (I:C) (25 mg/mL) was obtained from InVivogen.
Cell Culture, Infections, and Transfections.
UMNSAH/DF-1, a spontaneously immortalized chicken embryonic fibroblast cell line derived from East Lansing strain eggs (43), was maintained in DMEM plus 10% FBS. Cells (1.25 × 105) were seeded overnight in 24-well plates. Cells were cotransfected with 150 ng of pcDNA-RIG or empty pcDNA, 150 ng of pGL3-chIFNβ, and 10 ng of the constitutive renilla luciferase reporter phRG-TK (Promega). Thirty hours after plasmid transfection, cells were challenged with 21-mer 5′ppp RNA (800 ng), CIAP-treated RNA (800 ng), or poly (I:C) (25 μg/mL). Plasmids and RNA ligands were transfected using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) 15 h after challenge. For infection of transfected cells, DF-1 cells were maintained in DMEM plus 10% FBS and 1.25 × 105 cells were seeded overnight in 24-well plates. Cells were transfected with 1 μg of pcDNA-RIG or empty pcDNA. Twenty-four hours after transfection, cells were infected at a multiplicity of infection (MOI) of 1 with H5N2 A/mallard/BC/500/05 or H5N1 A/Vietnam/1203/04 virus. L-(tosylamido-2-phenyl) ethyl chloromethyl ketone–treated trypsin (Worthington Biochemicals) was used for infection with BC500 at a low concentration (0.1 μg/mL) in view of the trypsin sensitivity of DF-1 cells (44). Exogenous trypsin was not added for infection with VN1203. Fifteen hours after infection, RNA was extracted from cells for quantitative RT-PCR (qRT-PCR) and influenza virus titer was determined by plaque assay on MDCK cells.
Viruses and Duck Infections.
The H5N1 A/Vietnam/1203/04 HPAI was generated by reverse genetics (45) and H5N2 A/mallard/BC/500/05 LPAI was isolated by screening of environmental samples. The viruses were propagated in 10-d-old embryonated chicken eggs and handled at St. Jude Children's Research Hospital, with VN1203 handled in biosafety level 3+ facilities approved by the United States Department of Agriculture and Centers for Disease Control and Prevention. Outbred White Pekin ducks (A. platyrhynchos) were purchased from Ideal Poultry or Metzer Farms, and all animal experiments were approved by the Animal Care and Use Committee of St. Jude Research Hospital and performed in compliance with relevant institutional policies, National Institutes of Health regulations, and the Animal Welfare Act. A total of 106 of 50% egg infectious dose of BC500 and VN1203 were used to inoculate 6-week-old mallards via the natural route, in nares, eyes, and trachea. Mock infection was PBS only. Ducks were killed and tissues collected at d 1 and d 3 PI (n = 3, except for BC500 d 3 samples, n = 2). Tracheal and cloacal swabs were collected to monitor viral shedding.
Southern Hybridization.
Genomic DNA was extracted from blood of White Pekin ducks (A. platyrhynchos), White Leghorn chickens (G. gallus), and pigeon (Columba livia). Genomic DNA (10 μg) was digested to completion, separated on 0.8% agarose, and blotted to Nytran Supercharge (Schleicher & Schuell). DNA was immobilized by UV cross-linking and baking for 3 h at 80 °C. A multiple-exon 307-bp RIG-I probe in the helicase domain was amplified using the primers 5′-ACA GGT ATG ACC CTC CCA AGC CAG-3′ and 5′-CAT CCC ATT TCT GGA TCT TTT CAA CAG-3′ from a defined duck RIG-I clone. A 171-bp probe predicted to contain a single exon was amplified from the same template using 5′-GTA TGA CCC TCC CAA GCC AGA AGG -3′ and 5′-CTC TGA CTT GGA TCA TTT TGG TCA CG-3′. A 150-bp duck MDA5 probe was amplified with the following primers: 5′-GAG CAA AGG GAA GTC ATT GAT AAA TTC C-3′ and 5′-GGC CTG CAA CAT AGC AAT TTC ATT-3′ from a duck MDA5 clone. All probes were radiolabeled with 32P α-dCTP by random priming (PrimeIt Random Primer Labeling Kit; Stratagene). Blots were hybridized overnight at 42 °C in 50% formamide, 5× Denhardt solution, 4× SSPE, 5% dextran sulfate, 1% SDS, and 100 μg/mL salmon sperm DNA. Washes were carried out at low stringency in 1× sodium chloride/sodium phosphate/EDTA and 0.1% SDS at 52 °C. Film was exposed for 3 d.
Real-Time qRT-PCR.
RNA was extracted using TRIzol (Invitrogen), followed by purification from the final aqueous phase using the RNeasy Mini Kit (Qiagen). First strand cDNA synthesis was performed using the SuperScript III kit (Invitrogen) using OligodT (Invitrogen) and random primers (Invitrogen). To quantify RIG-I gene expression from duck tissues, 50 ng of cDNA was amplified in a 10-μL reaction using the Applied Biosystems 7500 real-time PCR system (Applied Biosystems). Duck primers and fluorogenic TaqMan FAM/TAMRA (6-carboxyfluorescein/6-carboxytetramethylrhodamine)–labeled hybridization probe mixes were obtained from Applied Biosystems and used with FastStart Universal Probe Master (Rox; Roche Applied Science). Duck GAPDH was used as an endogenous control. Primer and probe sequences were as follows: duck RIG-I primers 5′-GTG TAT GGA GGA AAA CCC TAT TTC TTA ACT-3′ and 5′-GGA GGG TCA TAC CTG TTG TTT GAT-3′, and probe 5′-TTC CGC GCC CCA TCA A-3′. Duck GAPDH primers were 5′-GCC TCT TGC ACC ACC AAC T-3′ and 5′-GGC ATG GAC AGT GGT CAT AAG AC-3′ and probe 5′-CAC AAT GCC AAA GTT G-3′. Changes in gene expression PI were expressed as a ratio of the level observed in a mock-infected animal. RT-PCR was performed for RIG-I and GAPDH in a single-plex format, with the following cycling conditions: 95 °C for 10 min for activation, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Quadruplicate cycle threshold CT values were analyzed with SDS software (Applied Biosystems) using the comparative CT (ΔΔCT) method.
To quantify gene expression from transfected DF-1 cells, 20 ng of cDNA was amplified in a 10-μL reaction using the Applied Biosystems 7500 real-time PCR system (Applied Biosystems). Chicken 28s RNA (endogenous control) and IFN-β probes and primer (46) were obtained from Applied Biosystems and used with TaqMan Fast Universal PCR Master Mix (Applied Biosystems). For Influenza A matrix gene expression, primers were designed according to CDC recommendations. InfA forward primer was 5′-GAC CRA TCC TGT CAC CTC TGA C-3′; InfA reverse was 5′-AGG GCA TTY TGG ACA AAK CGT CTA-3′; and InfA probe 5′-TGC AGT CCT CGC TCA CTG GGC ACG-3′ (Centers for Disease Control and Prevention). The primers and probes for chicken Mx1 (accession no. NM_204609) and PKR (accession no. NM_204487.1) were designed with the Roche online Universal Probe Library (UPL) system Assay Design Center. The primers for Mx1 were 5′-GTT TCG GAC ATG GGG AGT AA-3′ and 5′-GCA TAC GAT TTC TTC AAC TTT GG -3′ (UPL probe 80). The primers for PKR were 5′-TGC TTG ACT GGA AAG GCT ACT-3′ and 5′-TCA GTC AAG AAT AAA CCA TGT GTG-3′ (UPL probe 29). Cycling conditions were 95 °C for 20 s for activation, followed by 40 cycles at 95 °C for 3 s and 60 °C for 30 s. Changes in gene expression were expressed as a ratio of the level observed with cells transfected with RIG-I or vector only.
Supplementary Material
Acknowledgments
We thank Patrick Seiler for excellent technical assistance in BL3, John Pasick for the BC500 influenza isolate, Rick Wood-Samman for provision of avian blood samples, Troy Locke and John Franks for qRT-PCR technical assistance, Savita Shrivastava for bioinformatics help, and Brad Magor for helpful comments on the research and manuscript. This work was supported by the Canadian Institute for Health Research (K.E.M) and Natural Sciences and Engineering Research Council of Canada (NSERC) (K.E.M); Contract HHSN266200700005C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services; and American Lebanese Syrian Associated Charities (R.G.W). M.R.W.B. is supported by an NSERC Postgraduate Scholarship and a Canadian Poultry Research Council Postgraduate Scholarship Supplement.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU363349 and GU936632).
This article contains supporting information online at www.pnas.org/cgi/content/full/1001755107/DCSupplemental.
References
- 1.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Mol Biol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duan L, et al. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology. 2008;380:243–254. doi: 10.1016/j.virol.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to the World Health Organization. 2009. Available at http://www.who.int/csr/disease/avian_influenza/country/cases_table_2009_12_11/en/index.html. Accessed December 16, 2009.
- 4.Ellis TM, et al. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 2004;33:492–505. doi: 10.1080/03079450400003601. [DOI] [PubMed] [Google Scholar]
- 5.Sturm-Ramirez KM, et al. Reemerging H5N1 influenza viruses in Hong Kong in 2002 are highly pathogenic to ducks. J Virol. 2004;78:4892–4901. doi: 10.1128/JVI.78.9.4892-4901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulse-Post DJ, et al. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci USA. 2005;102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulse-Post DJ, et al. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J Virol. 2007;81:8515–8524. doi: 10.1128/JVI.00435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Negovetich NJ, Forrest HL, Webster RG. Ducks: the “Trojan horses” of H5N1 influenza. Influenza Other Respir Viruses. 2009;3:121–128. doi: 10.1111/j.1750-2659.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sturm-Ramirez KM, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 11.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 13.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 14.International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 15.Zou J, Chang M, Nie P, Secombes CJ. Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol. 2009;9:85. doi: 10.1186/1471-2148-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sarkar D, Desalle R, Fisher PB. Evolution of MDA-5/RIG-I-dependent innate immunity: independent evolution by domain grafting. Proc Natl Acad Sci USA. 2008;105:17040–17045. doi: 10.1073/pnas.0804956105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 19.Gack MU, et al. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc Natl Acad Sci USA. 2008;105:16743–16748. doi: 10.1073/pnas.0804947105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkins LE, Swayne DE. Pathogenicity of a Hong Kong-origin H5N1 highly pathogenic avian influenza virus for emus, geese, ducks, and pigeons. Avian Dis. 2002;46:53–63. doi: 10.1637/0005-2086(2002)046[0053:POAHKO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 21.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 24.Childs K, et al. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. 2007;359:190–200. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 25.van Tuinen M, Hedges SB. Calibration of avian molecular clocks. Mol Biol Evol. 2001;18:206–213. doi: 10.1093/oxfordjournals.molbev.a003794. [DOI] [PubMed] [Google Scholar]
- 26.Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology. 1978;84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, et al. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol. 2006;80:11115–11123. doi: 10.1128/JVI.00993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez-Sesma A, et al. Influenza virus evades innate and adaptive immunity via the NS1 protein. J Virol. 2006;80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 30.Egorov A, et al. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J Virol. 1998;72:6437–6441. doi: 10.1128/jvi.72.8.6437-6441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mibayashi M, et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 33.Opitz B, et al. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 34.Sirén J, et al. Retinoic acid inducible gene-I and mda-5 are involved in influenza A virus-induced expression of antiviral cytokines. Microbes Infect. 2006;8:2013–2020. doi: 10.1016/j.micinf.2006.02.028. [DOI] [PubMed] [Google Scholar]
- 35.Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarz H, Harlin O, Ohnemus A, Kaspers B, Staeheli P. Synthesis of IFN-beta by virus-infected chicken embryo cells demonstrated with specific antisera and a new bioassay. J Interferon Cytokine Res. 2004;24:179–184. doi: 10.1089/107999004322917025. [DOI] [PubMed] [Google Scholar]
- 37.Sarmento L, Afonso CL, Estevez C, Wasilenko J, Pantin-Jackwood M. Differential host gene expression in cells infected with highly pathogenic H5N1 avian influenza viruses. Vet Immunol Immunopathol. 2008;125:291–302. doi: 10.1016/j.vetimm.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Sick C, et al. Promoter structures and differential responses to viral and nonviral inducers of chicken type I interferon genes. J Biol Chem. 1998;273:9749–9754. doi: 10.1074/jbc.273.16.9749. [DOI] [PubMed] [Google Scholar]
- 39.Philbin VJ, et al. Identification and characterization of a functional, alternatively spliced Toll-like receptor 7 (TLR7) and genomic disruption of TLR8 in chickens. Immunology. 2005;114:507–521. doi: 10.1111/j.1365-2567.2005.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDonald MRW, Xia J, Smith AL, Magor KE. The duck toll like receptor 7: genomic organization, expression and function. Mol Immunol. 2008;45:2055–2061. doi: 10.1016/j.molimm.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 41.Pothlichet J, et al. Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS One. 2009;4:e7582. doi: 10.1371/journal.pone.0007582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shigemoto T, et al. Identification of loss of function mutations in human genes encoding RIG-I and MDA5: implications for resistance to type I diabetes. J Biol Chem. 2009;284:13348–13354. doi: 10.1074/jbc.M809449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer-Klein J, et al. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 44.Lee CW, Jung K, Jadhao SJ, Suarez DL. Evaluation of chicken-origin (DF-1) and quail-origin (QT-6) fibroblast cell lines for replication of avian influenza viruses. J Virol Methods. 2008;153:22–28. doi: 10.1016/j.jviromet.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 45.Salomon R, et al. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J Exp Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peters MA, Browning GF, Washington EA, Crabb BS, Kaiser P. Embryonic age influences the capacity for cytokine induction in chicken thymocytes. Immunology. 2003;110:358–367. doi: 10.1046/j.1365-2567.2003.01744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.